Applications of Machine Learning in Ambulatory ECG
Abstract
:Simple Summary
Abstract
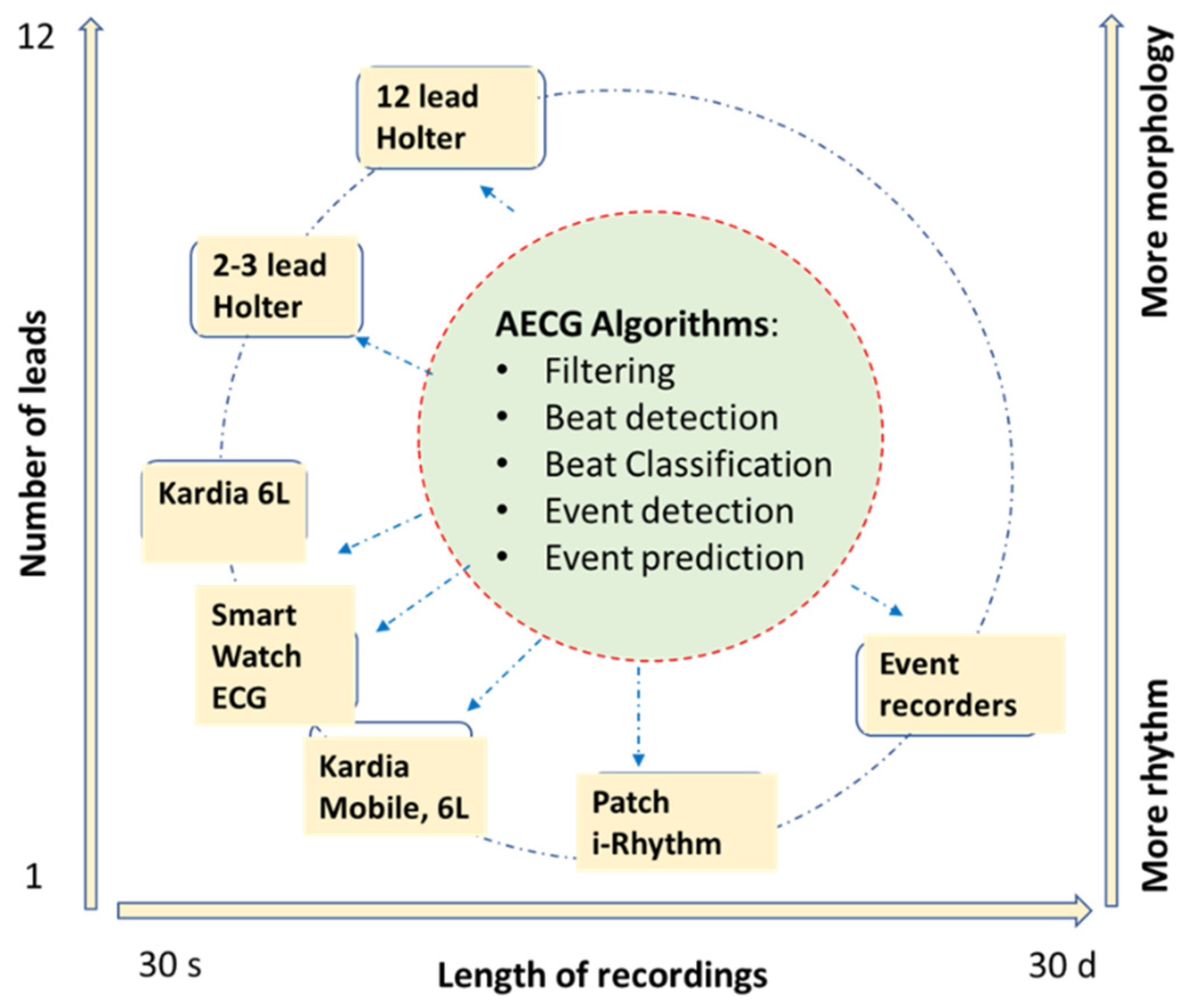
1. Introduction
2. A Summary of Machine Learning Algorithms Used for AECG
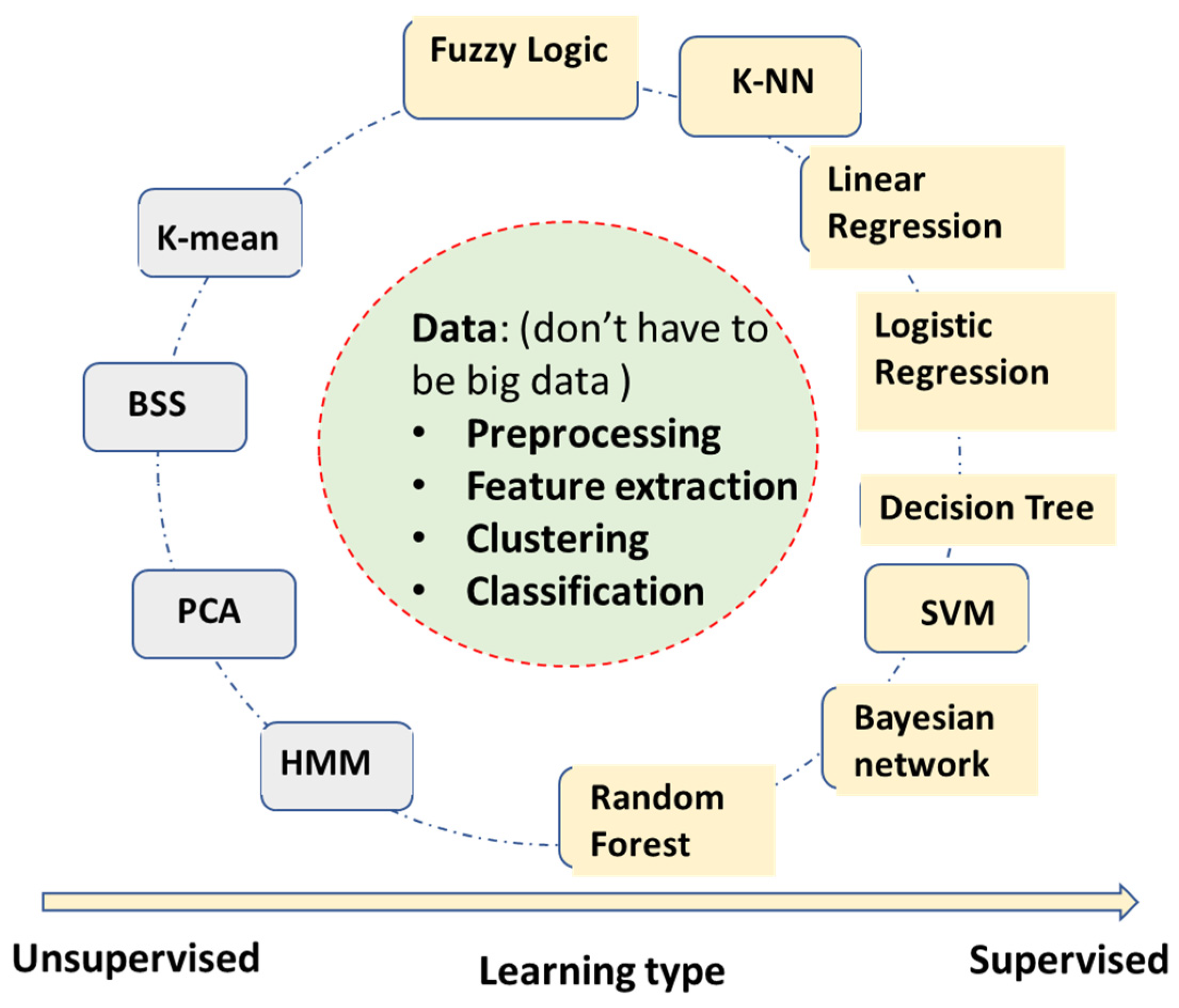
2.1. Machine Learning Algorithms without Deep Learning
- Fuzzy logic algorithm—Using fuzzy logic for rule-based classifications. The fuzzy logic uses a ‘soft edge’ decision boundary to replace the ‘crispy’ decision boundary on the basis of multiple criteria. It maintains its interpretable nature while being able to adapt to the complicated decision boundaries. However, the algorithm complexity can increase very fast with the increase of the number of input features.
