Abstract
The grooved carpet-shell clam is one of the most economically relevant shellfish species living in the Mediterranean and nearby Atlantic coasts. Previous studies using different types of genetic markers showed a remarkable genetic divergence of the eastern Mediterranean, western Mediterranean, and Atlantic populations, but important details remained unclear. Here, data from six nuclear introns scored for restriction fragment size polymorphisms in eight populations that have not been studied before have been pooled for the analysis with data scattered through three previous studies, totaling 32 samples from 29 locations. The results show lower levels of heterozygosity, higher mean number of alleles, and alleles with restricted distribution in the Mediterranean populations, suggesting the existence of a large, isolated population in the eastern Mediterranean at the middle Pleistocene. The data also confirm the similarity of populations from Tunisia to Western Mediterranean populations. Finally, a genetic mosaic is apparent in the Atlantic coasts of the Iberian Peninsula, with a divergence of Rias Baixas populations from more northern populations and Central Portugal populations. The effects of oceanic fronts, seasonal upwellings, river plumes, and/or fishery management operations could explain this and other features of the Atlantic populations.
1. Introduction
The grooved carpet-shell clam (Ruditapes decussatus) lives in estuaries, lagoons, and tidal flats along the coasts of the northeastern Atlantic Ocean and the Mediterranean Sea. It is one of the main commercial clam species native to this area. However, catches have decreased along this century in the European fishing grounds due to overfishing, recruitment failure, and mortality [1,2,3]. Clam fishery management practice has changed with the time. In the second half of the past century, when the market grew, spat was collected and translocated to areas where the environment favored survival and growth. In the 1970s, overfished stocks in Spain were replenished with juveniles taken in wild populations from other regions or other countries. European regulations now set limits to daily catches and a minimum size at harvest. Since the 1990s, bivalve hatcheries have been producing clam juveniles, especially in France, Spain, and Italy, making hatchery seed available for population restocking and grow-out in licensed coastal areas. These developments in management have raised an interest in the genetic aspects of wild and captive populations.
One of the tools to refine the management of clam stocks is the knowledge of the species’ population genetic structure. The population genetic data for R. decussatus are spread across several publications, each based on a handful of populations, often restricted to particular regions, and based on different types of genetic markers (protein polymorphisms, sequences of mitochondrial DNA fragments, restriction fragment length polymorphisms of introns in nuclear genes, and microsatellites), which, all together, make it difficult to reach general conclusions. Early studies based on protein polymorphisms indicated a high level of genetic variability and a low genetic differentiation of the populations [4,5]. With the availability of DNA-based genetic markers and increased population sampling, it was possible to detect a remarkable genetic differentiation among wide regions along the distribution range of the species. Using sequences of the mitochondrial cytochrome oxidase I gene (COI), and restriction fragment length polymorphisms (RFLPs) or size polymorphisms in sequences of introns of six genes, Cordero et al. reported a phylogeographic break (a sharp change in the sequences that appears in a DNA phylogeny between two or more geographic regions) located between the western Mediterranean Sea and the Adriatic and the Aegean seas [6]. The genetic structure reported by Cordero et al. from the mitochondrial marker was corroborated later by Sanna et al. [7]. Cordero et al. also reported a strong genetic differentiation between the populations located on the Atlantic coasts of southern Europe and the western Mediterranean Sea, but only for the intronic markers. The intronic markers also showed a strong differentiation between the populations in the eastern and western basins of the Mediterranean Sea. The pattern of differentiation between Atlantic populations and West and East Mediterranean populations has been reported in many species and is usually interpreted as the result of a secondary contact between Atlantic and Mediterranean subpopulations that were separated once or more times in the Pleistocene, when the sea level dropped because of the glaciations, cutting the connection between the Atlantic Ocean, the West Mediterranean Sea, and the East Mediterranean Sea. Present day oceanographic features such as the Almeria–Oran oceanographic front (AOOF) (Figure 1) and the patterns of marine circulation past the Siculo–Tunisian Strait are thought to contribute to the persistence of the genetic differentiation [8,9]. The effect of natural selection could favor the coincidence of allele frequency changes with oceanographic barriers [10,11].
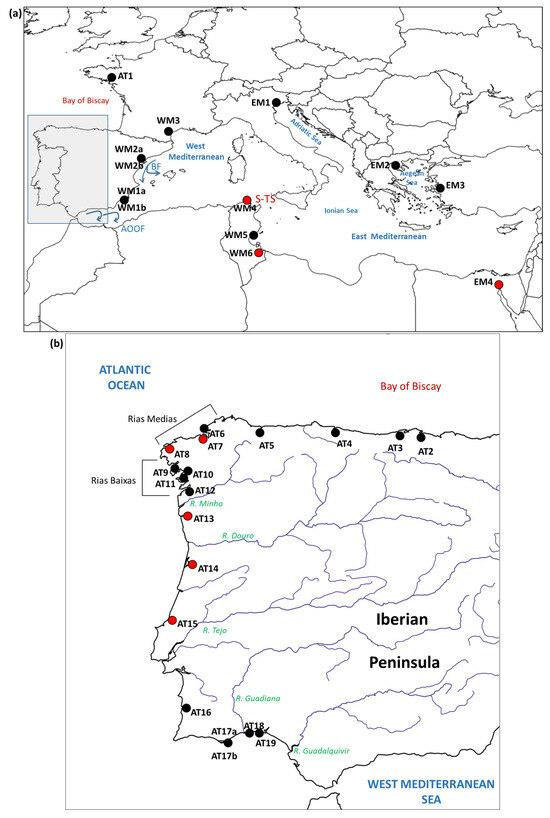
Figure 1.
Maps showing the locations considered in this study and the main geographic and oceanographic features cited in the text. Red dots show the new locations sampled for this study. Black dots show locations sampled in previous studies. (a) Locations outside of the Atlantic coasts of the Iberian Peninsula. BF: Balearic Front. AOOF: Almeria–Oran oceanographic front; S-TS: Siculo–Tunisian Strait. (b) Locations on the Atlantic coasts of the Iberian Peninsula.
The study of microsatellite markers in grooved carpet-shell clam populations from the Spanish Atlantic and Mediterranean coasts by Arias-Pérez et al. confirmed the genetic differentiation between the Atlantic and West Mediterranean populations and suggested the genetic differentiation of the populations from the central and northern Atlantic façade of the Iberian Peninsula with respect to those from the Bay of Biscay and the SW of the Iberian Peninsula [12]. Gharbi et al. studied 12 populations from Tunisia using sequences of COI and the internal transcribed spacers of the ribosomal RNA genes (ITS) and found little differentiation between populations [13]. Later, in a study focused on Atlantic Spanish and Portuguese coasts using microsatellites, Cruz et al. discovered an increased genetic differentiation of two populations located on the coasts of central Portugal, with respect to those from Rias Baixas and the Bay of Biscay (including one population from Rias Medias) [14]. Amane et al. [15] studied seven populations from Moroccan coasts with microsatellites and found a significant genetic differentiation of populations located south of parallel 25° N. Finally, Saavedra et al. [16] scored four populations for intron polymorphisms. They studied one population in southern France, an area that had not been studied before with nuclear markers, where they found alleles typical of the eastern Mediterranean in very low frequency.
