Abstract
Melanoma skin cancer is one of the most dangerous types of skin cancer, which, if not diagnosed early, may lead to death. Therefore, an accurate diagnosis is needed to detect melanoma. Traditionally, a dermatologist utilizes a microscope to inspect and then provide a report on a biopsy for diagnosis; however, this diagnosis process is not easy and requires experience. Hence, there is a need to facilitate the diagnosis process while still yielding an accurate diagnosis. For this purpose, artificial intelligence techniques can assist the dermatologist in carrying out diagnosis. In this study, we considered the detection of melanoma through deep learning based on cutaneous image processing. For this purpose, we tested several convolutional neural network (CNN) architectures, including DenseNet201, MobileNetV2, ResNet50V2, ResNet152V2, Xception, VGG16, VGG19, and GoogleNet, and evaluated the associated deep learning models on graphical processing units (GPUs). A dataset consisting of 7146 images was processed using these models, and we compared the obtained results. The experimental results showed that GoogleNet can obtain the highest performance accuracy on both the training and test sets (74.91% and 76.08%, respectively).
1. Background
At present, cancer diseases are among the most dangerous types of diseases that threaten human life. One of the most dangerous cancers is melanoma skin cancer, which may lead to death if not diagnosed early. Early diagnosis of melanoma skin cancer reduces mortality rates and reduces the complications of the treatment process. The diagnosis process is conducted by taking a biopsy sample from the patient, which is then examined by a dermatologist. The examination result depends on the doctor’s experience and the tools used. Figure 1 shows the various types of skin cancers [1,2].
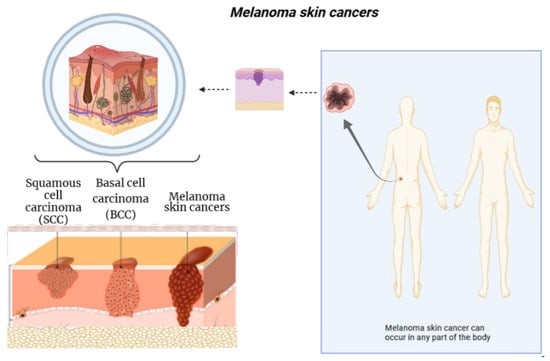
Figure 1.
An overview of melanoma skin cancers. Figure created with Biorender.com.
Accurately diagnosing such diseases may require experience and accurate techniques that improve the accuracy of the diagnosis. Therefore, the availability of precise instruments is of great importance to dermatologists, allowing for good diagnostic results, better treatment, and a reduction in the number of biopsies needed from patients. Hence, the role of artificial intelligence and its various techniques in this issue have been considered. Generally speaking, using AI techniques (and, particularly, deep learning) when diagnosing such diseases can help doctors to make diagnoses and make the process easier for patients [3,4,5,6]. From the above, it is clear that using these techniques in such cases can save much effort and time for doctors, while still allowing for accurate diagnosis, which is the main objective. However, we must not overlook the patients and their comfort. Deep learning has achieved great success in helping to solve such problems that require experience in the field and accuracy and speed in decision-making, which may not always be available to the doctor. Figure 2 presents a comparison between the stages of the manual diagnosis process and the diagnosis process based on deep learning technology.
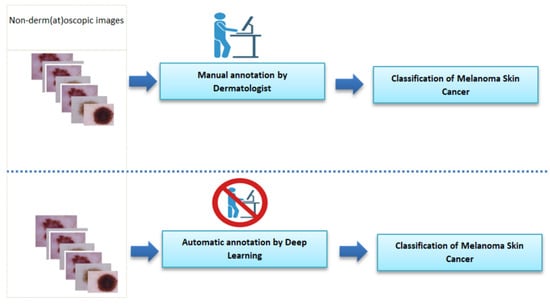
Figure 2.
Comparison of the stages of the manual diagnostic process and the diagnostic process based on deep learning technology. Figure created with Biorender.com.
Artificial intelligence plays a significant role in solving many problems at present, especially in the medical field. Its techniques involve machine learning and deep learning, but the latter may outperform machine learning in several cases when ample data are available [7,8], thus facilitating complex diagnostic operations that require many processes [9].
One type of deep learning technique is convolutional neural networks, which have achieved good results in studying images and finding relationships between them by extracting features [10,11,12,13]. Deep convolutional neural networks are often valuable for analyzing images in general and medical images in particular, one example being diagnosing melanoma from images in our field of study. However, more studies and experiments may be needed in order to reach highly accurate results. Therefore, employing convolutional neural networks for diagnosing tumors, especially melanoma skin cancer, can provide quality diagnostics for the associated disease and, thus, improve patient care. This study focuses on this technique, which can achieve good results. It is worth noting that several studies [13,14] have described convolutional neural networks as black-box models.