- Linear regression—It builds a linear correlation model between inputs and outputs, mostly used for continuous value output.
- Logistic regression—It also builds the correlation model between inputs and outputs, converging to binary level output by logistic functions such as sigmoid. It can be used for categorical classifications.
- Decision tree—Automatically generates a rule-based classification by a tree-like model. It usually has very intuitive flowchart symbols and rules, and therefore it is simple to understand and interpret. However, it can be relatively inaccurate compared to other predictors with the similar data.
- SVM (support vector machine)—A binary classification to maximize the distance between two classes. It has become one of the most robust binary classifiers. It also can perform nonlinear classification with its kernel functions.
- Bayesian network (naïve Bayesian network)—Applies simplified Bayesian theorem for classifications. It builds a probabilistic relationship between symptoms and measurements with the causes of diseases, and therefore is usually a better causality model than neural networks’ ‘black box’ models. However, it requires more known knowledge such as conditional/joint probabilities of input variables.
- k-NN (k-nearest neighbors)—A very intuitive classification algorithm wherein a sample is classified on the basis of a common majority rule of its k closest neighbors. It is a very effective classification algorithm with limited training samples. It works better with a small number of input features.
- Random forest—An ensemble learning method by constructing multiple decision trees. A test sample is classified on the basis of the selections made by most trees. It has some relationship with the decision tree algorithm but is better for avoiding overfitting. It has become one of the most widely used classification algorithms outside of neural networks models.
- K-mean—A clustering algorithm based on vector quantization that partitions n observations into K clusters, e.g., clustering ECG beats into templates in Holter analysis (not to be confused with k-NN, which is a supervised classification algorithm).
- BSS (blind signal separation)—Separates a set of source signals with little information about source signals, e.g., separate signal and noise of AECG.
- PCA (principal component analysis)—One of the most popular unsupervised algorithms for data reduction and feature optimization, e.g., determining which ECG features are more important for AFIB detection.
- HMM (hidden Markov model)—Assume signals to be a Markov process, current state, X(n) only depend on immediate previous state X(n-1). e.g., to describe R-R interval sequence of AFIB ECGs.
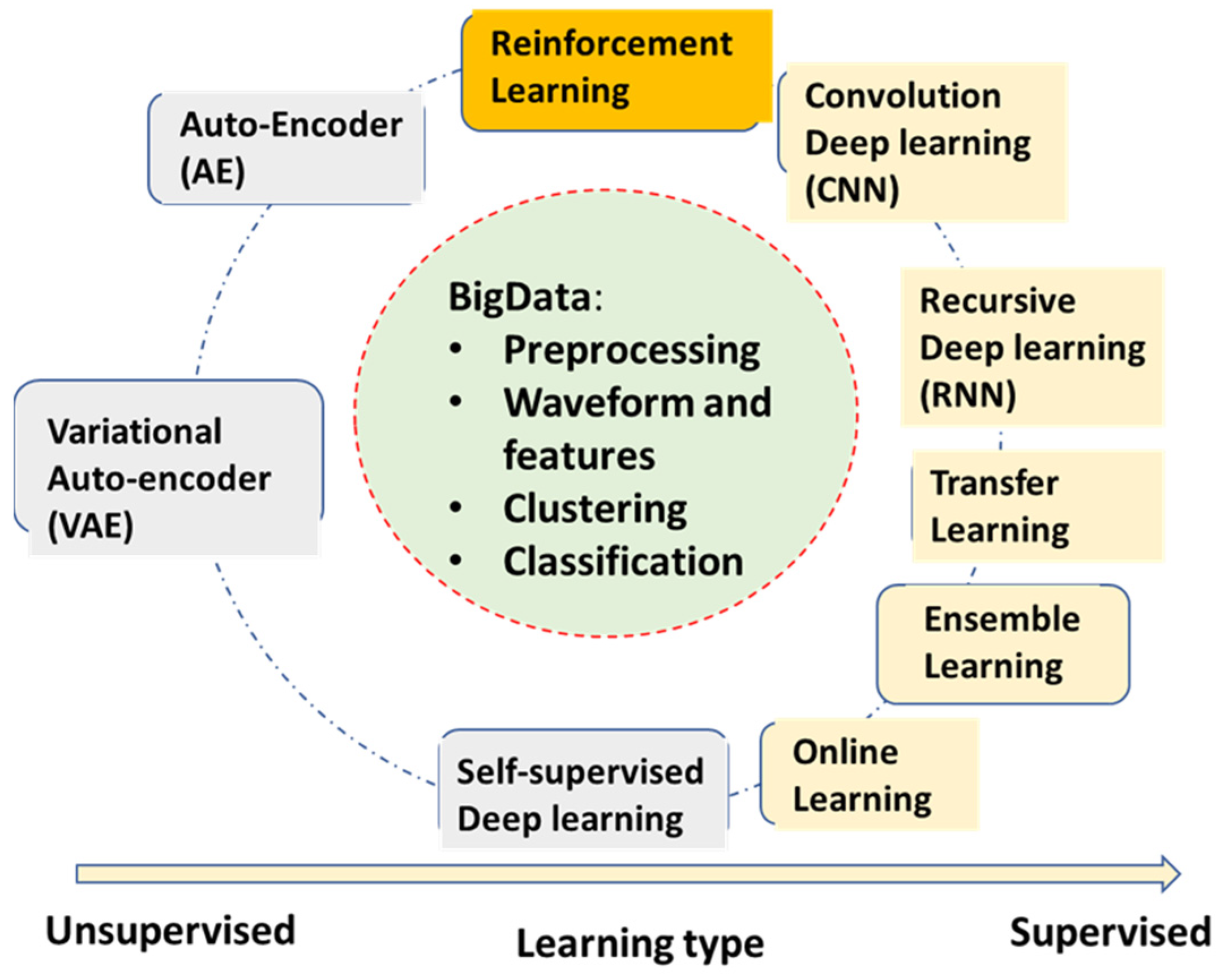
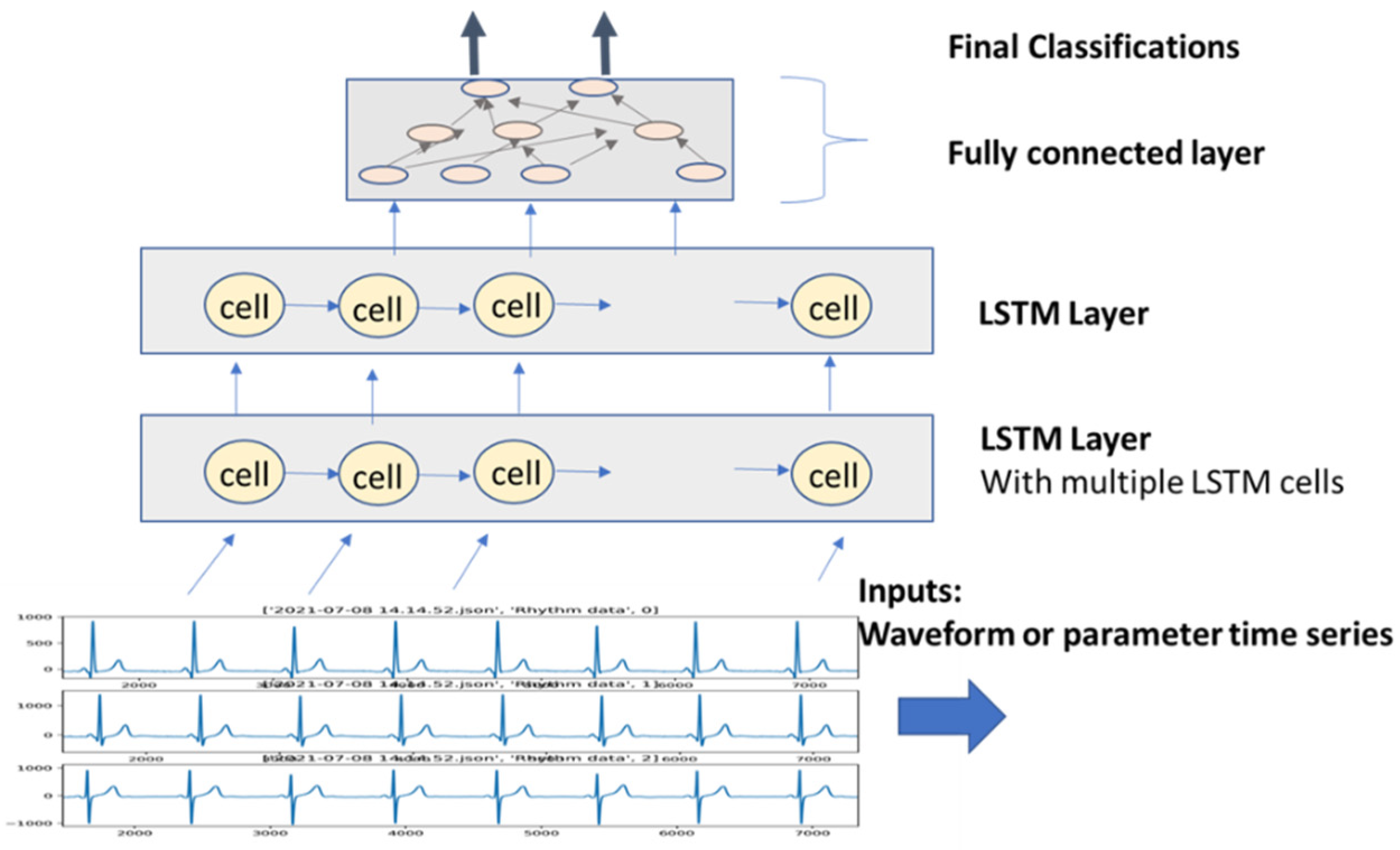
2.2. Neural Network Deep Learning Algorithms for AECG
- •
- Convolutional neural network deep learning (CNN)—The most popular DL algorithm with many applications in AECG. The multi-layer structures of CNN act as a filter bank, and the nonlinear activation functions of CNN act as feature extraction. Therefore, the CNN models are capable of handling original ECG waveforms directly.