There are several aspects of the previous studies that require more detailed scrutiny. Here, we will deal with three of them. Firstly, Cordero et al. found a strong genetic differentiation between the eastern and the western Mediterranean basins. This is a common observation in genetic studies of populations of many species of marine organisms [8,9,10]. However, in the study by Cordero et al., only populations from the northern coasts of the eastern Mediterranean (Adriatic and Aegean seas) were included. The observation of the Tunisian population joining the western Mediterranean populations in several analyses suggests that northern African populations could be more related to the western Mediterranean populations than to the Aegean and Adriatic ones. Therefore, it remains to be clarified if the clam populations from the southern part of the eastern Mediterranean are more similar to their western or to their northern counterparts. A second aspect that requires further scrutiny is related to the study of Gharbi et al., who found that populations from the northern and the eastern coasts of Tunisia did not differ markedly in the frequencies of COI and ITS haplotypes [6,13]. This observation agrees with the results reported by Cordero et al. for the Tunisian population, which clustered with the western Mediterranean populations. These results are in contrast to the majority of studies of other species, which usually report a genetic break placed at the Siculo–Tunisian strait (Figure 1), with the populations on the eastern coasts of Tunisia clustering with other eastern Mediterranean populations, and those from northern Tunisia clustering with the western Mediterranean ones (e.g., references [17,18]). Finally, the findings by Arias-Pérez et al. and Cruz et al. of a mosaic of genetically differentiated areas in central and N Portugal, and in the estuaries of NW Spain, are especially important from the point of view of fisheries management, as they affect one of the regions where exploitation of the species is most intensive. Therefore, it is interesting to characterize the genetics of the clam populations in that region in more detail.
The goals of this study are to clarify the aspects referred to above and to provide a comprehensive and up-to-date report of the genetic structure of Ruditapes decussatus populations and its potential causes, based on a much larger number of populations than those considered so far. We present new RFLP data from six introns in seven populations from NW Spain, Portugal, Tunisia, and Egypt that have not been studied before. For the analysis, these data have been pooled with those collected in our previous works [6,12,16], totaling 32 samples from 29 locations distributed from the eastern Mediterranean to NW France. This approach has led to the discovery of new aspects of the genetic structure of the grooved carpet shell clam that may be important for the management of the species’ genetic resources in fisheries and aquaculture, but had remained hidden due to the limited geographic sampling of previous studies.
2. Materials and Methods
2.1. Clam Sampling and DNA Extraction
Eight new locations from Spain (2), Portugal (3), Tunisia (2), and Egypt (1) have been sampled for this study (Figure 1 and Table 1). Clams from most locations were obtained from local fishermen and transported live to the IATS-CSIC facilities. Some cases of hybridization of R. decussatus with the introduced Manila clam (Ruditapes philippinarum) have been reported [19,20]. Special care was payed to not introduce in our study any Manila clam, which can be distinguished at the moment of collection or tissue sampling by the morphology of the shell and the siphons [21], or their hybrids, which are produced in very low amounts in nature and could be detected genetically by the fact that several introns can be amplified with our primers in the two species but result in specific PCR products of different size (e.g., reference [22] and our unpublished results).

Table 1.
Sampling data from the 32 populations included in this study. Population codes refer to Atlantic coasts (AT), West Mediterranean including eastern Tunisian coasts (WM), and eastern Mediterranean excluding Tunisian coasts (EM).
Upon collection, animals were euthanized and tissues of three organs (mantle, siphons, gills) were preserved in ethanol 90%. The samples from Egypt (EM4) and Tunisia (WM4 and WM6) were sent to the IATS as a piece of mantle or siphons preserved in eth-anol. DNA extraction was performed by using the EZNA Mollusc DNA extraction kit (Omega Bio-tek) following the protocol of the manufacturer. DNA quality and concentration were checked with agarose gel electrophoresis and by using a NanoDrop instrument.
After extraction, DNA was kept frozen at −20 °C until further use.
2.2. Intron Amplification and DNA Polymorphism Detection
The methods for amplification of the introns and restriction fragment length-polymorphism detection were exactly the same as used in our previous studies [6]. Six introns from different genes were studied: enoyl coenzyme A hydratase (Ech), fasciclin-like protein (Fas), signal recognition particle 54-kDa subunit (SRP54), TATA box binding protein/transcription factor IID (TBP), tRNA aspartic acid methyltransferase (Trdmt), and ubiquitin conjugating enzyme (Ubc). The introns were amplified in an Applied Biosystems GeneAmp PCR System 9700 thermocycler using intron-specific primer pairs. The digestion of the PCR products was conducted with a single restriction enzyme. The TBP PCR product showed a length polymorphism, so this marker did not need to be digested. DNA fragments were separated by electrophoresis in agarose gels, stained with GreenSafe Premium (Nzytech), and photographed under UV light. The sizes of the PCR products and the restriction fragments have been described previously [6]. Fragment sizes were estimated by comparison with the NZYDNA Ladder III (Nzytech). Typical results of electrophoresis for all introns and the conversion of DNA bands to genotypes are shown in Figure S1 (Supplementary Materials). Individuals of known genotypes from previous studies were run together with the newly scored samples as controls. Samples showing low-frequency alleles or potentially new alleles were amplified, digested, and run again all together at the end of the study to check for identical or different RFLP patterns.
2.3. Data Analysis
The data obtained from the eight new populations were pooled with those from our previous studies [6,12,16]. Table 1 gives the list of all localities and the acronyms employed in the original references and along this paper. This population set includes 29 locations and 32 population samples, as 3 locations have been sampled twice in different years. In total, 4 populations come from the eastern Mediterranean, 8 from the western Mediterranean, and 20 from the Atlantic coasts. Allele and genotype frequencies were calculated with the software Genepop 4.8.3 [23]. The goodness-of-fit test to the Hardy–Weinberg proportions was carried out with the software Arlequin ver. 3.11 [24,25]. Deviations from the expected Hardy–Weinberg proportions in each population were measured with the statistics FIS, which was estimated with the program GenPop 4.8.3 [23]. Statistical differences in allele frequencies among populations were tested with an exact G-test in GenPop 4.8.3. Null alleles were detected at one locus (Ech), and the allele frequencies were calculated by applying the correction derived from the Random Mutation Model [26,27] with the software ML—Null Freq [26]. Then, some Ech homozygote genotypes were recoded as heterozygotes for the null allele, in appropriate numbers to fit the null allele frequencies in each population. The recoded data were used for the estimation of the genetic parameters [27]. FST was used to measure the amount of genetic differentiation among populations in geographic regions and between pairs of populations. The statistical significance for the null hypothesis of FST = 0 was carried out by using a non-parametric permutation approach run for 20,000 replicates [28]. A hierarchical analysis of F-statistics was employed to estimate the amount of genetic differentiation among populations within regions, and among regions within the total area sampled. All F-statistics were estimated using the AMOVA routine in Arlequin with the option Locus by locus with the Pairwise differences distance method. AMOVA results are given as a weighted average of F-statistics over all loci.
FST values between pairs of populations were used to construct a neighbor-joining tree [29] with the software MEGA X [30].
A Bayesian analysis of genetic structure was also carried out using the software STRUCTURE version 2.3.3 [31]. The genotypic data of populations were analyzed under an admixture model of correlated allele frequencies among populations [32]. Sampling locations were assumed as prior information. Data were analyzed with clustering models from K = 1 to K = 10, with 10 replicates of 400,000 iterations and previous burn-in of 300,000 each one. Results from STRUCTURE were processed with CLUMPP and DIStruct, and a graphical image of ancestral gene clusters was generated [12,13]. The number of clusters that best explained the data was determined by computing the posterior probability of K supported by the estimates of the posterior probabilities of the data P(X|K) following Pritchard et al. [31] and the maximum posterior probability differences (ΔK) of contiguous K obtained by the Delta method of Evanno et al. [33].