After considering the problem of diagnosing skin cancer and the procedures that follow, based on experience, it is necessary to search for solutions to diagnose this disease with high accuracy, speed, and flexibility in dealing with the patient, for example avoiding medical biopsies. Therefore, the current study aims to improve the diagnosis process, compared to the current traditional method. Deep learning is one of the fields of artificial intelligence, being a modern area in data science and an extension of machine learning; however, the flexibility of deep learning approaches, compared to machine learning approaches, has been shown [15,16,17]. The advent of big data has created a fertile environment for growth and progress in this field [18,19]. Deep learning models are inspired by the human brain, for example based on the large number of deep retinal neurons [16,20,21,22]. Therefore, convolutional neural networks have emerged in many fields, due to their good performance in image processing. Among these areas is the medical field, where they have been considered in diagnosis or treatment. With the advancement of technology and the significant expansion in the use of deep learning techniques to perform many tasks with high quality results, at the forefront of these areas is the medical field, which relates to human life through the diagnosis and treatment of diseases, especially diseases that threaten human life. These include cancers in general and melanoma in particular, which requires significant effort to reach an accurate diagnosis [23]. Hence, there exists a need to employ rapid techniques for early and accurate prediction of melanoma skin cancer based on skin images. Convolutional neural network algorithms have shown better results than other algorithms in this field [24]. Therefore, many researchers have attempted to develop relevant computational methods for detecting skin cancer utilizing skin images.
For example, Hekler et al. [25] adapted ResNet50 to discriminate melanoma from nevi using histopathological images. Furthermore, experiments demonstrated the good performance of this architecture.
Jojoa et al. [26] proposed a deep learning approach composed of segmentation and classifying a cropped image as benign or malignant. Moreover, one architecture was applied in their experiment. The results of the experiment indicated the good performance of the proposed approach.
The objective of Brinker et al. [27] was to compare a convolutional neural network architecture with dermatologists (157) in classifying melanoma skin cancer. Experimental results showed that the deep learning approach outperformed 136 dermatologists from 12 university hospitals in Germany; however, it can be seen that the criteria for selecting dermatologists play a role, as experience and skill are two essential aspects in the diagnosis process.
In general, it can be said that the deep learning approach showed superior performance. In another study [25], conducted in Germany by selecting eleven physicians with different areas of expertise, the results also indicated the superiority of a deep learning approach over physicians. However, doctors diagnosed the images through desktop screens, which may have played a role through the lower-than-normal resolution.
Another work [28] used 600 images divided into 117, 90, and 393 melanoma, seborrheic keratoses, and nevus images. The accuracy obtained by the deep learning approach was 81.6%, while two dermatologists obtained accuracies of 65.56% and 66.0%.
In another study [29], the authors proposed a new DermoDeep system consisting of five layers, namely a construction of visual features feature layer (VF-L), a deep feature layer (DF-L), a feature fusion layer (FF-L), an optimization of features layer (OF-L), and a prediction layer (FF-PL). They used a dataset of 2800 images divided equally between nevus and malignant lesions. This study compared the proposed method with other techniques and demonstrated that it can ensure better accuracy.
Other approaches have been proposed to detect and analyze melanoma skin cancer [25,30,31,32,33,34,35]. Table 1 lists the differences between the most important studies in this field.

Table 1.
Summary of the most important previous studies.
Contributions: We list the main contributions of this work as follows:
- Unlike existing studies, we include recent (and previous state-of-the-art) deep learning (DL) architectures to comprehensively investigate their performance differences. The utilized DL architectures in this study include DenseNet201, MobileNetV2, ResNet50V2, ResNet152V2, Xception, VGG16, VGG19, and GoogleNet.
- As the accurate prediction performance is of great importance in the melanoma skin cancer classification task, therefore, we aimed to identify the best-performing deep learning model to assist dermatologists in finding the appropriate AI tool.
- We report training results of all DL architectures in this study. Moreover, we record the generalization performance results using the ISIC benchmark dataset pertaining to the melanoma skin cancer classification task.
- Although large networks such as DenseNet201 and ResNet152V2 have notably more deep layers than GoogleNet and the other ones, experimental results demonstrate that GoogleNet generated the best performance results when measured using the standard performance measures. These results show the superiority of GoogleNet when tackling the melanoma skin cancer classification task.
2. Materials and Methods
2.1. Dataset
The International Skin Imaging Collaboration (ISIC) Dataset 2019 version was used in our experiment [36,37,38] (data downloaded from https://challenge.isic-archive.com/ accessed on 6 October 2021). It contains 25,333 images, 7146 of which are relevant to the current field of study, with 4522 images of melanoma skin cancer.
2.2. Deep Learning Approach
We employed eight popular deep learning architectures to investigate their performance differences. VGG16 consists of 13 convolutional layers interleaved with 5 max-pooling layers for feature extraction, followed by 3 fully connected layers for the classification [39]. VGG19 consists of 16 convolutional layers interleaved with 5 max-pooling layers for the feature extraction, followed by 3 fully connected layers for the classification [39]. GoogleNet contains nine inception modules (each consisting of two layers of six convolutional layers and a max-pooling layer) for feature extraction, as well as fully connected layers for classification [40]. In Xception, the inception module is replaced by depthwise separable convolutions, to be finally followed by fully connected layers for classification [41]. DenseNet201 starts with a convolutional layer, a max-pooling layer, followed by four dense blocks consisting of [6, 12, 48, 32] layers, transition layers between a pair of dense blocks, and a classification layer [42]. Unlike ResNet50 (consisting of 49 convolutional layers, 1 max-pooling, 1 average pooling layer, and 1 fully connected layer for classification) [43], ResNet50V2 is a modified version using a different residual unit [44]. Similarly, ResNet152V2 is a modified version of ResNet152 (consisting of 151 convolutional layers, 1 max-pooling, 1 average pooling layer, 1 fully connected layer for classification) [43,44]. MobileNetV2 is different from MobileNet, as it has a different layer module, leading to efficient models on mobile apps [45]. We used pre-trained models to extract features from the images, and we modified the fully connected layers to deal with the binary classification problem in our study.