- •
- Recursive neural network deep learning (RNN)—RNN models add links among their hidden units (cells) to add memories for time series signals, similar to the concept of IIR filters used in signal processing. AECG signals are time-series signals whose features such as P-R and R-R intervals can be well fitted to certain time-series models.
- •
- Transfer learning—This algorithm is very useful for DL models without enough training data for the current application. A pre-trained DL model for another domain is transferred to the current application. It only re-trains fewer layers of the transferred model or adds fewer new layers for retraining.
- •
- Ensemble learning—This algorithm is not only useful for DL models. Random forest learning mentioned above is a type of ensemble learning. General ensemble learning can be extended to a wide range of groups of model collections and voting, suitable for complicated detection and prediction applications. Statistically, ensemble learning can also be viewed as an application of the central limit theorem. However, in practical use, it needs to know if computation power is enough to handle multiple models running.
- •
- Self (semi)-supervised learning—This learning method deals with limited labeled data and large amounts of unlabeled data, which almost all applications are facing. It can be very useful in applications of AECG, although there are not too many mentioned thus far.
- •
- Online learning—It allows for a pre-trained large model to be updated with the new coming samples, while still maintaining good performance for previously trained samples. This is a particularly good concept but lacks matured algorithms thus far. Some of the transfer learning examples can be viewed as special cases for online learning with only new training samples updated from a pre-trained model, but ‘old samples’ are most likely forgotten.
- Auto-encoder (AE)—This algorithm is a DL model for encoding the original input, such as ECG waveforms. After training, it can represent ECG waveform with a much-condensed latent variable vector.
- Variational auto-encoder (VAE)—It has the same structure and training as AE. However, during the applications, one can adjust its latent vector to form different variation patterns of the original waveforms, for example, to synthesize different noise patterns.
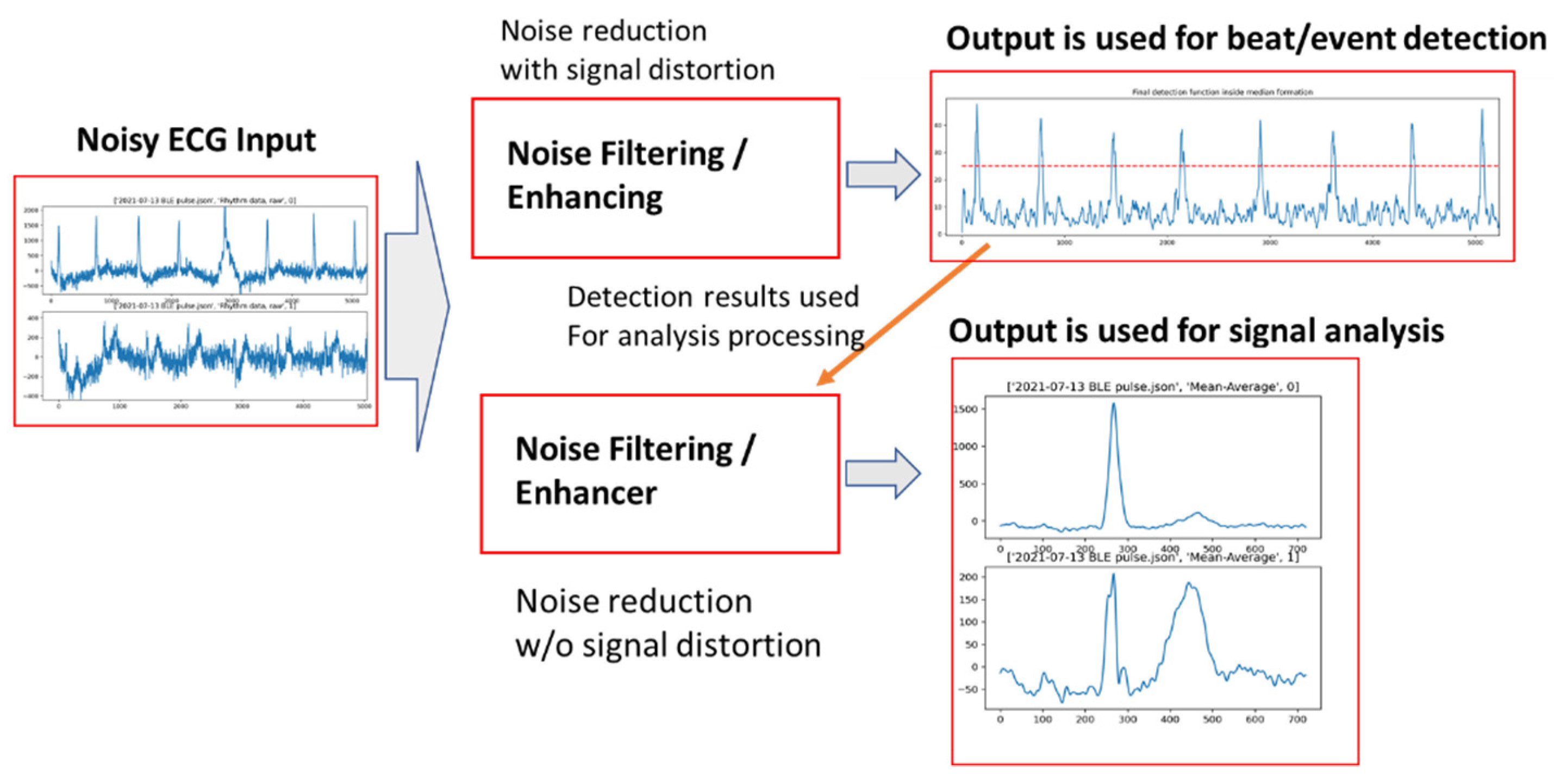
3. AECG Signal Preprocessing—Noise Filtering
3.1. AECG Signal Processing-Noise Reduction
3.2. Early Stage of ML Filtering of AECG
3.3. Using Deep Learning Models for ECG Denoising
