Comment on Arulananthan et al. The Status of the Coral Reefs of the Jaffna Peninsula (Northern Sri Lanka), with 36 Coral Species New to Sri Lanka Confirmed by DNA Bar-Coding. Oceans 2021, 2, 509–529
Author Contributions
Acknowledgments
Conflicts of Interest
References
- Arulananthan, A.; Herath, V.; Kuganathan, S.; Upasanta, A.; Harishchandra, A. The Status of the Coral Reefs of the Jaffna Peninsula (Northern Sri Lanka), with 36 Coral Species New to Sri Lanka Confirmed by DNA Bar-Coding. Oceans 2021, 2, 509–529. [Google Scholar] [CrossRef]
- Bonito, V.E.; Baird, A.H.; Bridge, T.; Cowman, P.F.; Fenner, D. Types, topotypes and vouchers are the key to progress in coral taxonomy: Comment on Wepfer et al. (2020). Mol. Phylogenetics Evol. 2021, 159, 107104. [Google Scholar] [CrossRef]
- Kelley, R. Indo Pacific Coral Finder, 3rd ed.; BYO Guides: Townsville, Australia, 2016; ISBN 978-0-646-52326-2. [Google Scholar]
- Veron, J.E.N. Corals of the World; Australian Institute of Marine Science: Townsville, Australia, 2000; Volume 1–3, ISBN 978-0-642-32236-4.
- Benzoni, F.; Arrigoni, R.; Stefani, F.; Stolarski, J. Systematics of the coral genus Craterastrea (Cnidaria, Anthozoa, Scleractinia) and description of a new family through combined morphological and molecular analyses. Syst. Biodivers. 2012, 10, 417–433. [Google Scholar] [CrossRef]
- Budd, A.F.; Romano, S.L.; Smith, N.D.; Barbeitos, M.S. Rethinking the phylogeny of scleractinian corals: A review of morphological and molecular data. Integr. Comp. Biol. 2010, 50, 411–427. [Google Scholar] [CrossRef]
- Kitahara, M.V.; Fukami, H.; Benzoni, F.; Huang, D. The New Systematics of Scleractinia: Integrating Molecular and Morphological Evidence. In The Cnidaria, Past, Present and Future: The World of Medusa and Her Sisters; Goffredo, S., Dubinsky, Z., Eds.; Springer International Publishing: Cham, Switzerland, 2016; pp. 41–59. ISBN 978-3-319-31305-4. [Google Scholar]
- Ramírez-Portilla, C.; Baird, A.H.; Cowman, P.F.; Quattrini, A.M.; Harii, S.; Sinniger, F.; Flot, J.F. Solving the coral species delimitation conundrum. Syst. Biol. 2022, 71, 461–475. [Google Scholar] [CrossRef]
- Fukami, H.; Niimura, A.; Nakamori, T.; Iryu, Y. Species composition and mitochondrial molecular phylogeny of Acropora corals in Funakoshi, Amami-Oshima Island, Japan: A proposal for its new taxonomic grouping. Galaxea J. Coral Reef Stud. 2021, 23, 17–35. [Google Scholar] [CrossRef]
- Rosser, N.L.; Thomas, L.; Stankowski, S.; Richards, Z.T.; Kennington, W.J.; Johnson, M.S. Phylogenomics provides new insight into evolutionary relationships and genealogical discordance in the reef-building coral genus Acropora. Proc. R. Soc. B 2017, 284, 20162182. [Google Scholar] [CrossRef]
- Kelley, R. Indo Pacific Coral Finder, 5th ed.; BYO Guides: Townsville, Australia, 2022; ISBN 978-0-646-52326-2. [Google Scholar]
- Hall, T. BioEdit: An important software for molecular biology. GERF Bull. Biosci. 2011, 2, 60–61. [Google Scholar]
- Hickerson, M.J.; Meyer, C.P.; Moritz, C. DNA barcoding will often fail to discover new animal species over broad parameter space. Syst. Biol. 2006, 55, 729–739. [Google Scholar] [CrossRef]
- Huang, D.; Meier, R.; Todd, P.A.; Chou, L.M. Slow mitochondrial COI sequence evolution at the base of the metazoan tree and its implications for DNA barcoding. J. Mol. Evol. 2008, 66, 167–174. [Google Scholar] [CrossRef]