In Figure 3, we show the computational deep learning approach for distinguishing between melanoma skin cancer images and non-melanoma skin cancer images [9]. We used the python script TensorFlow, Colaboratory, and convolutional neural networks as deep learning architectures to develop an efficient network for diagnosing melanoma skin cancer. Our objective was to train a specialized convolutional neural network model to detect whether an image showed melanoma skin cancer or not. The primary responsibility of this function is to draw common characteristics from the images and predict which category they belong to. This approach extracts features from the image and converts them into a new image, which is more efficient than the previous one for classification. Furthermore, it reduces the dimensions of the images to a good representation. The classification in our subject is binary; therefore, (1) indicated the case of melanoma skin cancer, and the reference (0) denoted non-melanoma skin cancer. At first, the data were read from the dataset file, and then, the image size was converted to 224 × 224 for the extraction of features. We explored and applied the model, then evaluated and analyzed the effect of the convolutional neural network architectures on the prognosis of melanoma skin cancer. The eight architectures that were used in this study, which are reviewed below, were as follows: DenseNet201, MobileNetV2, ResNet50V2, ResNet152V2, Xception, VGG16, VGG19, and GoogleNet. The data were divided into training, validation, and test sets. The first set was used to train the networks, and the second set was used to monitor and improve the model performance during the training process. Training, validation, and testing were performed using all the images in each architecture. At the same time, the third set was used to test the trained model; see Figure 4. The implementation of the project was carried out in Google Colaboratory. Python libraries including TensorFlow, Keras, pandas, NumPy, matplotlib, sklearn, scipy, torch, and seaborn were used, among others. Finally, we obtained performance measures for the trained models, including the accuracy, precision, recall, F1-score, training and validation losses, accuracy graphs, and confusion matrix.
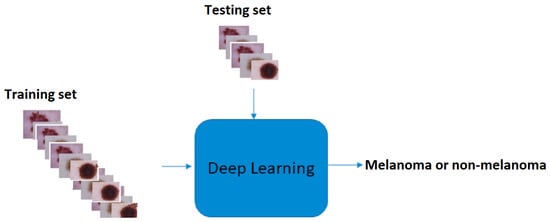
Figure 3.
The computational deep learning framework for distinguishing between melanoma skin cancer images and non-melanoma skin cancer images. Skin cancer images from [36,37,38].
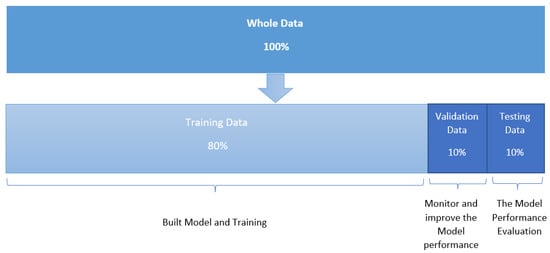
Figure 4.
Division of the dataset into training, verification, and testing sets.
The stages of the experiment can be summarized in six stages. The first stage began by taking data from the dataset separately. Then, processing of the images was carried out, followed by data division. Then, the model building stage was conducted for several architectures, and the associated results were assessed in the following two stages. Finally, the testing phase was carried out, where the testing operations were performed on the testing set sample. Several measures were recorded in order to compare the architectures used in building the models, as well as the implementation process after saving the model, through checking the best obtained values after making comparisons between them.
3. Results
We display the analysis results of the experiment in three main parts, as follows.
3.1. Classification Methodology
We adopted eight deep learning architectures, including DenseNet201, MobileNetV2, ResNet50V2, ResNet152V2, Xception, VGG16, VGG19, and GoogleNet. All of these previous architectures operate under a supervised learning framework. First, the model trains using a subset of the full dataset, verifying and adjusting the model’s performance in the process. Then, the third set (i.e., the testing set) was used to test the model. The model can generate specific predictions from 1 (positive) or 0 (negative). Several performance measures were used to evaluate the model performance, including accuracy, precision, recall, and F1-score. In addition, the training loss, validation loss, accuracy graphs, and confusion matrix were also considered, in order to carry out comprehensive evaluation of the models.
3.2. Implementation Details
We used Anaconda Python Version 3.7.4 with a Jupyter Notebook and several libraries, such as Keras and Sklearn. In addition, Google Colaboratory GPU was used to process the execution of the experiment.
3.3. Classification Results
The summary of the experimental results for all the architectures used on the dataset is provided in Table 2. It can be seen that GoogleNet generated the best performance results, as shown in bold. Its training accuracy was 74.91%, and the test accuracy was 76.08%. Then, DenseNet201 and ResNet50V2 achieved the next-highest accuracy results, with slightly lower training accuracy (73.96% and 73.74%, respectively) and test accuracy (74.68% and 73.42%, respectively). The worst accuracy performance was obtained by VGG16 and VGG19. The rest of the architectures achieved average results, ranging between 70and 74% for both training and test accuracy.

Table 2.
Performance comparison between different convolutional neural network models.