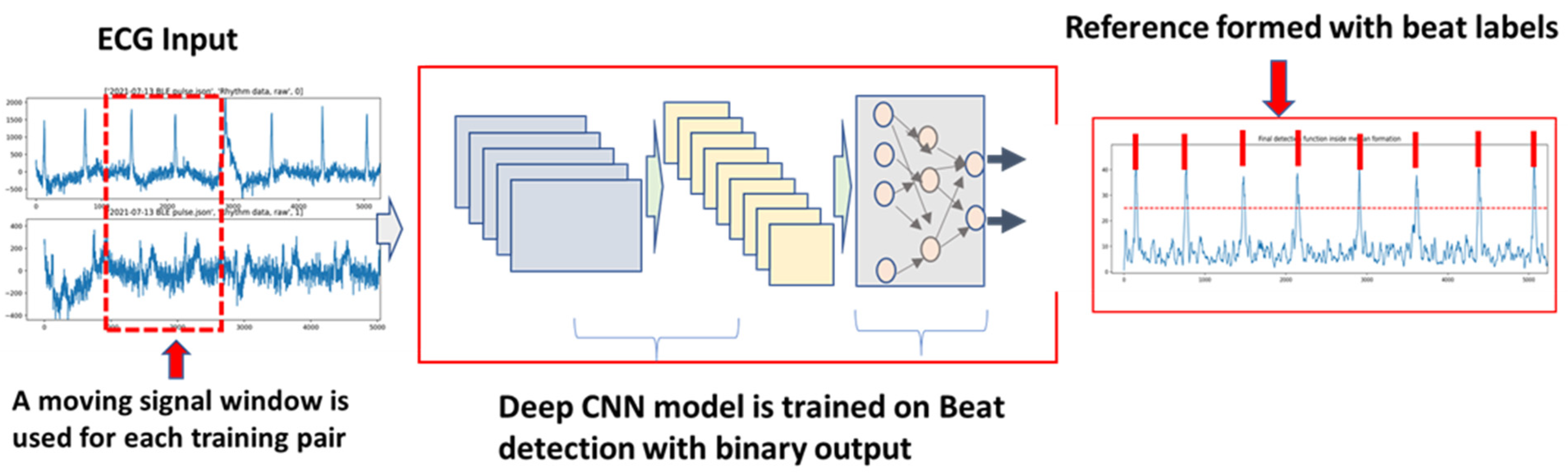
4. AECG Beat Detection and Classification
4.1. Conventional Algorithms for Beat Detection and Classification
4.1.1. Use Both Thresholding and Template Pattern Matching
4.1.2. Time Series Analysis
4.2. ML/DL-Based Beat Detection and Classification
4.2.1. DL Supervised Learning for Beat Detection and Classification
- •
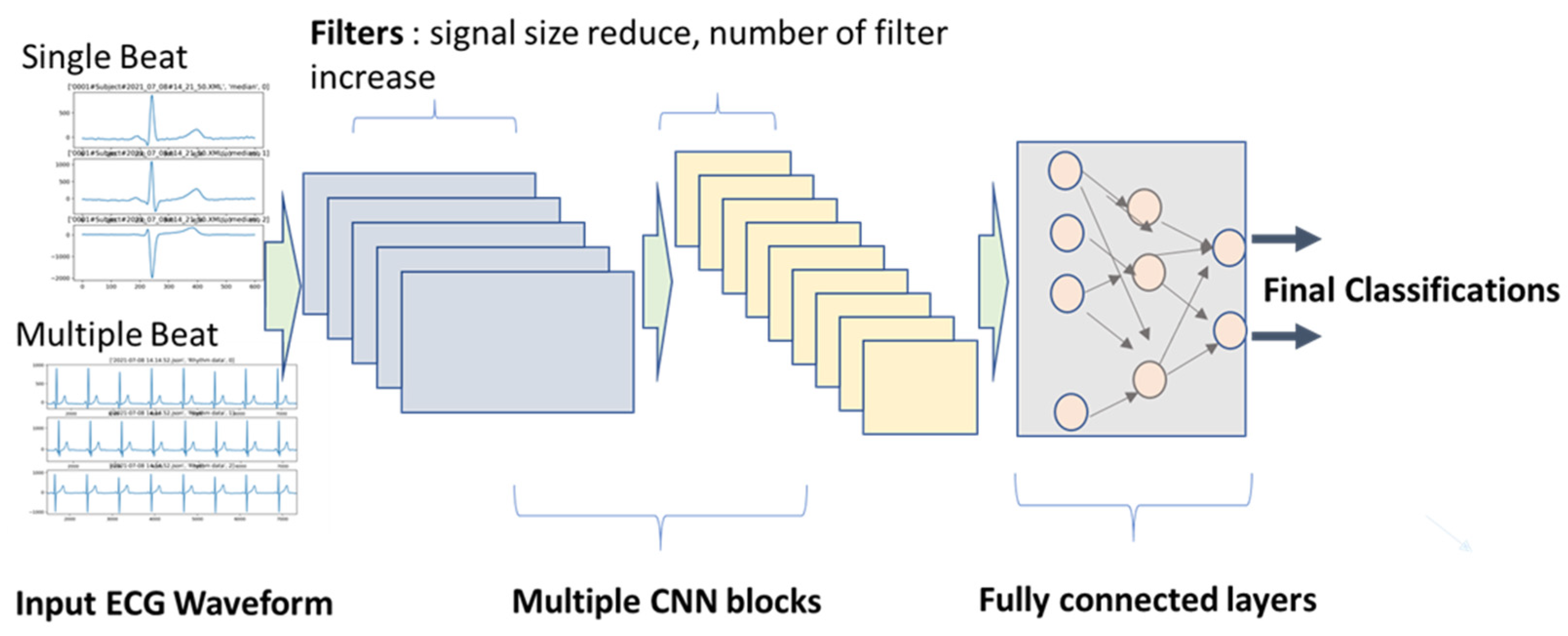
- Input data, length, and dimension: Some studies used one beat cycle, and some used multiple beat cycles. The advantage of using multiple beat cycles is to add time-series information. It is also beneficial for differentiating signal and noise. However, using multiple segments require larger training sets, since the variation is increased.
- •
- CNN kernel filter length and number for each layer: Length of the kernel filter seems not to affect performance by much, but the number of filters usually increases from layer to layer, such as 16, 32, and 64. For AECG processing, the number of filters increased with the complexity of the input signals. For example, if the input signals are rhythm signals in original sampling frequency, the number of kernel filters needed can go up to 128 for deep layers. However, if the input signals are shorter averaged/median beats, the number of filters can be reduced to 32. A large number of filters generate more latent variables. If there are too many latent variables than needed, the model learning speed can be unnecessarily slow.
- •
- Regular convolution or residual convolution (ResCNN) layer: ResCNN with a skip layer for each unit can increase training speed. Therefore, most studies adopted the ResCNN model structure.
- •
- Batch size: Usually, large batch size is preferred for a big data set and multiple classes, such as 256 or 512. For binary classification and limited data size, a smaller batch size can be used, such as 32 or 64. Another consideration is for GPU memory size, since the whole batch is loaded into GPU memory as one block to speed up the training. The larger batch will take more GPU memory. For AECG processing, the selection of the batch size is also heavily dependent on the available training samples. If the training set is very large, such as 1 million plus, the batch size can be set at 512 to generate a smoother gradient search path by avoiding too much fluctuation of a smaller batch size, and most importantly to avoid being ‘trapped’ in a local minimum. However, if the training set is relatively small, the batch size has to be reduced, with the reduction of the model layers.