3. Results
3.1. Newly Sampled Populations
Overall, 17 to 59 individuals were scored in each locality (median = 42). Allelic frequencies, heterozygosity, deviations from Hardy–Weinberg equilibrium (FIS), and sample sizes are shown in Tables S1–S3 and Figure S2 (Supplementary Materials), together with those from all populations from previous studies. No new RFLP alleles were found in the new locations. All loci were polymorphic in all populations with the exception of Fas in the Egyptian population EM4. Cordero et al. [6] found that most intronic alleles were present in the majority of the populations, but four showed restricted geographic distributions, either to the Mediterranean (Ech-3) or to the Adriatic and Aegean seas (Ech-4 and TBP-3). Later, Arias-Perez et al. found Ech-3 in an Atlantic population in very low frequency [12]. The data from the new populations do not change these observations. In previous studies, two private alleles were found (TBP-4 in WM5, and TBP-5 in AT5) (Table S2, Supplementary Materials). Allele TBP-4 has been found in the new Egyptian population EM4 (2 individuals), so it is no longer a private allele, but an allele restricted to the southern Mediterranean. Average heterozygosity (0.333–0.464) showed similar levels as in previous studies (0.353–0.482). A total of 9 out of 65 tests for Hardy–Weinberg equilibrium were significant, in all cases due to a deficit of heterozygotes (positive values of FIS). Significant heterozygote deficiencies were concentrated at locus Ech (five cases) where a null allele is segregating over the whole studied area [6]. The frequency of the Ech null varies from 0 to 0.500, and it is absent from the eastern Mediterranean (Figure S3 and Table S1, Supplementary Materials).
3.2. Analysis of the Pooled Set of Populations
3.2.1. Genetic Variability
The addition of data from new populations and pooling them with those already published has clarified the distribution of genetic variability across clam populations (Figure 2 and Table S1). The mean heterozygosity per locus (H) ranged from 0.334 to 0.482. The lowest value was found in the EM4 population, and the maximum in AT6. The mean number of alleles per locus (Na) ranged from 2.17 to 3.00, with minimum values registered at WM2a and maximum values registered at WM3 and EM2. The plot of Na against H clearly shows that Mediterranean populations have lower H values and higher Na values than the Atlantic ones. Only two West Mediterranean populations (WM1b and WM2a) and one East Mediterranean population (EM4) showed Na values equal to or lower than the lowest values registered in the Atlantic samples, and these populations were the ones with the lowest sample sizes in the entire study. However, the correlation coefficients of sample sizes with H (r = 0.19) and Na (r = 0.50) were not significantly different from zero, suggesting a negligible contribution of sample size differences to the variation of the estimates of genetic variability across populations.
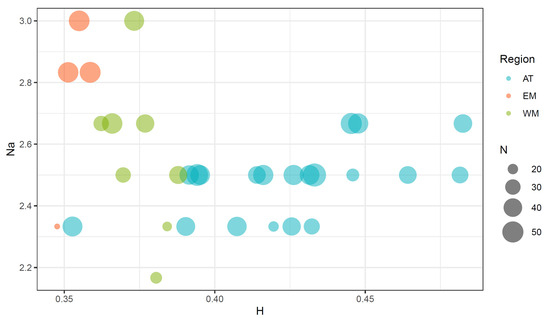
Figure 2.
Plot of the mean number of alleles per locus (Na) and the mean heterozygosity per locus (H) in the clam populations from the Atlantic (AT), West Mediterranean (WM), and East Mediterranean (EM) regions. Dot sizes are proportional to sample sizes (N).
3.2.2. FST Statistics and Population Neighbor-Joining Tree
The results of the study of genetic differentiation by means of F-statistics are presented in Table 2. The overall FST for all populations was 0.132. Mediterranean populations showed a higher overall FST (0.080) than the Atlantic populations (FST = 0.065). Within the Mediterranean, the eastern basin showed higher within-basin differentiation (FST = 0.101) than the western basin (FST = 0.036).

Table 2.
Hierarchical F-statistics analysis based on six intron RFLP markers in the grooved carpet-shell clam. Regional groupings of the populations are as in Table 1, except for Bay of Biscay (AT1, AT2, AT3, AT4, AT5), NW Spain (AT6, AT7, AT8, AT9, AT10, AT11, AT12), Central Portugal (AT14, AT15), SW Iberian Peninsula (AT16, AT17a, AT17b, AT18, AT19), and “a” (AT17a, WM1a, WM2a) and “b” (AT17b, WM1b, WM2b) temporal groups in model 11.
FST values between pairs of populations ranged between 0 and 0.345 (Table S4, Supplementary Materials). Zero values were found between geographically close populations (WM3 in SE France and WM2a in NE Spain), or between samples taken from the same location in different years (WM2a and WM2b). However, zero values were also observed between pairs of distant populations, such as EM1 (northern Adriatic Sea) and EM4 (Timsah Lake, Egypt). The highest value resulted from the comparison of AT14 (Aveiro, central Portugal) and EM3 (Izmir, Turkey). In the Atlantic populations, pairwise FST values ranged from 0 to 0.209. Two pairs of populations showed FST values higher than 0.200. The first pair was composed of two distant populations: AT1 from NW France and AT12 from NW Spain. The second pair, however, was composed of two nearby locations in northern Portugal: AT13 and AT14. The FST values for pairs of Mediterranean populations ranged from 0 to 0.198. The maximum divergence in the Mediterranean was observed between EM2 in Turkey and EM3 in Egypt. FST distances between pairs of populations have been used to draw an unrooted tree (Figure 3). Geographically close populations tend to be positioned close to each other also in the tree, with very few exceptions (see below). Mediterranean populations occupy the right part of the tree, and Atlantic populations appear in the left part. A group of populations from the SW Mediterranean, that includes the two samples from the WM1 population (SE Spain) and the three populations from Tunisia (WM4, WM5, and WM6), occupy the center of the tree. This SW Mediterranean group is characterized by low (0.000–0.050), and usually nonsignificant (Bonferroni-corrected, table-wide p-value = 0.001), pairwise FST values (Table S4, Supplementary Materials). The SW Mediterranean group is connected with the other Mediterranean populations through a node common to the northern Tunisian WM4 population, but this node then leads to two separate branches. One branch includes the northwestern Mediterranean populations WM2a, WM2b, and WM3. The other branch includes the eastern Mediterranean populations, excluding EM3 (Smyrna, Turkey). This population appears, surprisingly, at a very long branch that joins the SW Mediterranean group, indicating a high differentiation of EM3 from all the rest of the Mediterranean populations. Actually, the lowest pairwise FST value for population pairs including EM3 is 0.089, which results from the comparison of EM3 with the Atlantic AT19 population in south Spain, not with another Mediterranean population.
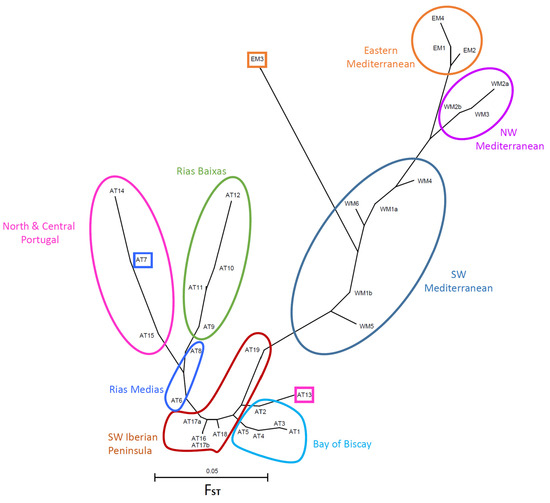
Figure 3.
Neighbor-joining tree of the clam populations based on FST distances. Colored lines group populations according to the geographic regions cited in the text. Note the lack of correspondence between genetic distance and geographic position of some populations, marked in squares.