A confusion matrix is needed to measure the overall classification performance. In Figure 5, we show confusion matrices for the eight architectures used in this study. The test data included 466 positive melanoma skin cancer images and 249 negative melanoma skin cancer images. In the GoogleNet model, the number of correctly predicted examples was 538; thus, this CNN model misclassified 177 out of 715 images. When examining the results obtained by the DenseNet201 model, 535 images were classified correctly and 180 images were classified incorrectly. ResNet50V2 had good results, similar to the two above-mentioned architectures. In detail, Table 3 displays the confusion matrices for all the architectures performed in this experiment, in addition to the number of images rated by the model that were correct or miscategorized (whether false positive or false negative). TP denotes true positive, referring to the number of melanoma skin cancer images that were correctly predicted as melanoma skin cancer images. FN denotes false negative, referring to the number of melanoma skin cancer images that were incorrectly predicted as non-melanoma skin cancer images. TN denotes true negative, referring to the number of non-melanoma skin cancer images that were correctly predicted as non-melanoma skin cancer images. FP denotes false positive, referring to the number of non-melanoma skin cancer images that were incorrectly predicted as melanoma skin cancer images. Furthermore, Table 4 presents the performance results for all architectures, based on the metrics of balanced accuracy, F1-score, precision, and recall.
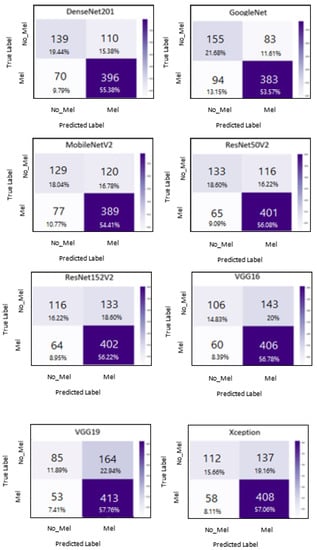
Figure 5.
Confusion matrices for all architectures used in the experiment.

Table 3.
Confusion matrix data for all architectures used in the experiment. TP, true positive. FN, false negative. TN, true negative. FP, false positive.

Table 4.
Performance results for all architectures based on various metrics.
Additionally, we display the training and validation loss of the CNN models used in the experiment, as shown in Figure 6, Figure 7, Figure 8 and Figure 9.
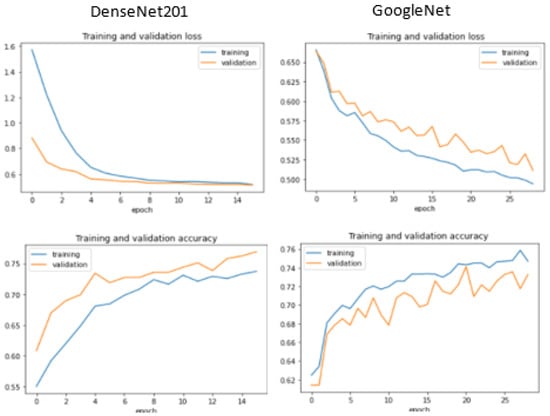
Figure 6.
Training and validation performance for DenseNet 201 and GoogleNet.
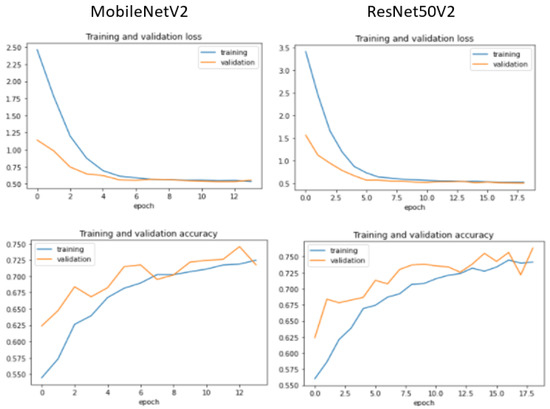
Figure 7.
Training and validation performance for MobileNetV2 and ResNet50V2.
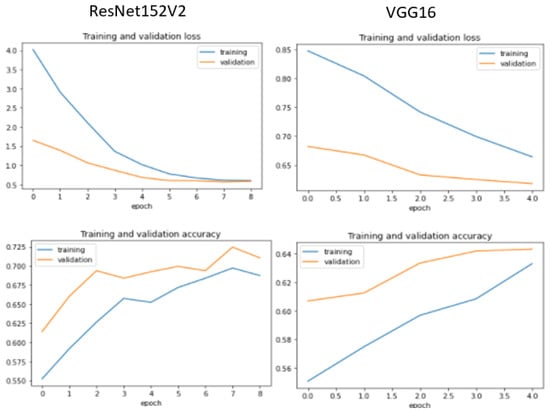
Figure 8.
Training and validation performance for ResNet152V2 and VGG16.
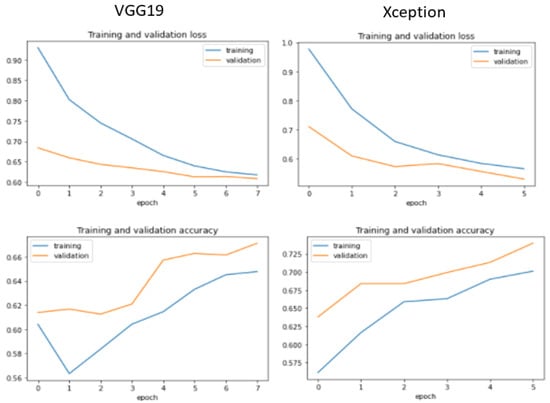
Figure 9.