- Johnston, E.C.; Forsman, Z.H.; Flot, J.F.; Schmidt-Roach, S.; Pinzón, J.H.; Knapp, I.S.; Toonen, R.J. A genomic glance through the fog of plasticity and diversification in Pocillopora. Sci. Rep. 2017, 7, 5991. [Google Scholar] [CrossRef]
- Shearer, T.L.; Coffroth, M.A. DNA BARCODING: Barcoding corals: Limited by interspecific divergence, not intraspecific variation. Mol. Ecol. Resour. 2008, 8, 247–255. [Google Scholar] [CrossRef]
- Hellberg, M.E. No variation and low synonymous substitution rates in coral mtDNA despite high nuclear variation. BMC Evol. Biol. 2006, 6, 24. [Google Scholar] [CrossRef]
- van Oppen, M.J.; Willis, B.L.; Miller, D.J. Atypically low rate of cytochrome b evolution in the scleractinian coral genus Acropora. Proc. R. Soc. Lond. Ser. B Biol. Sci. 1999, 266, 179–183. [Google Scholar] [CrossRef]
- Wares, J.P. Mitochondrial cytochrome b sequence data are not an improvement for species identification in scleractinian corals. PeerJ 2014, 2, 564. [Google Scholar] [CrossRef]
- Quattrini, A.M.; Faircloth, B.C.; Dueñas, L.F.; Bridge, T.C.; Brugler, M.R.; Calixto-Botía, I.F.; DeLeo, D.M.; Forêt, S.; Herrera, S.; Lee, S.M.; et al. Universal target-enrichment baits for anthozoan (Cnidaria) phylogenomics: New approaches to long-standing problems. Mol. Ecol. Resour. 2018, 18, 281–295. [Google Scholar] [CrossRef]
- Quattrini, A.M.; Rodríguez, E.; Faircloth, B.C.; Cowman, P.F.; Brugler, M.R.; Farfan, G.A.; Hellberg, M.E.; Kitahara, M.V.; Morrison, C.L.; Paz-García, D.A.; et al. Palaeoclimate ocean conditions shaped the evolution of corals and their skeletons through deep time. Nat. Ecol. Evol. 2020, 4, 1531–1538. [Google Scholar] [CrossRef]
- Cowman, P.F.; Quattrini, A.M.; Bridge, T.C.; Watkins-Colwell, G.J.; Fadli, N.; Grinblat, M.; Roberts, T.E.; McFadden, C.S.; Miller, D.J.; Baird, A.H. An enhanced target-enrichment bait set for Hexacorallia provides phylogenomic resolution of the staghorn corals (Acroporidae) and close relatives. Mol. Phylogenetics Evol. 2020, 153, 106944. [Google Scholar] [CrossRef]
- Erickson, K.L.; Pentico, A.; Quattrini, A.M.; McFadden, C.S. New approaches to species delimitation and population structure of corals: Two case studies using ultraconserved elements and exons. Mol. Ecol. Res. 2020, 21, 78–92. [Google Scholar] [CrossRef]
- Flot, J.F.; Couloux, A.; Tillier, S. Haplowebs as a graphical tool for delimiting species: A revival of Doyle’s” field for recombination” approach and its application to the coral genus Pocillopora in Clipperton. BMC Evol. Biol. 2010, 10, 372. [Google Scholar] [CrossRef]
- Sheets, E.A.; Warner, P.A.; Palumbi, S.R. Accurate population genetic measurements require cryptic species identification in corals. Coral Reefs 2018, 37, 549–563. [Google Scholar] [CrossRef]
- Wepfer, P.H.; Nakajima, Y.; Sutthacheep, M.; Radice, V.Z.; Richards, Z.; Ang, P.; Terraneo, T.; Sudek, M.; Fujimura, A.; Toonen, R.J.; et al. Evolutionary biogeography of the reef-building coral genus Galaxea across the Indo-Pacific ocean. Mol. Phylogenetics Evol. 2020, 151, 106905. [Google Scholar] [CrossRef]
- Suzuki, G.; Fukami, H. Evidence of genetic and reproductive isolation between two morphs of subtropical-dominant coral Acropora solitaryensis in the non-reef region of Japan. Zool. Sci. 2012, 29, 134–140. [Google Scholar] [CrossRef]
- Hoeksema, B.W.; Cairns, S. World List of Scleractinia. 2023. Available online: http://www.marinespecies.org/scleractinia (accessed on 8 March 2024).