AT19 is precisely the Atlantic population that is closest to the Mediterranean populations in the tree. The connection of AT19 with the remaining Atlantic populations proceeds through several nodes, which lead to two branches that include, respectively, one population from northern Portugal (AT13) and the populations from the Bay of Biscay (northern Spain and NW France). The tree continues with nodes connecting other populations from SW Spain, southern Portugal, and then one population from NW Spain, AT6. At this point, the tree divides in two branches. One contains one population from NW Spain (AT7) and two populations from northern Portugal, which are relatively distant from it (AT15 and AT16). The other branch contains the remaining populations from NW Spain estuaries, specifically the group of estuaries known as Rias Baixas (AT9, AT10, AT11, and AT12), plus a small estuary located a bit further to the north (AT8), which belongs to the group of estuaries known as Rias Medias.
3.2.3. Hierarchical FST Analysis
The hierarchical FST analyses using different models of genetic subdivision are shown in Table 2. The subdivision of the whole area in Atlantic, western Mediterranean (including Tunisia), and eastern Mediterranean regions (model 6) gives an overall FST = 0.18. This model also shows that among-regions differentiation (FCT = 0.131) is double than differentiation within regions (FSC = 0.063). Within the Atlantic, one population (AT13, in northern Portugal) showed pairwise FST values (Table S4) and cluster distribution profiles in the Bayesian analysis of genetic structure (see below) that deviate strongly from those of their neighbor populations and suggest a higher similarity to the populations of the Bay of Biscay (AT1–AT5). Therefore, we tested three models with four subdivisions, in which AT13 was included alternatively in the Central Portugal group, the NW Spain group, and the Bay of Biscay group (models 8–10 in Table 2). Model 10 gives the highest FCT (0.050), supporting the higher similarity of AT13 to the Bay of Biscay group.
3.2.4. Temporal Genetic Differentiation
The data set contains three locations that were sampled twice, with samplings separated by four (AT17) or seven years (WM1 and WM2) (Table 1). The data obtained from these samplings allow an empirical evaluation of the magnitude of the differences that can be expected to appear among samples taken at different time points. These samples gave an overall FST of 0.129 (Table 2, Model 11), and between-locations pairwise FST values comprised between 0.036 and 0.271 (Table S4, Supplementary Materials). The FST values for the between-year comparisons within locations ranged from 0 to 0.007, which are at least five times lower. Comparison of these numbers suggests that the changes in allele frequencies in populations over time are very small, compared to the variation in allele frequencies across geographically separated populations. Even in populations separated by relatively short distances in the same marine basin, such as those in the Mar Menor lagoon (WM1a and WM1b) and the Ebro Delta (WM2a and WM2b) in Mediterranean Spain, FST between pairs of locations (FST = 0.036–0.176) is at least five times higher than between temporal samples within each location (FST = 0.00–0.007) (Table S4).
3.2.5. Bayesian Analysis of Genetic Structure
The results of the estimation of the optimal number of ancestral genetic clusters resulting from the Bayesian analysis of genetic structure are shown in Figure 4. The optimal number according to the method of Pritchard et al. is K = 6 (Figure 4a). The method of Evanno et al. gives a very high value for K = 2 and values near 0 for other K values, with the exception of a slightly higher value for K = 6 (Figure 4b). Pritchard et al.’s method suggests substructuring at different geographic levels, as K = 10 has a similarly high posterior probability as K = 6, and gives low posterior probability to K = 2.
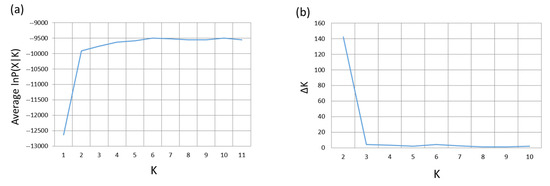
Figure 4.
(a) Plot of the posterior probabilities for each K value from the Bayesian analysis of genetic structure. (b) Plot of Evanno et al.’s ∆K for each K value from the Bayesian analysis of genetic structure.
The plots showing the clusters’ proportions for all studied individuals are presented in Figure 5 for K = 2 and K = 6. The plot for K = 2 shows that one cluster is almost restricted to the Mediterranean populations, and the other is almost the only one present in the Atlantic coasts. However, this cluster appears in the Mediterranean Sea in frequencies up to 50% in populations from SE Spain and Tunisia, becoming less frequent in the remaining western Mediterranean and Adriatic populations, and being almost absent from the Aegean Sea populations and Egypt.
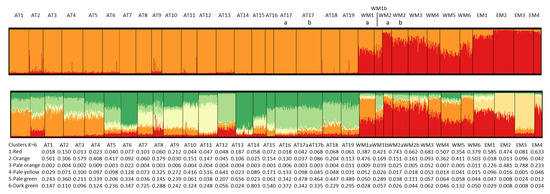
Figure 5.
Plots of cluster frequencies from the Bayesian analysis of genetic structure for K = 2 (above) and K = 6 (below), for all individuals and populations. Clusters are defined by colors. The table below the plots gives the average proportions of each cluster in each population for K = 6.
The plot for K = 6 shows the same overall picture but with more substructuring within each basin. Dark green to yellow-colored clusters shown in Figure 5 are more frequent in the Atlantic, and red to pale orange-colored clusters are more frequent in the Mediterranean. Cluster 3 (pale orange) is present only in the Aegean and the Adriatic seas and is especially abundant in the EM3 (Izmir) and EM2 (Halkidiki) populations. However, it is absent from the remaining Mediterranean and Atlantic populations. Cluster 1 (red) is the most abundant in most Mediterranean populations, especially in the north of the western Mediterranean (WM2a WM2b and WM3), but also in the Adriatic population EM1 (Venice). This cluster is present in very low frequency in the Atlantic populations but increases slightly in a few of them: AT2, AT9, and AT13. The third most common cluster in the Mediterranean populations is cluster 2 (orange), especially in the south of the western Mediterranean (WM1a, WM4, WM5 and WM6) but also in the north of the western Mediterranean in lower frequency. It is absent from the eastern Mediterranean populations, with the exception of EM3, where it appears in very low frequency. In the Atlantic populations, three clusters are especially abundant. Cluster 6 (dark green) appears in all populations at variable frequencies, being the most abundant in AT7 and in AT14. This cluster is present also in the Mediterranean Sea in very low frequencies. Another cluster (5, pale green) is very frequent in southern Portugal and SW Spain (AT16–AT19) and also in the Bay of Biscay populations (AT1–AT6) but reaches its highest abundance in AT13 (Viana), in strong contrast with the neighbor populations AT12 and AT14, where this cluster is almost absent. Another typical Atlantic cluster is cluster 4 (pale yellow), which is abundant in the estuaries of NW Spain, especially in the Rias Baixas estuaries (AT9–AT12) but also in the neighbor estuary of Camariñas (AT8). This cluster is present in very low frequencies in the remaining Atlantic and in the Mediterranean populations. The most surprising result of the Bayesian analysis of genetic structure is the presence of cluster 2 (orange) in intermediate to high frequencies in the Bay of Biscay populations (AT1–AT5). This cluster is present also in intermediate frequencies in the western Mediterranean populations (see above) and appears in low frequencies along the regions that separate the Bay of Biscay from the western Mediterranean: south Atlantic Spain, Portugal, and the NW Spain estuaries. The populations from the Bay of Biscay and the SW of the Iberian Peninsula differ especially in the frequency of this cluster.