Training and validation performance for VGG19 and Xception.
Table 5 presents the best number of epochs obtained for all architectures used in the experiment. A total of 50 epochs was applied for each architecture; however, if the accuracy decreased, the implementation process was exited and the best epoch was selected.

Table 5.
The best epoch obtained for the architectures used in the experiment.
4. Discussion
The main objective of this study was to develop various deep learning techniques for the classification of melanoma skin cancer and then to compare the performance results of these architectures. We adapted several deep learning architectures, including a custom CNN for the binary classification of melanoma. We resized the 7146 images in the dataset to 224 × 224 pixels. ImageDataGenerator technology was used to overcome the data volume problem. We then evaluated and compared the model performance for all of the architectures that were considered, based on common performance measures. It is worth noting that GoogleNet performed the best, when compared to the other deep learning models. When comparing this study with previous empirical studies, such as [27,28], we found that the distinction of this study is that we used several CNN architectures (i.e., eight architectures). The results differed between studies, and the results of this study may be good, to some extent. We may also be more comprehensive in adapting several architectures, as the results may be improved by using a larger dataset. Some research studies [46,47,48] have indicated that deep learning approaches requires massive data in order to train the network well and, thus, provide more accurate results. However, one of the challenges associated with this is the availability of high-quality hardware, such as GPUs. For example, our experiment on a modestly sized dataset (7146 images) took about six and a half hours to perform all operations with eight architectures, even though we used GPUs using Colaboratory. The larger the dataset, the more time that must be spent. Some studies [49] have indicated that CNNs should be trained on high-quality data, as noisy image data may increase misclassification rates. We noticed, throughout the course of the experiment, that some images contained some noise, such as shadows and hair, which may have reduced their quality. Thus, the extraction of essential features may be affected when training the network, which can lead to the wrong training on some features. Thus, ensuring only high-quality images are available in the dataset is a critical matter.
It can be seen from Table 4 and Figure 5 that ResNet-based architectures utilized (and promoted) in previous studies [25,26,27,28] did not outperform GoogleNet used in this study. GoogleNet performed better than MobileNetV2, which was promoted and used in [35]. It is worth noting that GoogleNet was not utilized in [25,26,27,28,35], and we utilized improved versions of ResNet-based architectures such as ResNet50V2 and ResNet152V2.
We used a moderate amount of the training and evaluation data in the experiment, which may play a role in training and testing deep neural network models. Therefore, we intend to expand upon this experiment through the use of larger image datasets, as several studies have indicated that convolutional neural networks should be fed extensive data to obtain results with very high accuracy.
Through the results of this study, we aimed to develop technological systems for use in daily life and to assist dermatologists in effectively detecting skin cancer. Moreover, the more images of melanoma of different types that the CNN is exposed to, the higher its accuracy in recognizing different features. This topic may be interesting in future research. However, unlike previous studies, we performed a detailed experimental study by adopting a range of deep learning architectures to comparatively assess their performance behavior and to identify the best-performing deep learning model for the classification task of melanoma skin cancer using medical images. As each DL architecture had a different number of processing layers, it can be observed that GoogleNet (with 22 layers) performed better than the others with more (or less) deep layers considered in this study, when considering the melanoma skin cancer classification task [50].
Deep learning (DL) requires hardware resources such as GPU to speed up the training time [51]. Unlike existing studies, we included recent DL architectures using the ISIC benchmark dataset to identify the appropriate DL architecture when specifically tackling the melanoma skin cancer classification task. It is worth noting that training was performed on an end-to-end basis using melanoma and non-melanoma skin cancer images. Therefore, we show the feasibility of DL when finding whether skin has melanoma or not without the need for the domain expert’s intervention for the feature engineering part.
5. Conclusions and Future Work
At present, the health field is one of the most prominent areas that has adopted the use of deep learning techniques. In this work, we performed binary classification of skin images using convolutional neural networks for the diagnosis of melanoma skin cancer. Several CNN architectures were adapted, including DenseNet201, MobileNetV2, ResNet50V2, ResNet152V2, Xception, VGG16, VGG19, and GoogleNet. One dataset was used for training these models, a second set to validate and adjust the training process, and finally, a third set to test each trained model, in order to assess the validity of its predictions. The experiment included 7146 images as the whole dataset. The experimental results on this dataset demonstrated that GoogleNet outperformed all the other models.
Future work will include: (1) proposing a hybrid approach based on machine learning and deep learning to improve the prediction performance; (2) incorporating different data augmentation techniques to enhance the prediction performance; (3) testing the performance under different learning settings, including active learning and transfer learning; and (4) boosting the performance of GoogleNet, as detailed in [51].
Author Contributions
Conceptualization, K.A. and T.T.; methodology, K.A.; validation, K.A. and T.T.; formal analysis, K.A. and T.T.; investigation, K.A. and T.T.; resources, K.A. and T.T.; data curation, K.A. and T.T.; writing—original draft preparation, K.A.; writing—review and editing, K.A. and T.T. All authors have read and agreed to the published version of the manuscript.
