Prostaglandin F2α Affects the Cycle of Clock Gene Expression and Mouse Behavior
Abstract
1. Introduction
2. Results
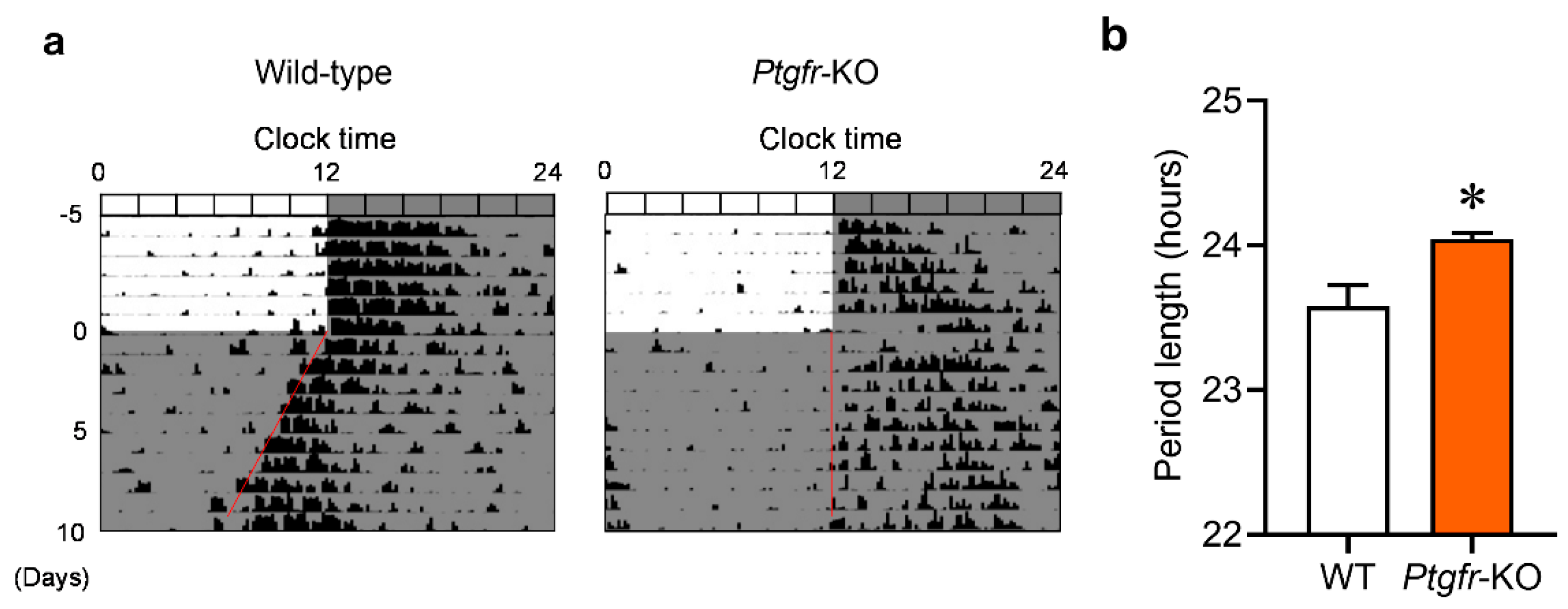
2.1. Ptgfr-KO Mice Exhibit