4. Discussion
4.1. Genetic Variability
The high number of samples included in this study has uncovered a pattern of genetic variability which had not been appreciated in previous studies. The mean heterozygosity per locus (H) is higher in Atlantic populations and lower in Mediterranean populations, especially in the eastern Mediterranean region (EMED). However, the mean number of alleles per locus (Na) is higher in the Mediterranean populations and increases to the East. These contrasting patterns of H and Na are due to the presence of several alleles in the populations from the Mediterranean, especially in the EMED, that are not present in the Atlantic populations. Lower heterozygosity associated to high number of alleles in low frequency is the expected outcome of mutation and genetic drift when a population experiences population size contractions followed by population expansions. The Pleistocene glacial–interglacial provides a framework to explain these observations. If most of the alleles that are restricted to the eastern Mediterranean were in very low frequencies, a contraction followed by expansion associated to Pleistocene glacial–interglacial cycles would be a possible explanation for the observed pattern. However, several alleles restricted to the EMED appear in relatively high frequencies (Ech-3, up to 0.35; Ech-4, up to 0.28; TBP-3, up to 0.38), and therefore, other processes are needed to account for the results. It is possible that the populations in EMED are remnants from an ancient population with a large effective size (Ne), and glacial–interglacial cycles decreased H, but alleles still remain. Cordero et al. [8] dated the origin of alleles restricted to the western Mediterranean (WEMED) and EMED at the loci Ech and TBP. Alleles Ech-D, TBP-A1, and TBP-C (corresponding to Ech-4, TBP-1, and TBP-3 in this study), gave best estimates for their divergence from the phylogenetically closest alleles of 206, 273, and 416 thousand years before present (KYBP). Allele Ech-A2 (which was a subgroup of sequences of the allele Ech-A (Ech-2 in this study)) was present in the whole Mediterranean and diverged at 424 KYBP. Finally, Ech-C (Ech-3 in this study), which is also widespread in the Mediterranean, diverged at 1078 KYBP. The divergence of the Mediterranean alleles present in both WMED and EMED therefore seems to have occurred earlier than the divergence of the alleles that are exclusive to the EMED. The most probable date for this divergence seems to be second half of the Pleistocene period but much earlier than the last glacial maximum (20 KYBP). Therefore, it is probable that, between less than 500 KYBP and the last glacial maximum, at two separated times, the eastern Mediterranean harbored a large population of clams that lost part of their genetic variability but did not lose many alleles. The oldest alleles would have expanded to the whole Mediterranean, while the more recent alleles would have remained restricted to the EMED. Another factor that could have influenced the levels of genetic variability in the Mediterranean Sea is natural selection, which could have favored one or more EMED-restricted alleles due to their linkage disequilibrium with other variants in the coding regions of the genes or in nearby genomic locations.
4.2. Temporal Genetic Differentiation
Studies of population genetic differentiation usually are based on data obtained from contemporary sampling conducted in the same year or just a few years apart. The 32 samples studied in this work have been taken along a period of 17 years, from 2004 to 2021, which is a longer-than-usual sampling period. An evaluation of the effect of the temporal change in gene frequencies is necessary in order to know whether distant sampling time affects the results. If temporally separated samples from the same locality differ as much, or more, than they do with respect to samples from other locations, the effect of temporal variation and geographic variation would be confounded, and no robust conclusions could be taken from the data. In this study, three locations have been sampled twice, at time points separated by four or seven years. In all three cases, the pairwise FST values between temporal samples from each location were not significantly different from zero. Moreover, the hierarchical FST analysis of the six samples also gave a nonsignificant between-years component (FCT). Therefore, we can conclude that the detected differences among geographic populations in this study are not affected by the time distance that separates their collection.
4.3. Geographic Genetic Differentiation
The results of this study agree with previous studies using mtDNA, intron-RFLP and microsatellite markers in showing a subdivision of the species in an Atlantic and two Mediterranean groups, corresponding to the western and eastern basins [6,12]. The genetic differentiation found between the Atlantic and the Mediterranean populations is consistent with many other studies showing the same result in other species using several types of genetic markers (enzyme polymorphisms, mitochondrial DNA, microsatellites, SNP) and have been related to the Pleistocene glacial–interglacial cycles and present-day restriction to gene flow at some points [8,9].
The inclusion in this study of new samples, and their analysis together with those from previous studies, has allowed us to discover new features of the genetic structure of the clam populations living the Mediterranean Sea. In the eastern Mediterranean, Cordero et al. [6] examined populations from the northern part of the basin only, i.e., the Adriatic and the Aegean seas. They used the initial letters of both seas to create an acronym (AEGAD) to refer to that group of populations. The inclusion of a sample of clams from Egypt in this study has allowed us to prove that the southern part of the eastern Mediterranean basin shares the genetic characteristics of the northern part, and therefore it is possible to speak of an eastern Mediterranean subpopulation or race (EMED). Further research could help to decide if the EMED populations represent a subspecies of R. decussatus.
Cordero et al. [6] also observed that the Tunisian population WM5 (Sfax), which geographically is located in the eastern Mediterranean basin, was genetically more similar to western Mediterranean populations than to the remaining eastern Mediterranean populations. Gharbi et al. [13] examined the genetics of twelve Tunisian populations with sequences of the mitochondrial gene COI and the nuclear ITS and found no important differences in haplotype frequencies between the populations from the northern coast of the country, which geographically belong in the western Mediterranean basin, and the populations from eastern coast, which belong in the eastern basin. In this study, two of the population samples analyzed by Gharbi et al. have been included, so three Tunisian populations, coming from the northern and eastern coasts of the country, have been available for analysis. All analyses carried out in this study clearly show that these populations fall on the WMED group. Studies have reported that populations of marine species from the eastern coast of Tunisia can be genetically more similar to western Mediterranean populations in some cases, or to the eastern Mediterranean in others. The reasons for this variability are not well known. A combination of factors might be acting, including the east to west flow of the northern African current favoring dispersal past the Siculo–Tunisian Strait and the effect of endogenous barriers to gene flow [6,18]. Specific studies, including populations beyond the Tunisian borders to the east, should be carried out in order to understand the mechanisms that explain this situation.
Some aspects of the allelic distributions in the Mediterranean could be due to historical and/or environmental factors, or to anthropogenic causes. For example, the presence in very low frequency of allele TBP-4 in Tunisia (WM4) was interpreted as a private allele by Cordero et al., but in this study, it appeared in slightly higher frequency in a population near the Suez Canal (Timsah Lake) in Egypt, some 2500 km away. This finding points to the presence of this allele in low frequency along the northern African coasts but is also compatible with a spread of the allele from Egypt to Tunisia through ballast waters along one of the busiest maritime routes in the world. This seems also the most plausible explanation for the presence of typical eastern Mediterranean alleles Ech-5 and TBP-3 in the Thau lagoon, near the port of Sète in southeast France (WM3 population), but more data from the NW Mediterranean are necessary to be conclusive.
The inclusion of data from a sample from SW France reported by Saavedra et al. [16], along with the addition of two new samples from Tunisia, has allowed us to confirm the relative homogeneity of the populations of the southern part of the western Mediterranean. However, a slightly higher differentiation of the northern populations in that basin, here represented by samples from the Ebro Delta in NE Spain and from the mentioned Thau lagoon in SW France, was detected. These populations formed a separate branch in the NJ tree and showed a higher frequency of cluster 1 and a lower frequency of cluster 2 in the Bayesian analysis of genetic structure. Genetic differentiation of populations from the north and the south of the WMED has been shown in other marine species living in that region, and it has been related to restrictions to gene flow due to the Balearic oceanographic front [20]. It would be interesting to study samples from other French and Italian populations, as well as from the western Mediterranean Islands, in order to better characterize the genetic structure of the WMED.
This study has increased considerably the sampling along the Atlantic coasts of the Iberian Peninsula and has found out interesting novelties. Previously, Arias-Pérez et al. had found that the populations from the SW of the Iberian Peninsula were more similar to the populations of the Bay of Biscay than to other populations on the Atlantic façade of the Iberian Peninsula, which are geographically closer [12]. Here, we have shown that these two regions are not so similar, as they rendered significant pairwise-FST values and showed different frequencies of clusters in the Bayesian analysis of genetic structure (especially of clusters 2 and 6 for K = 6).