Funding
This project was funded by the Deanship of Scientific Research (DSR) at King Abdulaziz University, Jeddah, under grant no. D-087-611-1440. The authors, therefore, thank DSR for technical and financial support.
Institutional Review Board Statement
Not applicable.
Informed Consent Statement
Not applicable.
Data Availability Statement
The dataset was downloaded from: https://challenge.isic-archive.com/data (accessed on 11 April 2022).
Conflicts of Interest
The authors declare no conflict of interest.
References
- Argenziano, G.; Longo, C.; Cameron, A.; Cavicchini, S.; Gourhant, J.Y.; Lallas, A.; McColl, I.; Rosendahl, C.; Thomas, L.; Tiodorovic-Zivkovic, D.; et al. Blue-black rule: A simple dermoscopic clue to recognize pigmented nodular melanoma. Br. J. Dermatol. 2011, 165, 1251–1255. [Google Scholar] [CrossRef] [PubMed]
- Vestergaard, M.; Macaskill, P.; Holt, P.; Menzies, S. Dermoscopy compared with naked eye examination for the diagnosis of primary melanoma: A meta-analysis of studies performed in a clinical setting. Br. J. Dermatol. 2008, 159, 669–676. [Google Scholar] [CrossRef] [PubMed]
- Charoenpong, J.; Pimpunchat, B.; Amornsamankul, S.; Triampo, W.; Nuttavut, N. A Comparison of Machine Learning Algorithms and their Applications. Int. J. Simul.–Syst. Sci. Technol. 2019, 20, 1–17. [Google Scholar]
- Rezayi, S.; Mohammadzadeh, N.; Bouraghi, H.; Saeedi, S.; Mohammadpour, A. Timely Diagnosis of Acute Lymphoblastic Leukemia Using Artificial Intelligence-Oriented Deep Learning Methods. Comput. Intell. Neurosci. 2021, 2021, 5478157. [Google Scholar] [CrossRef]
- Tufail, A.B.; Ma, Y.K.; Kaabar, M.K.; Martínez, F.; Junejo, A.; Ullah, I.; Khan, R. Deep learning in cancer diagnosis and prognosis prediction: A minireview on challenges, recent trends, and future directions. Comput. Math. Methods Med. 2021, 2021, 9025470. [Google Scholar] [CrossRef]
- Li, X.; Jiao, H.; Wang, Y. Edge detection algorithm of cancer image based on deep learning. Bioengineered 2020, 11, 693–707. [Google Scholar] [CrossRef]
- Gheisari, M.; Wang, G.; Bhuiyan, M.Z.A. A survey on deep learning in big data. In Proceedings of the 2017 IEEE International Conference on Computational Science and Engineering (CSE) and IEEE International Conference on Embedded and Ubiquitous Computing (EUC), Guangzhou, China, 21–24 July 2017; Volume 2, pp. 173–180. [Google Scholar]
- Banka, A.A.; Mir, R.N. Current Big Data Issues and Their Solutions via Deep Learning: An Overview. Iraqi J. Electr. Electron. Eng. 2018, 14, 127–138. [Google Scholar] [CrossRef]
- Alzubaidi, L.; Zhang, J.; Humaidi, A.J.; Al-Dujaili, A.; Duan, Y.; Al-Shamma, O.; Santamaría, J.; Fadhel, M.A.; Al-Amidie, M.; Farhan, L. Review of deep learning: Concepts, CNN architectures, challenges, applications, future directions. J. Big Data 2021, 8, 1–74. [Google Scholar] [CrossRef]
- Chen, X.; Weng, J.; Lu, W.; Xu, J.; Weng, J. Deep manifold learning combined with convolutional neural networks for action recognition. IEEE Trans. Neural Netw. Learn. Syst. 2017, 29, 3938–3952. [Google Scholar] [CrossRef]
- Ghods, A.; Cook, D.J. A survey of deep network techniques all classifiers can adopt. Data Min. Knowl. Discov. 2021, 35, 46–87. [Google Scholar] [CrossRef]
- Liu, W.; Wang, Z.; Liu, X.; Zeng, N.; Liu, Y.; Alsaadi, F.E. A survey of deep neural network architectures and their applications. Neurocomputing 2017, 234, 11–26. [Google Scholar] [CrossRef]
- Yamashita, R.; Nishio, M.; Do, R.K.G.; Togashi, K. Convolutional neural networks: An overview and application in radiology. Insights Imaging 2018, 9, 611–629. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Khan, A.; Sohail, A.; Zahoora, U.; Qureshi, A.S. A survey of the recent architectures of deep convolutional neural networks. Artif. Intell. Rev. 2020, 53, 5455–5516. [Google Scholar] [CrossRef] [Green Version]
- Dong, Y.N.; Liang, G.S. Research and discussion on image recognition and classification algorithm based on deep learning. In Proceedings of the 2019 International Conference on Machine Learning, Big Data and Business Intelligence (MLBDBI), Taiyuan, China, 8–10 November 2019; pp. 274–278. [Google Scholar]
- Sharma, O. Deep challenges associated with deep learning. In Proceedings of the 2019 International Conference on Machine Learning, Big Data, Cloud and Parallel Computing (COMITCon), Faridabad, India, 14–16 February 2019; pp. 72–75. [Google Scholar]