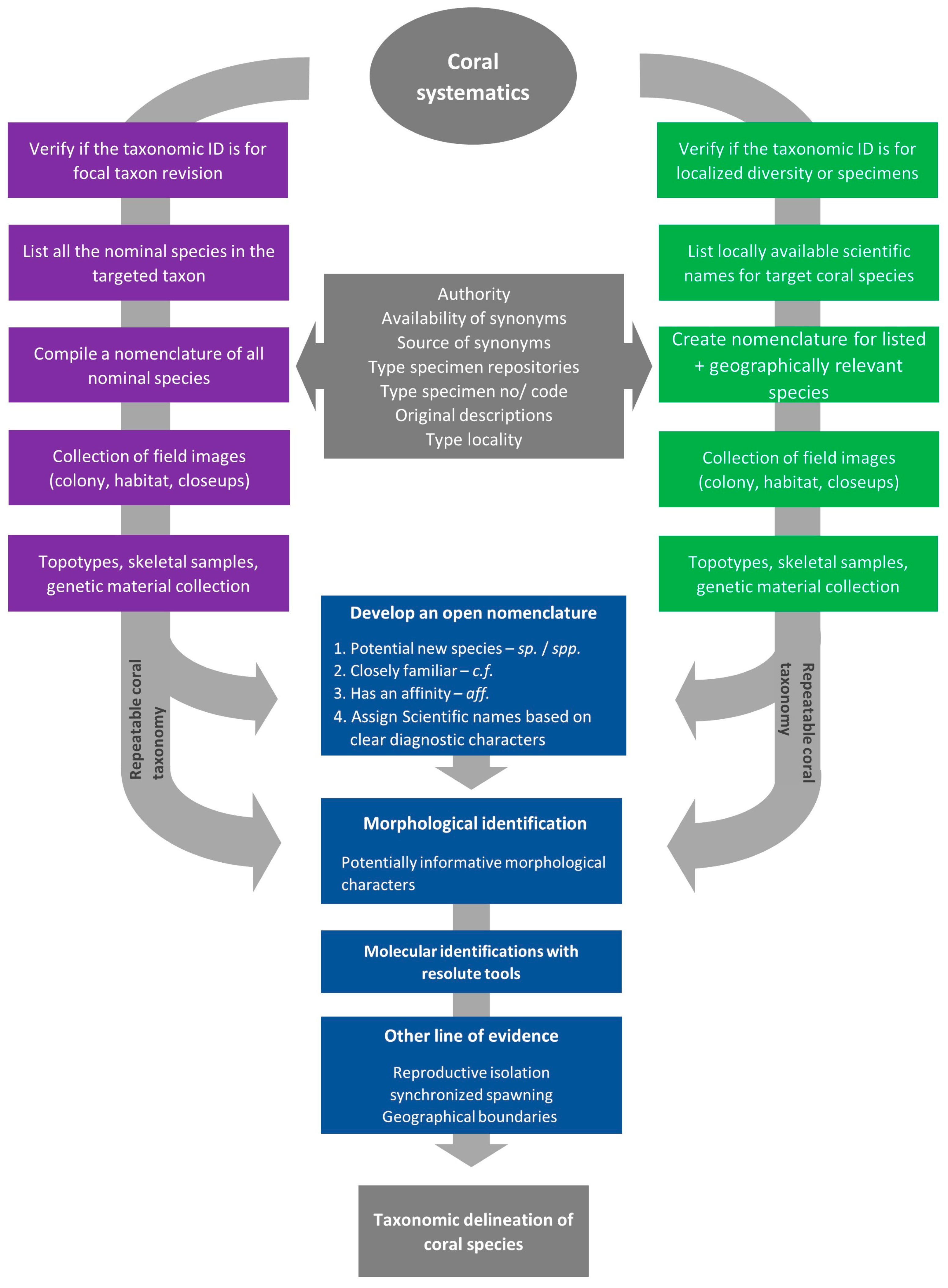
| Species Names with Authorities | Basis of the Identification | Availability of Voucher Material for Cross-Checking | Type Specimen Repository | Code/ Type No |
|---|---|---|---|---|
| Acroporidae Verrill, 1901 | ||||
| Acropora aspera Dana, 1846 | DNA marker COI | Not collected | USNM | 287 |
| Acropora digitifera Dana, 1846 | DNA marker COI | Not collected | MTQ | G37980 |
| Acropora gemmifera Brook, 1892 | DNA marker COI | Not collected | NHM | 1892.6.8.114 |
| Acropora latistella Brook, 1891 | Morphology based on [4] | Not collected | NHM | 1892.6.8.275 |
| Acropora pulchra Brook, 1891 | Morphology based on [4] | Not collected | NHM | 1884.2.16.1 |
| Alveopora allingi Hoffmeister, 1925 | Morphology based on [4] | Not collected | USNM | 62494 |
| Astreopora myriophthalma Lamarck, 1816 | Morphology based on [4] | Not collected | MNHN | 219c |
| Astreopora listeri Bernard, 1896 | Morphology based on [4] | Not collected | NMH | 1891-3-6-20 |
| Astreopora ocellata Bernard, 1896 | Morphology based on [4] | Not collected | NHM | 1892-12-1-150 |
| Montipora flabellata Studer,1901 | DNA marker COI | Not collected | N/A | Not available |
| Montipora informis Bernard, 1897 | Morphology based on [4] | Not collected | NHM | 1885-6-30-3 |
| Merulinidae Verrill, 1865 | ||||
| Coelastrea palauensis Yabe & Sugiyama, 1936 | Morphology based on [4] | Not collected | NHMTU | 56631 |
| Cyphastrea japonica Yabe & Sugiyama, 1936 | Morphology based on [4] | Not collected | NHMTU | 40323 |
| Cyphastrea microphthalma Lamarck, 1816 | Morphology based on [4] | Not collected | MNHN? | Not available |
| Platygyra acuta Veron, 2000 | Morphology based on [4] | Not collected | MQT | G55845 |
| Echinopora gemmacea Lamarck, 1816 | DNA marker COI | Not collected | MNHN? | Not available |
| Dipsastraea amicorum Milne Edwards & Haime, 1850 | Morphology based on [4] | Not collected | MNHN? | Not available |
| Dipsastraea lizardensis Veron, Pichon & Wijsman-Best, 1977 | Morphology based on [4] | Not collected | NHM | 1977.1.1.2 |
| Pachyseridae Benzoni & Hoeksema, 2023 | ||||
| Pachyseris gemmae Nemenzo, 1955 | Morphology based on [4] | Not collected | UP | UP C-123 |
| Oulastreidae Vaughan,1919 | ||||
| Oulastrea crispata Lamarck, 1816 | Morphology based on [4] | Not collected | MNHN | scle15 |
| Poritidae Gray, 1842 | ||||
| Porites evermanni Vaughan, 1907 | Morphology based on [4] | Not collected | USNM | 21627 |