Cruz et al. [14], using microsatellites, described a set of two populations in central Portugal (Obidos lagoon and Aveiro) that showed moderate genetic differentiation with respect to their neighbor Portuguese and Spanish populations. In this study, we have presented a more detailed data set of that part of the Iberian Peninsula. We have confirmed the differentiation of the same two populations from central Portugal (AT14 and AT15) with respect to their neighbor populations, not only the Spanish ones but also one Portuguese population located in northern Portugal (AT13, from Viana do Castelo), which has been studied here for the first time. Surprisingly, AT13 is neither more similar to its closest neighbor in NW Spain (AT12, from the Ría de Vigo estuary) nor to other Portuguese populations, but rather to the populations of the Bay of Biscay. Moreover, the clams from Obidos and Aveiro have the highest similarity with those from A Coruña (AT7), a more northern population ca. 500 km apart, which is also different from their closest neighbor populations AT6 and AT8. Finally, between A Coruña and Viana do Castelo, there is a group of populations (AT8–AT12) which appear closer to one another in the NJ tree and have in common the sharing of STRUCTURE cluster 4 in higher frequency than any other populations included in this study. These populations belong in the set of estuaries known as Rias Baixas but also include the northern neighbor population AT8, which is part of the Rias Medias (Figure 1).
In summary, there is a genetic mosaic along the NW corner of the Iberian Peninsula that had not been described before. This mosaic is partially related to the geographic framework of the different sets of rias and requires specific explanations. A similar pattern of genetic differentiation between populations has been reported for species such as snails, algae, and littoral plants from that region and has been explained as the result of the Finisterre oceanographic front [34,35,36]. This front is formed in summer off the coasts of Cape Finisterre due to the encounter of eastern north Atlantic central waters (ENACW) of subtropical and subpolar origins (Figure 6) [37,38]. The front could affect the transport of the grooved carpet-shell clam larvae, which reproduce in summer months in that region [39,40]. Disruption of larval transport could result in limited gene flow, which could account for the differentiation between the AT8–AT12 group from the AT6–AT7 and Bay of Biscay populations. But this would leave unexplained the similarity of A Coruña (AT7) in the Rias Medias, with Aveiro (AT14) and Obidos (AT15) in central Portugal, and the similarity of Viana in northern Portugal (AT13), with the populations from the Bay of Biscay. A possible explanation for this inconsistency is just a higher isolation and lower effective size in these populations, which would result in the fact that distant populations would look alike just by chance. Another possible explanation is the effect of river plumes, which have been proposed as causes of genetic differentiation of populations located on both sides of the plume in some marine species due to the limitation of gene flow by means of restricting larval transport across the plume [41,42]. In this study, the plumes of rivers Minho, Douro, and Tejo (Figure 1b), which show some of the highest discharge rates among the Iberian rivers flowing to the Atlantic (340, 660, and 550 m3/s, respectively), could act as barriers to gene flow [43]. The differentiation of AT13 with respect to the populations from Rias Baixas and to AT14 and AT15 would fit this model.
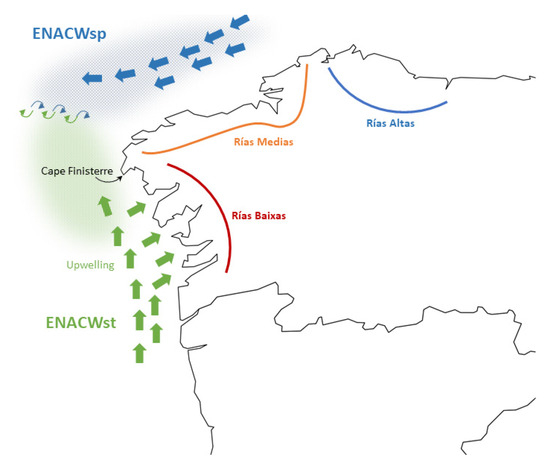
Figure 6.
Flow of water masses (thick arrows), upwelling, and front (thin arrows) near the NW coasts of the Iberian Peninsula in summer. See Discussion for explanations.
However, since intronic markers are located in protein coding regions (genes), which may be sensitive to natural selection, non-neutral (selective or adaptive) explanations cannot be excluded. One adaptive explanation for the differentiation of Rias Baixas is the upwelling of cold and nutrient-rich eastern north Atlantic central water (ENACW) inside these estuaries. The upwelling is favored by N component winds in spring and summer. Upwelling episodes are more frequent in the Rias Baixas than in the more northern estuaries and induce important differences between the two groups of estuaries in temperature, primary production, and phytoplankton communities that may affect both larval and adult clams [44]. Another plausible adaptive explanation is that AT7, AT13, and AT14 locations are characterized by a more oceanic than estuarine environment, and the differences with respect to their neighbor locations are due to the adaptation to this type of environment.
Finally, anthropogenic causes cannot be left aside, as the concerned populations in the NW Iberian Peninsula are among the most commercially exploited in Europe. The exchange of juvenile clams between locations for fishery management purposes, and the introduction of clam spat obtained in hatcheries, could also result in the observed pattern of differentiation. The similarity of the Viana (AT13) population with those of the Bay of Biscay would fit this human intervention, as hatcheries in N Spain and Atlantic France are usual suppliers of clam seed for the fishers guilds.
5. Conclusions
This study, based on the largest population genetic data set obtained so far from the grooved carpet-shell clam, has confirmed previous results showing a remarkable differentiation between the Atlantic, the western Mediterranean, and the eastern Mediterranean groups of populations and has added new details to this general framework. The Mediterranean populations showed less heterozygosity but a higher number of alleles than the Atlantic populations. The difference was especially intense with regard to the eastern Mediterranean samples. Population contractions and gene-flow restrictions during the Pleistocene seem the most reasonable cause of this observation. The study has also shown that one population from Egypt belongs in the eastern Mediterranean group, which strongly suggests that the AEGAD genetic subdivision described by Cordero et al. [6] actually comprises all or a great extension of the eastern Mediterranean (EMED) basin. Moreover, the data showed clearly that the Tunisian populations should be considered as part of the western Mediterranean group (WMED) described by Cordero et al. The results obtained also indicate that there is some genetic differentiation between the north and the south of the western Mediterranean that could be related to the Balearic front, but more populations from the entire region should be studied to be conclusive. As for the populations in the Atlantic, they appeared as a genetic mosaic, with the clams from the SW of the Iberian Peninsula showing limited similarity to the populations of the Bay of Biscay, and a group of rather more genetically subdivided set of populations occupying the north of Portugal and the NW of Spain. In that region, the Rias Baixas estuaries appear as a more homogeneous unit with respect to their northern neighbor populations and to the populations in central Portugal. Some populations in this region show high contrast with their geographic neighbors, which could be due to oceanographic features (oceanic fronts, upwellings), river plumes, environmental differences (oceanic vs. estuarine), or fishery management activities (translocations, restocking with hatchery seed). Further studies based on much larger numbers of genetic markers and more intensive geographic sampling will be necessary to decide which of these explanations, or their combinations, account best for the observed genetic population structure of R. decussatus.
Supplementary Materials
The following supporting tables and figures can be downloaded at: https://www.mdpi.com/article/10.3390/oceans5020016/s1. Figure S1: Examples of polymorphisms scored in agarose gels for the six introns employed in this study; SM: size marker. Figure S2: Bar charts showing allelic frequency variation across the study area; allele 5 in Ech corresponds to the null allele(s). Figure S3: allelic contributions to clusters in the Bayesian analysis of genetic structure. (a) K = 2. (b) K = 6. Table S1: Allele frequencies in the populations considered in this study; Table S2: Heterozygosity estimates and mean number of alleles per locus, by locus and population. Table S3: FIS per locus and population; Table S4: FST estimates between pairs of populations (above) and associated p-values (below).