- Rajput, D.S.; Reddy, T.S.K.; Raju, D.N. Investigation on Deep Learning Approach for Big Data: Applications and Challenges. In Deep Learning and Neural Networks: Concepts, Methodologies, Tools, and Applications; IGI Global: Hershey, PA, USA, 2020; pp. 1016–1029. [Google Scholar]
- Jan, B.; Farman, H.; Khan, M.; Imran, M.; Islam, I.U.; Ahmad, A.; Ali, S.; Jeon, G. Deep learning in big data analytics: A comparative study. Comput. Electr. Eng. 2019, 75, 275–287. [Google Scholar] [CrossRef]
- Zhang, Q.; Yang, L.T.; Chen, Z.; Li, P. A survey on deep learning for big data. Inf. Fusion 2018, 42, 146–157. [Google Scholar] [CrossRef]
- Qian, Y. Exploration of machine algorithms based on deep learning model and feature extraction. Math. Biosci. Eng. 2021, 18, 7602–7618. [Google Scholar] [CrossRef]
- Pouyanfar, S.; Sadiq, S.; Yan, Y.; Tian, H.; Tao, Y.; Reyes, M.P.; Shyu, M.L.; Chen, S.C.; Iyengar, S.S. A survey on deep learning: Algorithms, techniques, and applications. ACM Comput. Surv. (CSUR) 2018, 51, 1–36. [Google Scholar] [CrossRef]
- Cullell-Dalmau, M.; Otero-Viñas, M.; Manzo, C. Research techniques made simple: Deep learning for the classification of dermatological images. J. Investig. Dermatol. 2020, 140, 507–514. [Google Scholar] [CrossRef] [Green Version]
- Schadendorf, D.; van Akkooi, A.C.; Berking, C.; Griewank, K.G.; Gutzmer, R.; Hauschild, A.; Stang, A.; Roesch, A.; Ugurel, S. Melanoma. Lancet 2018, 392, 971–984. [Google Scholar] [CrossRef]
- Chassagnon, G.; Vakalopolou, M.; Paragios, N.; Revel, M.P. Deep learning: Definition and perspectives for thoracic imaging. Eur. Radiol. 2020, 30, 2021–2030. [Google Scholar] [CrossRef]
- Hekler, A.; Utikal, J.S.; Enk, A.H.; Berking, C.; Klode, J.; Schadendorf, D.; Jansen, P.; Franklin, C.; Holland-Letz, T.; Krahl, D.; et al. Pathologist-level classification of histopathological melanoma images with deep neural networks. Eur. J. Cancer 2019, 115, 79–83. [Google Scholar] [CrossRef] [Green Version]
- Acosta, M.F.J.; Tovar, L.Y.C.; Garcia-Zapirain, M.B.; Percybrooks, W.S. Melanoma diagnosis using deep learning techniques on dermatoscopic images. BMC Med. Imaging 2021, 21, 6. [Google Scholar] [CrossRef] [PubMed]
- Brinker, T.J.; Hekler, A.; Enk, A.H.; Berking, C.; Haferkamp, S.; Hauschild, A.; Weichenthal, M.; Klode, J.; Schadendorf, D.; Holland-Letz, T.; et al. Deep neural networks are superior to dermatologists in melanoma image classification. Eur. J. Cancer 2019, 119, 11–17. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Bisla, D.; Choromanska, A.; Berman, R.S.; Stein, J.A.; Polsky, D. Towards automated melanoma detection with deep learning: Data purification and augmentation. In Proceedings of the IEEE/CVF Conference on Computer Vision and Pattern Recognition Workshops, Long Beach, CA, USA, 16–20 June 2019. [Google Scholar]
- Abbas, Q.; Celebi, M.E. DermoDeep-A classification of melanoma-nevus skin lesions using multi-feature fusion of visual features and deep neural network. Multimed. Tools Appl. 2019, 78, 23559–23580. [Google Scholar] [CrossRef]
- Tognetti, L.; Bonechi, S.; Andreini, P.; Bianchini, M.; Scarselli, F.; Cevenini, G.; Moscarella, E.; Farnetani, F.; Longo, C.; Lallas, A.; et al. A new deep learning approach integrated with clinical data for the dermoscopic differentiation of early melanomas from atypical nevi. J. Dermatol. Sci. 2020, 101, 115–122. [Google Scholar] [CrossRef] [PubMed]
- Pennig, L.; Shahzad, R.; Caldeira, L.; Lennartz, S.; Thiele, F.; Goertz, L.; Zopfs, D.; Meißner, A.K.; Fürtjes, G.; Perkuhn, M.; et al. Automated Detection and Segmentation of Brain Metastases in Malignant Melanoma: Evaluation of a Dedicated Deep Learning Model. Am. J. Neuroradiol. 2021, 42, 655–662. [Google Scholar] [CrossRef]
- Zhou, Q.; Shi, Y.; Xu, Z.; Qu, R.; Xu, G. Classifying melanoma skin lesions using convolutional spiking neural networks with unsupervised stdp learning rule. IEEE Access 2020, 8, 101309–101319. [Google Scholar] [CrossRef]
- Pollastri, F.; Bolelli, F.; Paredes, R.; Grana, C. Augmenting data with GANs to segment melanoma skin lesions. Multimed. Tools Appl. 2020, 79, 15575–15592. [Google Scholar] [CrossRef] [Green Version]