- •
- Loss function and output function: These strategies need to be clarified to avoid misuse. The classification task can be divided into three categories (as shown in Table 1): (1) binary classification, e.g., QRS complex vs. noise; 2) multi-class classification that is mutually exclusive, e.g., classify QRS beats into N, S, V, F, Q beat types; (3) multi-class classification but not mutually exclusive, e.g., morphology-related ECG abnormal: LBBB, ischemia, sinus, etc. Different loss functions and output functions are selected accordingly. The suggestions are also listed in Table 1.
- •
- Balance of different class types in each batch of training samples: Very often, we can have a very different distribution of classes. For example, there are many more normal beats than PVC or other abnormal beats. If the same distribution is used in the training batch, it is very likely that the sensitivity of PVC beat detection will be poor. Therefore, the number of PVC beats can be augmented in each training batch, which can be in their original form or with variations. Another method is to use weight balancing. Popular machine learning frameworks, such as Keras, provide a ‘class weight’ parameter for this purpose [43].
- •
- Prevent overfitting: Theoretically, there are concerns for both underfitting and overfitting. However, in most DNN studies, the model size/layer is so large that we might only need to worry about overfitting problems. The overfitting is usually caused by a lack of training samples with too many model parameters that need to be trained. There are several methods that can be used. The first one is to increase the training; for example, by adding certain noise to the original ECG recording to avoid simple repetition of the same data. The second method is to apply drop-off to the training process, which randomly ‘disconnects’ the weights to the output. The third method is to apply transfer learning [44,45].
4.2.2. Unsupervised Learning for Beat Detection and Classification
4.2.3. Transfer Learning
4.2.4. Ensemble Learning
5. AECG Event Detection and Classification
5.1. AFIB/AFLUT
5.2. PVC/VT
5.3. QT Analysis
5.4. Noise Segment
6. ECG Risk Stratification/Prediction
7. Discussion
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Steinberg, J.S.; Varma, N.; Cygankiewicz, I.; Aziz, P.; Balsam, P.; Baranchuk, A.; Cantillon, D.J.; Dilaveris, P.; Dubner, S.J.; El-Sherif, N.; et al. 2017 ISHNE-HRS expert consensus statement on ambulatory ECG and external cardiac monitoring/telemetry. Heart Rhythm 2017, 14, 55–96, Epub 2017 May 8. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Rosenberg, M.A.; Samuel, M.; Thosani, A.; Zimetbaum, P.J. Use of a Noninvasive Continuous Monitoring Device in the Management of Atrial Fibrillation: A Pilot Study. Pacing Clin. Electrophysiol. 2013, 36, 328. [Google Scholar] [CrossRef] [PubMed]
- Isakadze, N.; Martin, S.S. How useful is the smartwatch ECG? Trends Cardiovasc. Med. 2020, 30, 442–448. [Google Scholar] [CrossRef]
- Hall, A.; Mitchell, A.R.J.; Wood, L.; Holland, C. Effectiveness of a single lead AliveCor electrocardiogram application for the screening of atrial fibrillation: A systematic review. Medicine 2020, 99, e21388. [Google Scholar] [CrossRef] [PubMed]
- Pan, J.; Tompkins, W.J. A Real-Time QRS Detection Algorithm. IEEE Trans. Biomed. Eng. 1985, 32, 230–236. [Google Scholar] [CrossRef] [PubMed]
- Manisha; Dhull, S.K.; Singh, K.K. ECG Beat Classifiers: A Journey from ANN to DNN. Procedia Comput. Sci. 2020, 167, 747–759. [Google Scholar] [CrossRef]
- Somani, S.; Russak, A.J.; Richter, F.; Zhao, S.; Vaid, A.; Chaudhry, F.; De Freitas, J.K.; Naik, N.; Miotto, R.; Nadkarni, G.N.; et al. Deep learning and the electrocardiogram: Review of the current state-of-the-art. Europace 2021, 23, 1179–1191. [Google Scholar] [CrossRef]
- Ribeiro, A.L.P.; Ribeiro, M.H.; Paixão, G.M.M.; Oliveira, D.M.; Gomes, P.R.; Canazart, J.A.; Ferreira, M.P.S.; Andersson, C.R.; Macfarlane, P.W.; Meira, W.; et al. Automatic diagnosis of the 12-lead ECG using a deep neural network. Nat. Commun. 2020, 11, 1760. [Google Scholar] [CrossRef] [Green Version]
- Kashou, A.H.; Ko, W.-Y.; Attia, Z.I.; Cohen, M.S.; Friedman, P.A.; Noseworthy, P.A. A comprehensive artificial intelligence–enabled electrocardiogram interpretation program. Cardiovasc. Digit. Health J. 2020, 1, 62–70. [Google Scholar] [CrossRef]
- Hannun, A.Y.; Rajpurkar, P.; Haghpanahi, M.; Tison, G.H.; Bourn, C.; Turakhia, M.P.; Ng, A.Y. Cardiologist-level arrhythmia detection and classification in ambulatory electrocardiograms using a deep neural network. Nat. Med. 2019, 25, 65–69. [Google Scholar] [CrossRef]
- Lu, W.; Shuai, J.; Gu, S.; Xue, J. Method to Annotate Arrhythmias by Deep Network. In Proceedings of the 2018 IEEE International Conference on Internet of Things (iThings), Halifax, NS, Canada, 30 July–3 August 2018. [Google Scholar] [CrossRef] [Green Version]
- Hong, S.; Zhou, Y.; Shang, J.; Xiao, C.; Sun, J. Opportunities and Challenges of Deep Learning Methods for Electrocardiogram Data: A Systematic Review. Comput. Biol. Med. 2020, 122, 103801. [Google Scholar] [CrossRef] [PubMed]
- Ebrahimi, Z.; Loni, M.; Daneshtalab, M.; Gharehbaghi, A. A review on deep learning methods for ECG arrhythmia classification. Expert Syst. Appl. X 2020, 7, 100033. [Google Scholar] [CrossRef]
- Serhani, M.A.; El Kassabi, H.T.; Ismail, H.; Navaz, A.N. ECG Monitoring Systems: Review, Architecture, Processes, and Key Challenges. Sensors 2020, 20, 1796. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wasimuddin, M.; Elleithy, K.; Abuzneid, A.S.; Faezipour, M.; Abuzaghleh, O. Stages-based ECG signal analysis from traditional signal processing to machine learning approaches: A survey. IEEE Access 2020, 8, 177782–177803. [Google Scholar] [CrossRef]
- Hoffmann, J.; Mahmood, S.; Fogou, P.S.; George, N.; Raha, S.; Safi, S.; Schmailzl, K.J.; Brandalero, M.; Hubner, M. A Survey on Machine Learning Approaches to ECG Processing. In Signal Processing—Algorithms, Architectures, Arrangements, and Applications; IEEE: Piscataway, NJ, USA, 2020; pp. 36–41. [Google Scholar] [CrossRef]
- Hastie, T.; Tibshirani, R.; Friedman, J. The Elements of Statistical Learning—Data Mining, Inference, and Prediction; Springer Series in Statistics; Springer: New York, NY, USA, 2008. [Google Scholar]
- Goodfellow, I.; Bengio, Y.; Courville, A. Deep Learning; MIT Press: Cambridge, MA, USA, 2016; Available online: http://www.deeplearningbook.org (accessed on 17 July 2021).