Longer Behavioral Cycles under Constant Dark Conditions
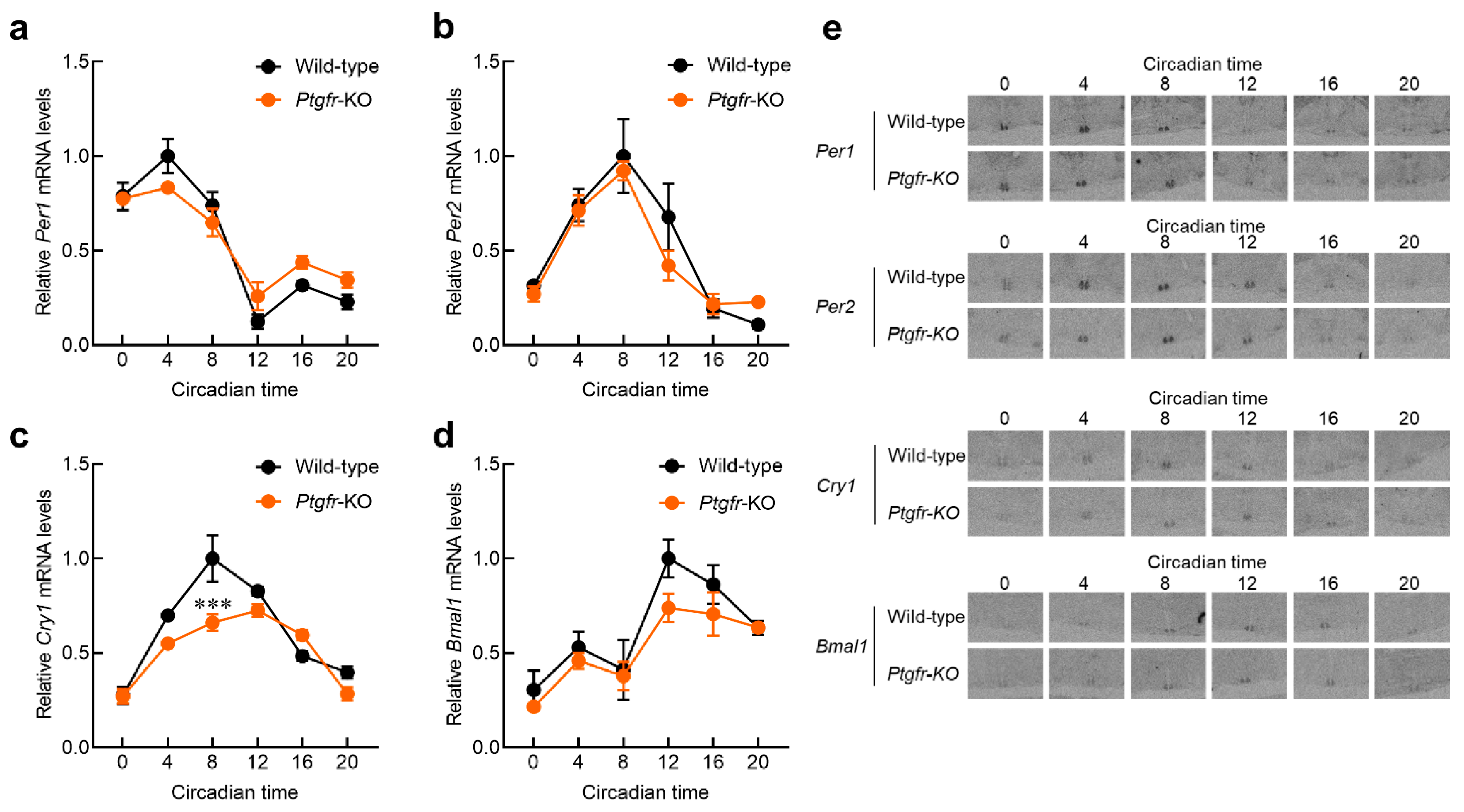
2.2. Time-Dependent Changes in Expression of Clock Genes in the Suprachiasmatic Nucleus (SCN) of Ptgfr-KO Mice