| Porites murrayensis Vaughan, 1918 | Morphology based on [4] | Not collected | USNM | 47237 |
| Porites pukoensis Vaughan, 1907 | Morphology based on [4] | Not collected | USNM | 22236 |
| Goniopora lobata Milne Edwards, 1860 | Morphology based on [4] | Not collected | MNHN? | Not available |
| Goniopora minor Crossland, 1952 | Morphology based on [4] | Not collected | NHM | 56 |
| Goniopora somaliensis Vaughan, 1907 | Morphology based on [4] | Not collected | MNHN | Not available |
| Goniopora tenuidens Quelch, 1886 | Morphology based on [4] | Not collected | NHM | Not available |
| Dendrophylliidae Gray, 1847 | ||||
| Turbinaria frondens Dana, 1846 | Morphology based on [4] | Not collected | USNM | 214 |
| Turbinaria reniformis Bernard, 1896 | Morphology based on [4] | Not collected | NHM | Not available |
| Turbinaria stelluta Lamarck, 1816 | Morphology based on [4] | Not collected | MNHN | Not available |
| Lobophylliidae Dai & Horng, 2009 | ||||
| Acanthastrea ishigakiensis Veron, 1990 | Morphology based on [4] | Not collected | MTQ | G32484 |
| Micromussa amakusensis Veron, 1990 | Morphology based on [4] | Not collected | MTQ | G32485 |
| Euphylliidae Milne Edwards & Haime, 1857 | ||||
| Coeloseris mayeri Vaughan, 1918 | Morphology based on [4] | Not collected | USNM | 45546 (syntype series (a series of multiple types) but no holotype) |
| Siderastreidae Vaughan & Wells, 1943 | ||||
| Siderastrea savignyana Milne Edwards & Haime, 1849 | Morphology based on [4] | Not collected | NHM | Not available |
| Species Name with Authorities | Type Locality | Potential Junior Synonyms in Sri Lanka |
|---|---|---|
| Acroporidae Verrill, 1901 | ||
| Acropora aspera Dana, 1846 * | Fiji | Acropora yeayamaensis (Eguchi & Shirai, 1977), Japan; Madrepora manni (Quelch, 1886), Zamboanga, Philippines |
| Acropora digitifera Dana, 1846 * | Marshall Islands | Acropora schmitti (Wells, 1950), Cocos (Keeling) Islands, |
| Acropora gemmifera Brook, 1892 | Rocky Islands, Great Barrier Reef, Australia | |
| Acropora latistella Brook, 1891 * | Port Denison, Great Barrier Reef, Australia | Acropora imperfecta (Nemenzo, 1971), Puerto Galera, Philippines; Acropora loricata (Nemenzo, 1967), Cebu, Philippines |
| Acropora pulchra Brook, 1891 | Cocos (Keeling) Islands, Indian Ocean | |
| Alveopora allingi Hoffmeister, 1925 | Pago Pago Harbour, American Samoa | |