- Xue, J.Q.; Hu, Y.H.; Tompkins, W.J. Neural-Network-Based Adaptive matched Filtering for QRS Detection. IEEE Trans. Biomed. Eng. 1992, 39, 317–329. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Xuel, J.Q.; Hu, Y.H.; Tompkins, W.J. Training of ECG signals in nueral network pattern recognition. In Proceedings of the Internaional Conference of the IEEE Engineering in Medicine and Biology Society, Philadelphia, PA, USA, 1–4 November 1990; Volume 12, pp. 1465–1466. [Google Scholar]
- Xue, Q.; Reddy, B.R.S. Late Potential Recognition by Artificial Neural Networks. IEEE Trans. Biomed. Eng. 1997, 44, 132–143. [Google Scholar] [CrossRef] [PubMed]
- LeCun, Y.; Haffner, P.; Bottou, L.; Bengio, Y. Object recognition with gradient-based learning. In Shape, Contour and Grouping in Computer Vision; Lecture Notes in Computer Science (Including Subseries Lecture Notes in Artificial Intelligence and Lecture Notes in Bioinformatics); Springer: Berlin/Heidelberg, Germany, 1999; Volume 1681, pp. 319–345. [Google Scholar] [CrossRef]
- Krizhevsky, A.; Sutskever, I.; Hinton, G.E. ImageNet classification with deep convolutional neural networks. Commun. ACM 2017, 60, 84–90. [Google Scholar] [CrossRef]
- Arulkumaran, K.; Deisenroth, M.P.; Brundage, M.; Bharath, A.A. A Brief Survey of Deep Reinforcement Learning. IEEE Signal Process. Mag. 2017, 34, 26–38. [Google Scholar] [CrossRef] [Green Version]
- Silver, D.; Huang, A.; Maddison, C.J.; Guez, A.; Sifre, L.; Driessche, G.V.D.; Schrittwieser, J.; Antonoglou, I.; Panneershelvam, V.; Lanctot, M.; et al. Mastering the game of Go with deep neural networks and tree search. Nature 2016, 529, 484–489. [Google Scholar] [CrossRef]
- Insani, A.; Jatmiko, W.; Sugiarto, A.T.; Jati, G.; Wibowo, S.A. Investigation Reinforcement learning method for R-Wave detection on Electrocardiogram signal. In Proceedings of the International Seminar on Research of Information Technology and Intelligent Systems, Yogyakarta, Indonesia, 5–6 December 2019. [Google Scholar] [CrossRef]
- IEC 60601-2-47:2012 | IEC Webstore. Available online: https://webstore.iec.ch/publication/2666 (accessed on 17 July 2021).
- IEC 60601-2-25:2011 | IEC Webstore. Available online: https://webstore.iec.ch/publication/2636 (accessed on 17 July 2021).
- Thakor, N.V.; Zhu, Y.-S. Adaptve filtering to ECG analysis. IEEE Trans. Biomed. Eng. 1991, 38, 785–794. [Google Scholar] [CrossRef]
- Awal, M.A.; Mostafa, S.S.; Ahmad, M.; Rashid, M.A. An adaptive level dependent wavelet thresholding for ECG denoising. Biocybern. Biomed. Eng. 2014, 34, 238–249. [Google Scholar] [CrossRef]
- PhysioBank Databases. Available online: https://archive.physionet.org/physiobank/database/ (accessed on 17 July 2021).
- Arsene, C. Design of Deep Convolutional Neural Network Architectures for Denoising Electrocardiographic Signals. In Proceedings of the IEEE Conference on Computational Intelligence in Bioinformatics and Computational Biology, Viña del Mar, Chile, 27–29 October 2020. [Google Scholar] [CrossRef]
- Xiong, P.; Wang, H.; Liu, M.; Zhou, S.; Hou, Z.; Liu, X. ECG signal enhancement based on improved denoising auto-encoder. Eng. Appl. Artif. Intell. 2016, 52, 194–202. [Google Scholar] [CrossRef]
- Xiong, P.; Wang, H.; Liu, M.; Liu, X. Denoising auto-encoder for eletrocardiogram signal enhancement. J. Med Imaging Health Inform. 2015, 5, 1804–1810. [Google Scholar] [CrossRef]
- Xiong, P.; Wang, H.; Liu, M.; Lin, F.; Hou, Z.; Liu, X. A stacked contractive denoising auto-encoder for ECG signal denoising. Physiol. Meas. 2016, 37, 2214–2230. [Google Scholar] [CrossRef] [PubMed]
- Garus, J.; Pabian, M.; Wisniewski, M.; Sniezynski, B. Electrocardiogram Quality Assessment with Auto-encoder. In Proceedings of the Internarional Conference on Computational Science, 2021 ICCS, Krakow, Poland, 16–18 June 2021; Springer: Cham, Switzerland, 2021; pp. 693–706. [Google Scholar] [CrossRef]
- Antczak, K. A Generative Adversarial Approach to ECG Synthesis and Denoising. arXiv 2020, arXiv:2009.02700v1. [Google Scholar]
- Yildirim, O.; Tan, R.S.; Acharya, U.R. An efficient compression of ECG signals using deep convolutional auto-encoders. Cogn. Syst. Res. 2018, 52, 198–211. [Google Scholar] [CrossRef]
- Wang, F.; Ma, Q.; Liu, W.; Chang, S.; Wang, H.; He, J.; Huang, Q. A novel ECG signal compression method using spindle convolutional auto-encoder. Comput. Methods Programs Biomed. 2019, 175, 139–150. [Google Scholar] [CrossRef]
- Bekiryazici, T.; Gurkan, H. ECG Compression method based on convolutional auto-encoder and discrete wavelet transform. In Proceedings of the 2020 28th Signal Processing and Communications Applications Conference (SIU 2020), Gaziantep, Turkey, 5–7 October 2020. [Google Scholar] [CrossRef]
- Arsene, C. Complex Deep Learning Models for Denoising of Human Heart ECG signals. arXiv 2019, arXiv:1908.10417. [Google Scholar]
- Antczak, K. Deep Recurrent Neural Networks for ECG Signal Denoising. arXiv 2018, arXiv:1807.11551v3. [Google Scholar]
- Johnson, J.M.; Khoshgoftaar, T.M. Survey on deep learning with class imbalance. J. Big Data 2019, 6, 27. [Google Scholar] [CrossRef]
- Weimann, K.; Conrad, T.O.F. Transfer learning for ECG classification. Sci. Rep. 2021, 11, 5251. [Google Scholar] [CrossRef]
- Salem, M.; Taheri, S.; Yuan, J.-S. ECG Arrhythmia Classification Using Transfer Learning from 2-Dimensional Deep CNN Features. In Proceedings of the IEEE Biomedical Circuits and Systems Conference (BioCAS), Cleveland, OH, USA, 17–19 October 2018. [Google Scholar] [CrossRef] [Green Version]
- Xue, J.; Gao, W.; Chen, Y.; Han, X. Study of repolarization heterogeneity and electrocardiographic morphology with a modeling approach. J. Electrocardiol. 2008, 41, 581–587. [Google Scholar] [CrossRef]
- Romeo, I. PCA and ICA applied to noise reduction in multi-lead ECG. In Computer in Cardiology; IEEE: Piscataway, NJ, USA, 2011; pp. 613–616. [Google Scholar]
- Alickovic, E.; Subasi, A. Effect of Multiscale PCA De-noising in ECG Beat Classification for Diagnosis of Cardiovascular Diseases. Circuits Syst. Signal Process. 2015, 34, 513–533. [Google Scholar] [CrossRef]
- Koski, A. Modelling ECG signals with hidden Markov models. Artif. Intell. Med. 1996, 8, 453–471. [Google Scholar] [CrossRef]
- Andreão, R.V.; Dorizzi, B.; Boudy, J. ECG signal analysis through hidden Markov models. IEEE Trans. Biomed. Eng. 2006, 53, 1541–1549. [Google Scholar] [CrossRef]
- Liao, Y.; Xiang, Y.; Du, D. Automatic Classification of Heartbeats Using ECG Signals via Higher Order Hidden Markov Model. In Proceedings of the IEEE International Conference on Automation Science and Engineering, Hong Kong, China, 20–21 August 2020; pp. 69–74. [Google Scholar] [CrossRef]