- Winkler, J.K.; Fink, C.; Toberer, F.; Enk, A.; Deinlein, T.; Hofmann-Wellenhof, R.; Thomas, L.; Lallas, A.; Blum, A.; Stolz, W.; et al. Association between surgical skin markings in dermoscopic images and diagnostic performance of a deep learning convolutional neural network for melanoma recognition. JAMA Dermatol. 2019, 155, 1135–1141. [Google Scholar] [CrossRef]
- Almaraz-Damian, J.A.; Ponomaryov, V.; Sadovnychiy, S.; Castillejos-Fernandez, H. Melanoma and nevus skin lesion classification using handcraft and deep learning feature fusion via mutual information measures. Entropy 2020, 22, 484. [Google Scholar] [CrossRef] [Green Version]
- Combalia, M.; Codella, N.C.; Rotemberg, V.; Helba, B.; Vilaplana, V.; Reiter, O.; Carrera, C.; Barreiro, A.; Halpern, A.C.; Puig, S.; et al. BCN20000: Dermoscopic lesions in the wild. arXiv 2019, arXiv:1908.02288. [Google Scholar]
- Tschandl, P.; Rosendahl, C.; Kittler, H. The HAM10000 dataset, a large collection of multi-source dermatoscopic images of common pigmented skin lesions. Sci. Data 2018, 5, 180161. [Google Scholar] [CrossRef] [PubMed]
- Codella, N.C.; Gutman, D.; Celebi, M.E.; Helba, B.; Marchetti, M.A.; Dusza, S.W.; Kalloo, A.; Liopyris, K.; Mishra, N.; Kittler, H.; et al. Skin lesion analysis toward melanoma detection: A challenge at the 2017 international symposium on biomedical imaging (isbi), hosted by the international skin imaging collaboration (isic). In Proceedings of the 2018 IEEE 15th international symposium on biomedical imaging (ISBI 2018), Washington, DC, USA, 4–7 April 2018; pp. 168–172. [Google Scholar]
- Simonyan, K.; Zisserman, A. Very deep convolutional networks for large-scale image recognition. arXiv 2014, arXiv:1409.1556. [Google Scholar]
- Ragab, D.A.; Attallah, O.; Sharkas, M.; Ren, J.; Marshall, S. A framework for breast cancer classification using multi-DCNNs. Comput. Biol. Med. 2021, 131, 104245. [Google Scholar] [CrossRef]
- Chollet, F. Xception: Deep learning with depthwise separable convolutions. In Proceedings of the IEEE Conference on Computer Vision and Pattern Recognition, Honolulu, HI, USA, 21–26 July 2017; pp. 1251–1258. [Google Scholar]
- Huang, G.; Liu, Z.; Van Der Maaten, L.; Weinberger, K.Q. Densely connected convolutional networks. In Proceedings of the IEEE Conference on Computer Vision and Pattern Recognition, Honolulu, HI, USA, 21–26 July 2017; pp. 4700–4708. [Google Scholar]
- He, K.; Zhang, X.; Ren, S.; Sun, J. Deep residual learning for image recognition. In Proceedings of the IEEE Conference on Computer Vision and Pattern Recognition, Las Vegas, NV, USA, 27–30 June 2016; pp. 770–778. [Google Scholar]
- He, K.; Zhang, X.; Ren, S.; Sun, J. Identity mappings in deep residual networks. In Computer Vision—ECCV 2016, Proceedings of the European Conference on Computer Vision, Amsterdam, The Netherlands, 11–14 October 2016; Springer: Cham, Switzerland, 2016; pp. 630–645. [Google Scholar]
- Howard, A.G.; Zhu, M.; Chen, B.; Kalenichenko, D.; Wang, W.; Weyand, T.; Andreetto, M.; Adam, H. Mobilenets: Efficient convolutional neural networks for mobile vision applications. arXiv 2017, arXiv:1704.04861. [Google Scholar]
- Najafabadi, M.M.; Villanustre, F.; Khoshgoftaar, T.M.; Seliya, N.; Wald, R.; Muharemagic, E. Deep learning applications and challenges in big data analytics. J. Big Data 2015, 2, 1–21. [Google Scholar] [CrossRef] [Green Version]
- Christin, S.; Hervet, É.; Lecomte, N. Applications for deep learning in ecology. Methods Ecol. Evol. 2019, 10, 1632–1644. [Google Scholar] [CrossRef]
- Angermueller, C.; Pärnamaa, T.; Parts, L.; Stegle, O. Deep learning for computational biology. Mol. Syst. Biol. 2016, 12, 878. [Google Scholar] [CrossRef]
- Szegedy, C.; Zaremba, W.; Sutskever, I.; Bruna, J.; Erhan, D.; Goodfellow, I.; Fergus, R. Intriguing properties of neural networks. arXiv 2013, arXiv:1312.6199. [Google Scholar]
- Khan, S.; Muhammad, K.; Mumtaz, S.; Baik, S.W.; de Albuquerque, V.H.C. Energy-efficient deep CNN for smoke detection in foggy IoT environment. IEEE Internet Things J. 2019, 6, 9237–9245. [Google Scholar] [CrossRef]
- Turki, T.; Wei, Z. Improved Deep Convolutional Neural Networks via Boosting for Predicting the Quality of In Vitro Bovine Embryos. Electronics 2022, 11, 1363. [Google Scholar] [CrossRef]
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).