| Astreopora myriophthalma Lamarck, 1816 * | Red Sea | Astreopora stellae (Nemenzo, 1964), Cebu, Philippines; |
| Astreopora listeri Bernard, 1896 * | Tonga | Astreopora horizontalis (Bernard, 1896), Seychelles |
| Astreopora ocellata Bernard, 1896 | Torres Strait | |
| Montipora flabellata Studer,1901 | Hawaii | |
| Montipora informis Bernard, 1897 | Torres Strait | |
| Merulinidae Verrill, 1865 | ||
| Coelastrea palauensis Yabe & Sugiyama, 1936 | Palau | |
| Cyphastrea japonica Yabe & Sugiyama, 1936 | Tanabe-wan, Wakayama-ken, subtropical Japan | |
| Cyphastrea microphthalma Lamarck, 1816 * | the Ocean around New Holland— possibly Western Australia | Cyphastrea minuta (Nemenzo & Ferraris, 1982), Cebu, Philippines |
| Platygyra acuta Veron, 2000 | Mahe, Seychelles | |
| Echinopora gemmacea Lamarck, 1816 | Indian Ocean? | |
| Dipsastraea amicorum Milne Edwards & Haime, 1850 * | Tonga | Barabattoia modesta (Nemenzo, 1971), Cebu, Philippines |
| Dipsastraea lizardensis Veron, Pichon & Wijsman-Best, 1977 | Lizard Island, Great Barrier Reef | |
| Pachyseridae Benzoni& Hoeksema, 2023 (Authors have listed Pachyseris gemmae Nemenzo, 1955 under Scleractinia incertae sedis but it is now under Pachyseridae Benzoni& Hoeksema, 2023 [28]) | ||
| Pachyseris gemmae Nemenzo, 1955 | Bohol, Philippines | |
| Oulastreidae Vaughan, 1919 | ||
| Oulastrea crispata Lamarck, 1816 | Indian Ocean | |
| Poritidae Gray, 1842 | ||
| Porites evermanni Vaughan, 1907 | Oahu, Hawaii | |
| Porites murrayensis Vaughan, 1918 | Mer Island, Torres Strait | |
| Porites pukoensis Vaughan, 1907 | Molokai, Hawaii | |
| Goniopora lobata Milne Edwards, 1860 | Red Sea | |
| Goniopora minor Crossland, 1952 (Considered as a junior subjective synonym of G. pedunculata Quoy and Gaimard, 1833 [28]) | Great Barrier Reef | |
| Goniopora somaliensis Vaughan, 1907 | French Somaliland | |
| Goniopora tenuidens Quelch, 1886 | Zamboanga, Philippines | |
| Dendrophylliidae Gray, 1847 | ||
| Turbinaria frondens Dana, 1846 * | Fiji | Turbinaria carcarensis (Nemenzo, 1971), Cebu, Philippines; Turbinaria contorta (Bernard, 1896), South China Seas; Turbinaria ramosa (Yabe & Sugiyama, 1941), Meitu Minami-Naka-gun, Miyazaki-ken, Japan; Turbinaria rugosa (Bernard, 1896), Formosa |
| Turbinaria reniformis Bernard, 1896 * | Palm Islands, Great Barrier Reef | Turbinaria disparata (Bernard, 1896), Basay, Oriental Negros, Philippines; |