- Silipo, R.; Deco, G.; Schürmann, B.; Vergassola, R.; Gremigni, C. Investigating the underlying Markovian dynamics of ECG rhythms by information flow. Chaos Solitons Fractals 2001, 12, 2877–2888. [Google Scholar] [CrossRef]
- Hajimolahoseini, H.; Hashemi, J.; Redfearn, D. ECG Delineation for Qt Interval Analysis Using an Unsupervised Learning Method. In Proceedings of the ICASSP, IEEE International Conference on Acoustics, Speech and Signal Processing, Calgary, AB, Canada, 15–20 April 2018; pp. 2541–2545. [Google Scholar] [CrossRef]
- Tighiouart, B.; Rubel, P.; Bedda, M. Improvement of QRS boundary recognition by means of unsupervised learning. Comput. Cardiol. 2003, 30, 49–52. [Google Scholar] [CrossRef]
- Lagerholm, M.; Peterson, G. Clustering ECG complexes using hermite functions and self-organizing maps. IEEE Trans. Biomed. Eng. 2000, 47, 838–848. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zhang, C.; Wang, G.; Zhao, J.; Gao, P.; Lin, J.; Yang, H. Patient-specific ECG classification based on recurrent neural networks and clustering technique. In Proceedings of the 13th IASTED International Conference on Biomedical Engineering (BioMed 2017), Innsbruck, Austria, 20–21 February 2017; pp. 63–67. [Google Scholar] [CrossRef]
- Osowski, S.; Linh, T.H. ECG beat recognition using fuzzy hybrid neural network. IEEE Trans. Biomed. Eng. 2001, 48, 1265–1271. [Google Scholar] [CrossRef] [PubMed]
- Chudáček, V.; Petrík, M.; Georgoulas, G.; Čepek, M.; Lhotská, L.; Stylios, C. Comparison of seven approaches for holter ECG clustering and classification. In Proceedings of the Annual International Conference of the IEEE Engineering in Medicine and Biology, Lyon, France, 22–26 August 2007; pp. 3844–3847. [Google Scholar] [CrossRef]
- Korürek, M.; Nizam, A. A new arrhythmia clustering technique based on Ant Colony Optimization. J. Biomed. Inform. 2008, 41, 874–881. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Nurmaini, S.; Partan, R.U.; Caesarendra, W.; Dewi, T.; Rahmatullah, M.N.; Darmawahyuni, A.; Bhayyu, V.; Firdaus, F. An automated ECG beat classification system using deep neural networks with an unsupervised feature extraction technique. Appl. Sci. 2019, 9, 2921. [Google Scholar] [CrossRef] [Green Version]
- Hou, B.; Yang, J.; Wang, P.; Yan, R. LSTM-Based Auto-Encoder Model for ECG Arrhythmias Classification. IEEE Trans. Instrum. Meas. 2020, 69, 1232–1240. [Google Scholar] [CrossRef]
- Kuznetsov, V.V.; Moskalenko, V.A.; Zolotykh, N.Y. Electrocardiogram Generation and Feature Extraction Using a Variational Auto-encoder. arXiv 2020, arXiv:2002.00254. [Google Scholar]
- Rubel, P.; Fayn, J.; Macfarlane, P.W.; Pani, D.; Schlögl, A.; Värri, A. The History and Challenges of SCP-ECG: The Standard Communication Protocol for Computer-Assisted Electrocardiography. Hearts 2021, 2, 384–409. [Google Scholar] [CrossRef]
- Clifford, G.; Liu, C.; Moody, B.; Lehman, L.-W.; Silva, I.; Li, Q.; Johnson, A.; Mark, R. AF Classification from a Short Single Lead ECG Recording: The PhysioNet/Computing in Cardiology Challenge 2017. In Computing in Cardiology; IEEE: Piscataway, NJ, USA, 2017. [Google Scholar] [CrossRef]
- Zabihi, M.; Rad, A.B.; Katsaggelos, A.K.; Kiranyaz, S.; Narkilahti, S.; Gabbouj, M. Detection of atrial fibrillation in ECG hand-held devices using a random forest classifier. In Computing in Cardiology; IEEE: Piscataway, NJ, USA, 2017; Volume 44, pp. 1–4. [Google Scholar] [CrossRef]
- Lin, Z.; Ge, Y.; Tao, G. Algorithm for Clustering Analysis of ECG Data. In Proceedings of the Annual International Conference of the IEEE Engineering in Medicine and Biology Society. IEEE Engineering in Medicine and Biology Society. Annual Conference, Shanghai, China, 17–18 January 2006; pp. 3857–3860. [Google Scholar] [CrossRef]
- Xia, Y.; Han, J.; Wang, K. Quick detection of QRS complexes and R-waves using a wavelet transform and K-means clustering. Bio-Med Mater. Eng. 2015, 26, S1059–S1065. [Google Scholar] [CrossRef] [Green Version]
- Özbay, Y.; Ceylan, R.; Karlik, B. A fuzzy clustering neural network architecture for classification of ECG arrhythmias. Comput. Biol. Med. 2006, 36, 376–388. [Google Scholar] [CrossRef] [PubMed]
- Stridh, M.; Rosenqvist, M. Automatic screening of atrial fibrillation in thumb-ECG recordings. In Proceedings of the 39th Conference on Computing in Cardiology, Krakow, Poland, 9–12 September 2012; IEEE: Krakow, Poland, 2012; Volume 39, pp. 161–164. [Google Scholar]
- Plesinger, F.; Nejedly, P.; Viscor, I.; Halamek, J.; Jurak, P. Automatic detection of atrial fibrillation and other arrhythmias in holter ECG recordings using rhythm features and neural networks. In Computing in Cardiology; IEEE: Piscataway, NJ, USA, 2017; Volume 44, pp. 1–4. [Google Scholar] [CrossRef]
- Tan, W.W.; Foo, C.L.; Chua, T.W. Type-2 fuzzy system for ECG arrhythmic classification. In Proceedings of the IEEE International Conference on Fuzzy Systems, London, UK, 23–26 July 2007. [Google Scholar] [CrossRef]
- Teijeiro, T.; García, C.A.; Castro, D.; Félix, P. Arrhythmia classification from the abductive interpretation of short single-lead ECG records. In Computing in Cardiology; IEEE: Piscataway, NJ, USA, 2017; Volume 44, pp. 1–4. [Google Scholar] [CrossRef]
- Antink, C.H.; Leonhardt, S.; Walter, M. Fusing QRS detection and robust interval estimation with a random forest to classify atrial fibrillation. In Computing in Cardiology; IEEE: Piscataway, NJ, USA, 2017; Volume 44, pp. 1–4. [Google Scholar] [CrossRef]
- Yu, J.; Wang, X.; Chen, X.; Guo, J. Automatic Premature Ventricular Contraction Detection Using Deep Metric Learning and KNN. Biosensors 2021, 11, 69. [Google Scholar] [CrossRef]
- Kaya, Y.; Pehlivan, H. Feature selection using genetic algorithms for premature ventricular contraction classification. In Proceedings of the ELECO 2015—9th International Conference on Electrical and Electronics Engineering, Bursa, Turkey, 26–28 November 2015; pp. 1229–1232. [Google Scholar] [CrossRef]
- Boublenza, S.; Chikh, A.; Bouchikhi, M.A. Discrete hidden Markov model classifier for premature ventricular contraction detection. Int. J. Biomed. Eng. Technol. 2015, 17, 371–386. [Google Scholar]
- Casas, M.M.; Avitia, R.L.; Gonzalez-Navarro, F.F.; Cardenas-Haro, J.A.; Reyna, M.A. Bayesian Classification Models for Premature Ventricular Contraction Detection on ECG Traces. J. Healthc. Eng. 2018, 2018, 2694768. [Google Scholar] [CrossRef] [Green Version]
- Xie, T.; Li, R.; Shen, S.; Zhang, X.; Zhou, B.; Wang, Z. Intelligent Analysis of Premature Ventricular Contraction Based on Features and Random Forest. J. Healthc. Eng. 2019, 2019, 5787582. [Google Scholar] [CrossRef]