Author Contributions
Conceptualization, C.S.; methodology, C.S. and D.C.; validation, C.S. and D.C.; investigation, C.S. and D.C.; resources, C.S. and D.C.; data curation, D.C.; writing—original draft preparation, C.S.; writing—review and editing, C.S. and D.C.; supervision, C.S.; project administration, C.S.; funding acquisition, C.S. All authors have read and agreed to the published version of the manuscript.
Funding
This research was funded by CSIC.
Institutional Review Board Statement
According to the Spanish law, which is transposed from the European Union directives, studies with marine invertebrates do not require to be supervised by the Ethics Committee of the research institutions, with the only exception of Cephalopods (octopuses and the like), which is not the case.
Data Availability Statement
Original data are available on request to the corresponding author.
Acknowledgments
We are indebted to Andreia Cruz and Fiz da Costa for providing AT14 and AT15 samples; to Nahla El-Shenawy for providing the sample from Egypt; to Miguel B. Gaspar for providing the AT13 sample; to Aicha Gharbi for providing the Tunisian samples WM4 and WM6; and to Beatriz Yermo, and the Cofradía de Camariñas fisher guild, for providing the AT8 sample. We thank Martina Tremonti for her technical assistance with the molecular work and artwork.
Conflicts of Interest
The authors declare no conflicts of interest. The funders had no role in the design of the study; in the collection, analyses, or interpretation of data; in the writing of the manuscript; or in the decision to publish the results.
References
- Macho, G.; Woodin, S.A.; Wethey, D.S.; Vázquez, E. Impacts of Sublethal and Lethal High Temperatures on Clams Exploited in European Fisheries. J. Shellfish. Res. 2016, 35, 405–419. [Google Scholar] [CrossRef]
- Aranguren, R.; Gomez-León, J.; Balseiro, P.; Costa, M.M.; Novoa, B.; Figueras, A. Abnormal Mortalities of the Carpet Shell Clam Ruditapes decussatus (Linnaeus 1756) in Natural Bed Populations: A Practical Approach. Aquac. Res. 2014, 45, 1303–1310. [Google Scholar] [CrossRef]
- Bidegain, G.; Bárcena, J.F.; García, A.; Juanes, J.A. Predicting Coexistence and Predominance Patterns between the Introduced Manila Clam (Ruditapes philippinarum) and the European Native Clam (Ruditapes decussatus). Estuar. Coast. Shelf Sci. 2015, 152, 162–172. [Google Scholar] [CrossRef]
- Borsa, P.; Jarne, P.; Belkhir, K.; Bonhomme, F. Genetic Structure of the Palourde Ruditapes decussatus L. in the Mediterranean. In Genetics and Evolution of Aquatic Organisms; Chapman and Hall: London, UK, 1994; pp. 103–113. [Google Scholar]
- Gharbi, A.; Zitari-Chatti, R.; Van Wormhoudt, A.; Dhraief, M.N.; Denis, F.; Said, K.; Chatti, N. Allozyme Variation and Population Genetic Structure in the Carpet Shell Clam Ruditapes decussatus across the Siculo-Tunisian Strait. Biochem. Genet. 2011, 49, 788–805. [Google Scholar] [CrossRef]
- Cordero, D.; Peña, J.B.; Saavedra, C. Phylogeographic Analysis of Introns and Mitochondrial DNA in the Clam Ruditapes decussatus Uncovers the Effects of Pleistocene Glaciations and Endogenous Barriers to Gene Flow. Mol. Phylogenetics Evol. 2014, 71, 274–287. [Google Scholar] [CrossRef] [PubMed]
- Sanna, D.; Lai, T.; Cossu, P.; Scarpa, F.; Dedola, G.L.; Cristo, B.; Francalacci, P.; Curini-Galletti, M.; Mura, L.; Fois, N.; et al. Cytochrome c Oxidase Subunit I Variability in Ruditapes decussatus (Veneridae) from the Western Mediterranean. Eur. Zool. J. 2017, 84, 554–565. [Google Scholar] [CrossRef][Green Version]
- Patarnello, T.; Volckaert, F.A.M.J.; Castilho, R. Pillars of Hercules: Is the Atlantic-Mediterranean Transition a Phylogeographical Break? Mol. Ecol. 2007, 16, 4426–4444. [Google Scholar] [CrossRef] [PubMed]
- Pascual, M.; Rives, B.; Schunter, C.; MaCpherson, E. Impact of Life History Traits on Gene Flow: A Multispecies Systematic Review across Oceanographic Barriers in the Mediterranean Sea. PLoS ONE 2017, 12, e0176419. [Google Scholar] [CrossRef] [PubMed]
- Bierne, N.; Welch, J.; Loire, E.; Bonhomme, F.; David, P. The Coupling Hypothesis: Why Genome Scans May Fail to Map Local Adaptation Genes. Mol. Ecol. 2011, 20, 2044–2072. [Google Scholar] [CrossRef]
- El Ayari, T.; Trigui El Menif, N.; Hamer, B.; Cahill, A.E.; Bierne, N. The Hidden Side of a Major Marine Biogeographic Boundary: A Wide Mosaic Hybrid Zone at the Atlantic–Mediterranean Divide Reveals the Complex Interaction between Natural and Genetic Barriers in Mussels. Heredity 2019, 122, 770–784. [Google Scholar] [CrossRef]
- Arias-Pérez, A.; Cordero, D.; Borrell, Y.; Sánchez, J.A.; Blanco, G.; Freire, R.; Insua, A.; Saavedra, C. Assessing the Geographic Scale of Genetic Population Management with Microsatellites and Introns in the Clam Ruditapes decussatus. Ecol. Evol. 2016, 6, 3380–3404. [Google Scholar] [CrossRef] [PubMed]
- Gharbi, A.; Chatti, N.; Said, K.; Wormhoudt, A. Genetic Variation and Population Structure of the Carpet Shell Clam Ruditapes decussatus along the Tunisian Coast Inferred from mtDNA and ITS1 Sequence Analysis. Biologia 2010, 65, 688–696. [Google Scholar] [CrossRef][Green Version]
- Cruz, A.; da Costa, F.; Fernández-Pérez, J.; Nantón, A.; Fernández-Boo, S.; Insua, A.; Méndez, J. Genetic Variability in Ruditapes decussatus Clam Combined with Perkinsus Infection Level to Support Founder Population Selection for a Breeding Program. PeerJ 2020, 8, e9728. [Google Scholar] [CrossRef]
- Amane, Z.; Tazi, L.; Ouagajjou, Y.; Ouazzani, K.C.; Nabich, A.; Chlaida, M. Genetic Structuring in the Grooved Carpet Shell Clam Ruditapes decussatus along the Moroccan Coasts Revealed by Microsatellites. Reg. Stud. Mar. Sci. 2021, 46, 101888. [Google Scholar] [CrossRef]
- Saavedra, C.; Milan, M.; Leite, R.B.; Cordero, D.; Patarnello, T.; Cancela, M.L.; Bargelloni, L. Transcriptional Profiling of Populations in the Clam Ruditapes decussatus Suggests Genetically Determined Differentiation in Gene Expression along Parallel Temperature Gradients and between Races of the Atlantic Ocean and West Mediterranean Sea. Fishes 2023, 8, 203. [Google Scholar] [CrossRef]
- Nikula, R.; Väinölä, R. Phylogeography of Cerastoderma glaucum (Bivalvia: Cardiidae) across Europe: A Major Break in the Eastern Mediterranean. Mar. Biol. 2003, 143, 339–350. [Google Scholar] [CrossRef]