| Turbinaria stelluta Lamarck, 1816 * | American Ocean? | Turbinaria globularis (Bernard, 1896), Diego Garcia; Turbinaria titizimaensis (Yabe & Sugiyama, 1941), Ogasawa Islands, Japan; Turbinaria nitida (Nemenzo, 1960), Quezon, Philippines |
| Lobophylliidae Dai & Horng, 2009 | ||
| Acanthastrea ishigakiensis Veron, 1990 (Basionym Acanthastrea ishigakiensis is now accepted as Lobophyllia ishigakiensis [28]) | Ishigaki Island, Japan | |
| Micromussa amakusensis Veron, 1990 | Amaku Island, Japan | |
| Euphylliidae Milne Edwards & Haime, 1857 (Authors have listed Coeloseris mayeri Vaughan, 1918 under the Family Agariciidae Gray, 1847 but it should be Euphylliidae Milne Edwards & Haime, 1857 [28]) | ||
| Coeloseris mayeri Vaughan, 1918 | Mer Island, Torres Strait | |
| Siderastreidae Vaughan & Wells, 1943 (Siderastreidae Vaughan & Wells, 1943 is now accepted as Rhizangiidae d’Orbigny, 1851 [28]) | ||
| Siderastrea savignyana Milne Edwards & Haime, 1849 | Red Sea |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Hendawitharana, M.P.; Gittenberger, A.; Jayasinghe, P.K.; Herath, D.R. Comment on Arulananthan et al. The Status of the Coral Reefs of the Jaffna Peninsula (Northern Sri Lanka), with 36 Coral Species New to Sri Lanka Confirmed by DNA Bar-Coding. Oceans 2021, 2, 509–529. Oceans 2024, 5, 276-284. https://doi.org/10.3390/oceans5020017
Hendawitharana MP, Gittenberger A, Jayasinghe PK, Herath DR. Comment on Arulananthan et al. The Status of the Coral Reefs of the Jaffna Peninsula (Northern Sri Lanka), with 36 Coral Species New to Sri Lanka Confirmed by DNA Bar-Coding. Oceans 2021, 2, 509–529. Oceans. 2024; 5(2):276-284. https://doi.org/10.3390/oceans5020017
Chicago/Turabian StyleHendawitharana, Manuja Promodya, Adriaan Gittenberger, Prabath Krishantha Jayasinghe, and Deishini Rupika Herath. 2024. "Comment on Arulananthan et al. The Status of the Coral Reefs of the Jaffna Peninsula (Northern Sri Lanka), with 36 Coral Species New to Sri Lanka Confirmed by DNA Bar-Coding. Oceans 2021, 2, 509–529" Oceans 5, no. 2: 276-284. https://doi.org/10.3390/oceans5020017
APA StyleHendawitharana, M. P., Gittenberger, A., Jayasinghe, P. K., & Herath, D. R. (2024). Comment on Arulananthan et al. The Status of the Coral Reefs of the Jaffna Peninsula (Northern Sri Lanka), with 36 Coral Species New to Sri Lanka Confirmed by DNA Bar-Coding. Oceans 2021, 2, 509–529. Oceans, 5(2), 276-284. https://doi.org/10.3390/oceans5020017