- De Marco, F.; Finlay, D.; Bond, R.R. Classification of Premature Ventricular Contraction Using Deep Learning. In Computing in Cardiology; IEEE: Piscataway, NJ, USA, 2020. [Google Scholar] [CrossRef]
- Murugesan, B.; Ravichandran, V.; Ram, K.; S.P., P.; Joseph, J.; Shankaranarayana, S.M.; Sivaprakasam, M. ECGNet: Deep Network for Arrhythmia Classification. In Proceedings of the IEEE International Symposium on Medical Measurements and Applications (MeMeA), Rome, Italy, 11–13 June 2018. [Google Scholar] [CrossRef]
- Novotna, P.; Vicar, T.; Ronzhina, M.; Hejc, J.; Kolarova, J. Deep-Learning Premature Contraction Localization in 12-lead ECG from Whole Signal Annotations. In Computing in Cardiology; IEEE: Piscataway, NJ, USA, 2020. [Google Scholar] [CrossRef]
- Javadi, M.; Ebrahimpour, R.; Sajedin, A.; Faridi, S.; Zakernejad, S. Improving ECG classification accuracy using an ensemble of neural network modules. PLoS ONE 2011, 6, e24386. [Google Scholar] [CrossRef]
- Teplitzky, B.A.; McRoberts, M.; Ghanbari, H. Deep learning for comprehensive ECG annotation. Heart Rhythm 2020, 17, 881–888. [Google Scholar] [CrossRef]
- Yang, T.; Yu, L.; Jin, Q.; Wu, L.; He, B. Localization of Origins of Premature Ventricular Contraction by Means of Convolutional Neural Network from 12-Lead ECG. IEEE Trans. Biomed. Eng. 2018, 65, 1662–1671. [Google Scholar] [CrossRef] [PubMed]
- Prabhakararao, E.; Manikandan, M.S. Efficient and robust ventricular tachycardia and fibrillation detection method for wearable cardiac health monitoring devices. Healthc. Technol. Lett. 2016, 3, 239–246. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Xue, J.Q. Robust QT Interval Estimation—From Algorithm to Validation. Ann. Noninvasive Electrocardiol. 2009, 14 (Suppl. 1), S35. [Google Scholar] [CrossRef]
- Hnatkova, K.; Gang, Y.; Batchvarov, V.N.; Malik, M. Precision of QT interval measurement by advanced electrocardiographic equipment. Pacing Clin. Electrophysiol. PACE 2006, 29, 1277–1284. [Google Scholar] [CrossRef] [PubMed]
- Giudicessi, J.R.; Schram, M.; Bos, J.M.; Galloway, C.D.; Shreibati, J.B.; Johnson, P.W.; Carter, R.E.; Disrud, L.W.; Kleiman, R.; Attia, Z.I.; et al. Artificial Intelligence–Enabled Assessment of the Heart Rate Corrected QT Interval Using a Mobile Electrocardiogram Device. Circulation 2021, 143, 1274–1286. [Google Scholar] [CrossRef]
- Ansari, S.; Gryak, J.; Najarian, K. Noise Detection in Electrocardiography Signal for Robust Heart Rate Variability Analysis: A Deep Learning Approach. In Proceedings of the 2018 40th Annual International Conference of the IEEE Engineering in Medicine and Biology Society (EMBC), Honolulu, HI, USA, 18–21 July 2018. [Google Scholar]
- Sasmal, B.; Roy, S. ECG Guided Automated Diagnostic Intervention of Cardiac Arrhythmias with Extra-Cardiac Noise Detection using Neural Network. In Proceedings of the 7th International Conference on Optimization and Applications (ICOA), Wolfenbüttel, Germany, 19–20 May 2021. [Google Scholar] [CrossRef]
- Dhala, A.; Underwood, D.; Leman, R.; Madu, E.; Baugh, D.; Ozawa, Y.; Kasamaki, Y.; Xue, Q.; Reddy, S. Signal-Averaged P-Wave Analysis of Normal Controls and Patients with Paroxysmal Atrial Fibrillation: A Study in Gender Differences, Age Dependence, and Reproducibility. Clin. Cardiol. 2002, 25, 525. [Google Scholar] [CrossRef] [PubMed]
- Budeus, M.; Hennersdorf, M.; Felix, O.; Reimert, K.; Perings, C.; Sack, S.; Wieneke, H.; Erbel, R. Prediction of atrial fibrillation in patients with cardiac dysfunction: P wave signal-averaged ECG and chemoreflexsensitivity in atrial fibrillation. Europace 2007, 9, 601–607. [Google Scholar] [CrossRef] [PubMed]
- Raghunath, S.; Pfeifer, J.M.; Ulloa-Cerna, A.E.; Nemani, A.; Carbonati, T.; Jing, L.; Vanmaanen, D.P.; Hartzel, D.N.; Ruhl, J.A.; Lagerman, B.F.; et al. Deep Neural Networks Can Predict New-Onset Atrial Fibrillation from the 12-Lead ECG and Help Identify Those at Risk of Atrial Fibrillation-Related Stroke. Circulation 2021, 143, 1287–1298. [Google Scholar] [CrossRef] [PubMed]
- Luongo, G.; Azzolin, L.; Schuler, S.; Rivolta, M.W.; Almeida, T.P.; Martínez, J.P.; Soriano, D.C.; Luik, A.; Müller-Edenborn, B.; Jadidi, A.; et al. Machine learning enables noninvasive prediction of atrial fibrillation driver location and acute pulmonary vein ablation success using the 12-lead ECG. Cardiovasc. Digit. Health J. 2021, 2, 126–136. [Google Scholar] [CrossRef] [PubMed]
- Kwon, J.-M.; Jeon, K.-H.; Kim, H.M.; Kim, M.J.; Lim, S.; Kim, K.-H.; Song, P.S.; Park, J.; Choi, R.K.; Oh, B.-H. Deep-learning-based risk stratification for mortality of patients with acute myocardial infarction. PLoS ONE 2019, 14, e0224502. [Google Scholar] [CrossRef] [PubMed]
- Merchant, F.M.; Sayadi, O.; Moazzami, K.; Puppala, D.; Armoundas, A.A. T-wave Alternans as an Arrhythmic Risk Stratifier: State of the Art. Curr. Cardiol. Rep. 2013, 15, 398. [Google Scholar] [CrossRef] [Green Version]
- Xue, J.; Aufderheide, T.; Wright, R.S.; Klein, J.; Farrell, R.; Rowlandson, I.; Young, B. Added value of new acute coronary syndrome computer algorithm for interpretation of prehospital electrocardiograms. J. Electrocardiol. 2004, 37, 233–239. [Google Scholar] [CrossRef] [PubMed]
- Goh, G.S.W.; Lapuschkin, S.; Weber, L.; Samek, W.; Binder, A. Understanding Integrated Gradients with SmoothTaylor for Deep Neural Network Attribution. arXiv 2020, arXiv:2004.10484. [Google Scholar]
- Hoi, S.C.H.; Sahoo, D.; Lu, J.; Zhao, P. Online Learning: A Comprehensive Survey. Neurocomputing 2021, 459, 249–289. [Google Scholar] [CrossRef]
- PhysioNet Index. Available online: https://physionet.org/challenge/ (accessed on 17 July 2021).
| Classification Tasks | Binary Classification | Multi-Class Classifications (Mutually Exclusive) | Multi-Class Classifications (Mutually Nonexclusive) |
|---|---|---|---|
| Examples: | QRS beat vs. noise | N, S, V, F, Q beat types | LBBB, ischemia, sinus |
| Loss function | Binary cross-entropy | Categorical cross-entropy | Binary cross-entropy |
| Target function | One-hot vector (Softmax) | One-hot vector (Softmax) | Sigmoid, scalar target |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Xue, J.; Yu, L. Applications of Machine Learning in Ambulatory ECG. Hearts 2021, 2, 472-494. https://doi.org/10.3390/hearts2040037
Xue J, Yu L. Applications of Machine Learning in Ambulatory ECG. Hearts. 2021; 2(4):472-494. https://doi.org/10.3390/hearts2040037
Chicago/Turabian StyleXue, Joel, and Long Yu. 2021. "Applications of Machine Learning in Ambulatory ECG" Hearts 2, no. 4: 472-494. https://doi.org/10.3390/hearts2040037
APA StyleXue, J., & Yu, L. (2021). Applications of Machine Learning in Ambulatory ECG. Hearts, 2(4), 472-494. https://doi.org/10.3390/hearts2040037