- Bahri-Sfar, L.; Lemaire, C.; Hassine, O.K.B.; Bonhomme, F. Fragmentation of Sea Bass Populations in the Western and Eastern Mediterranean as Revealed by Microsatellite Polymorphism. Proc. R. Soc. Lond. Ser. B 2000, 267, 929–935. [Google Scholar] [CrossRef]
- Hurtado, N.S.; Pérez-García, C.; Morán, P.; Pasantes, J.J. Genetic and Cytological Evidence of Hybridization between Native Ruditapes decussatus and Introduced Ruditapes philippinarum (Mollusca, Bivalvia, Veneridae) in NW Spain. Aquaculture 2011, 311, 123–128. [Google Scholar] [CrossRef]
- Habtemariam, B.T.; Arias, A.; García-Vázquez, E.; Borrell, Y.J. Impacts of Supplementation Aquaculture on the Genetic Diversity of Wild Ruditapes decussatus from Northern Spain. Aquac. Environ. Interact. 2015, 6, 241–254. [Google Scholar] [CrossRef]
- Markaide, P.; Gairín, I.; Cordero, D.; Ibarrola, I.; Saavedra, C. No Hybridization and Marked Interspecific Differences in Individual Growth Rate in Mixed Cultures of Manila Clam (Ruditapes philippinarum) and Grooved Carpet-Shell Clam (R. decussatus). Aquaculture 2021, 541, 736824. [Google Scholar] [CrossRef]
- Cordero, D.; Peña, J.B.; Saavedra, C. Polymorphisms at Three Introns in the Manila Clam (Ruditapes philippinarum) and the Grooved Carpet-Shell Clam (R. decussatus). J. Shellfish. Res. 2008, 27, 301–306. [Google Scholar] [CrossRef]
- Rousset, F. GENEPOP’007: A Complete Re-Implementation of the GENEPOP Software for Windows and Linux. Mol. Ecol. Resour. 2008, 8, 103–106. [Google Scholar] [CrossRef]
- Guo, S.W.; Thompson, E.A. Performing the Exact Test of Hardy-Weinberg Proportion for Multiple Alleles. Biometrics 1992, 48, 361–372. [Google Scholar] [CrossRef]
- Excoffier, L.; Lischer, H.E.L. Arlequin Suite Ver 3.5: A New Series of Programs to Perform Population Genetics Analyses under Linux and Windows. Mol. Ecol. Resour. 2010, 10, 564–567. [Google Scholar] [CrossRef]
- Kalinowski, S.T.; Taper, M.L. Maximum Likelihood Estimation of the Frequency of Null Alleles at Microsatellite Loci. Conserv. Genet. 2006, 7, 991–995. [Google Scholar] [CrossRef]
- Chapuis, M.-P.; Estoup, A. Microsatellite Null Alleles and Estimation of Population Differentiation. Mol. Biol. Evol. 2007, 24, 621–631. [Google Scholar] [CrossRef]
- Excoffier, L.; Smouse, P.E.; Quattro, J.M. Analysis of Molecular Variance Inferred from Metric Distances among DNA Haplotypes: Application to Human Mitochondrial DNA Restriction Data. Genetics 1992, 131, 479–491. [Google Scholar] [CrossRef]
- Saitou, N.; Nei, M. The Neighbor-Joining Method: A New Method for Reconstructing Phylogenetic Trees. Mol. Biol. Evol. 1987, 4, 406–425. [Google Scholar] [CrossRef]
- Kumar, S.; Stecher, G.; Li, M.; Knyaz, C.; Tamura, K. MEGA X: Molecular Evolutionary Genetics Analysis across Computing Platforms. Mol. Biol. Evol. 2018, 35, 1547–1549. [Google Scholar] [CrossRef] [PubMed]
- Pritchard, J.K.; Stephens, M.; Donnelly, P. Inference of Population Structure Using Multilocus Genotype Data. Genetics 2000, 155, 945–959. [Google Scholar] [CrossRef] [PubMed]
- Falush, D.; Stephens, M.; Pritchard, J. Inference of Population Structure Using Multilocus Genotype Data: Linked Loci and Correlated Allele Frequencies. Genetics 2003, 164, 1567–1587. [Google Scholar] [CrossRef]
- Evanno, G.; Regnaut, S.; Goudet, J. Detecting the Number of Clusters of Individuals Using the Software STRUCTURE: A Simulation Study. Mol. Ecol. 2005, 14, 2611–2620. [Google Scholar] [CrossRef]
- Leys, M.; Petit, E.J.; El-Bahloul, Y.; Liso, C.; Fournet, S.; Arnaud, J. Spatial Genetic Structure in Beta vulgaris Subsp. maritima and Beta macrocarpa Reveals the Effect of Contrasting Mating System, Influence of Marine Currents, and Footprints of Postglacial Recolonization Routes. Ecol. Evol. 2014, 4, 1828–1852. [Google Scholar] [CrossRef]
- Piñeira, J.; Quesada, H.; Rolán-Alvarez, E.; Caballero, A. Genetic Discontinuity Associated with an Environmentally Induced Barrier to Gene Exchange in the Marine Snail Littorina saxatilis. Mar. Ecol. Prog. Ser. 2008, 357, 175–184. [Google Scholar] [CrossRef]
- Alberto, F.; Santos, R.; Leitão, J. Assessing Patterns of Geographic Dispersal of Gelidium sesquipedale (Rhodophyta) through RAPD Differentiation of Populations. Mar. Ecol. Prog. Ser. 1999, 191, 101–108. [Google Scholar] [CrossRef][Green Version]
- Fraga, F.; Mouriño, C.; Manríquez, M. Las masas de agua en las costas de Galicia: Junio-octubre. Result. Exped. Científicas 1982, 10, 51–77. [Google Scholar]
- Alvarez, I.; Gomez-Gesteira, M.; deCastro, M.; Lorenzo, M.N.; Crespo, A.J.C.; Dias, J.M. Comparative Analysis of Upwelling Influence between the Western and Northern Coast of the Iberian Peninsula. Cont. Shelf Res. 2011, 31, 388–399. [Google Scholar] [CrossRef]
- Rodríguez-Moscoso, E.; Arnaiz, R. Gametogenesis and Energy Storage in a Population of the Grooved Carpet-Shell Clam, Tapes decussatus (Linne, 1787), in Northwest Spain. Aquaculture 1998, 162, 125–139. [Google Scholar] [CrossRef]
- Ojea, J. Estudio del Desarrollo Gametogénico de la Almeja Fina, Ruditapes decussatus (Linné, 1758) en el Medio Natural y Optimización de las Condiciones del Acondicionamiento en Criadero. Ph.D. Thesis, Universidad de Santiago de Compostela, Santiago, Spain, 2013. [Google Scholar]
- Peres, P.A.; Bracken-Grissom, H.; Timm, L.E.; Mantelatto, F.L. Genomic Analyses Implicate the Amazon–Orinoco Plume as the Driver of Cryptic Speciation in a Swimming Crab. Genes 2022, 13, 2263. [Google Scholar] [CrossRef] [PubMed]
- Narváez-Barandica, J.C.; Quintero-Galvis, J.F.; Aguirre-Pabón, J.C.; Castro, L.R.; Betancur, R.; Acero Pizarro, A. A Comparative Phylogeography of Three Marine Species with Different PLD Modes Reveals Two Genetic Breaks across the Southern Caribbean Sea. Animals 2023, 13, 2528. [Google Scholar] [CrossRef] [PubMed]
- Instituto Geográfico Nacional, Ministerio de Fomento. Caudal Absoluto. Available online: https://www.ign.es/espmap/mapas_agua_bach/pdf/Hidro_Mapa_02texto.pdf (accessed on 20 April 2024).
- Prego, R.; Barciela, M.D.C.; Varela, M. Nutrient Dynamics in the Galician Coastal Area (Northwestern Iberian Peninsula): Do the Rias Bajas Receive More Nutrient Salts than the Rias Altas? Cont. Shelf Res. 1999, 19, 317–334. [Google Scholar] [CrossRef]
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).