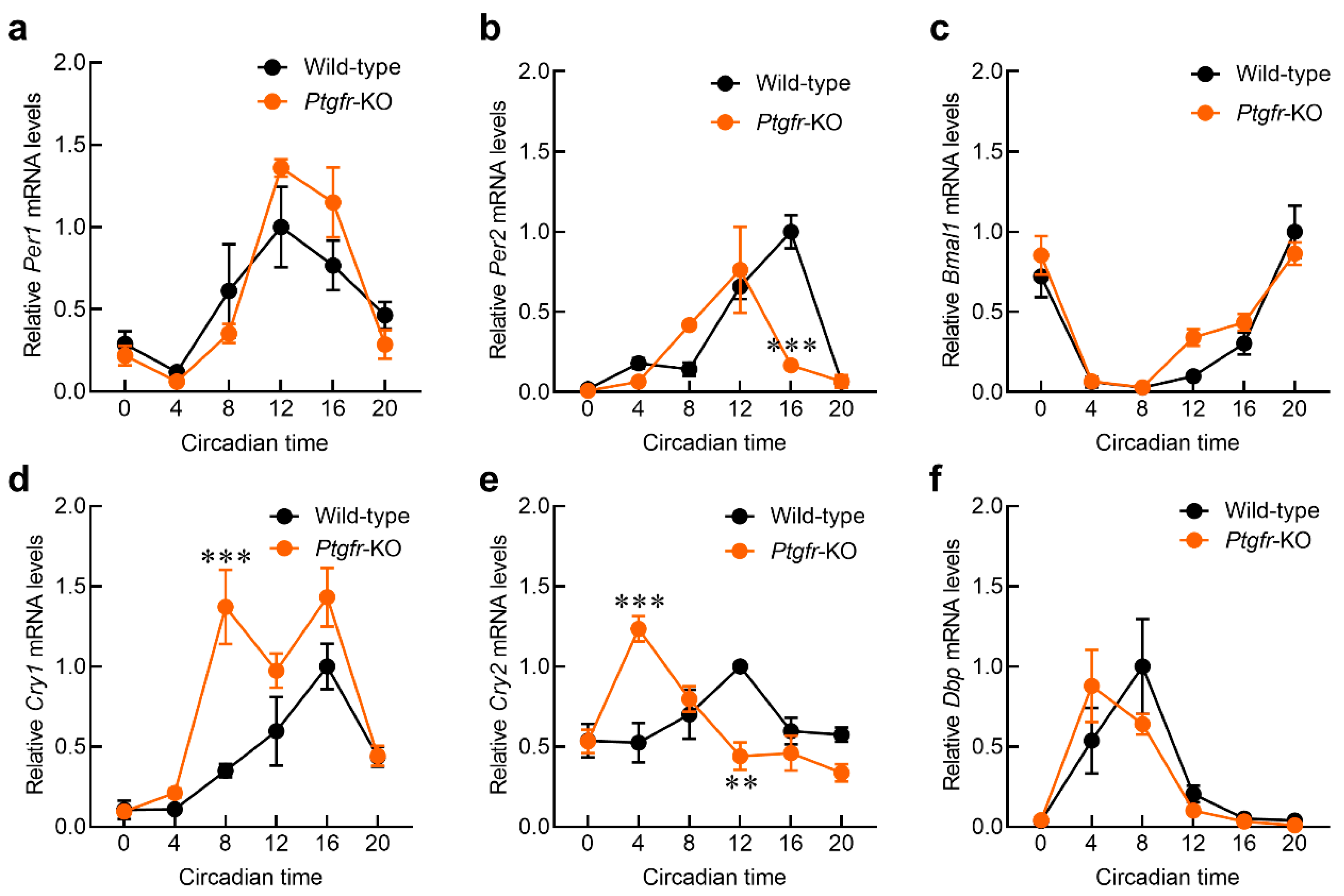
2.3. Time-Dependent Changes in Clock Gene Expression in the Liver of Ptgfr-KO Mice
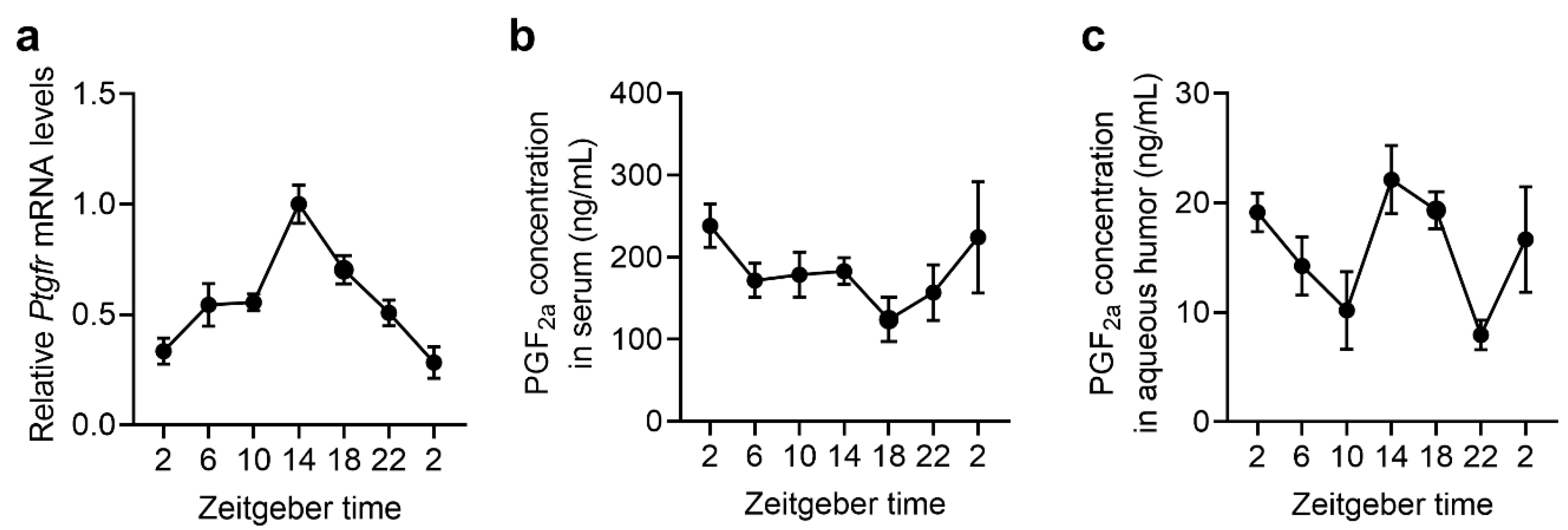
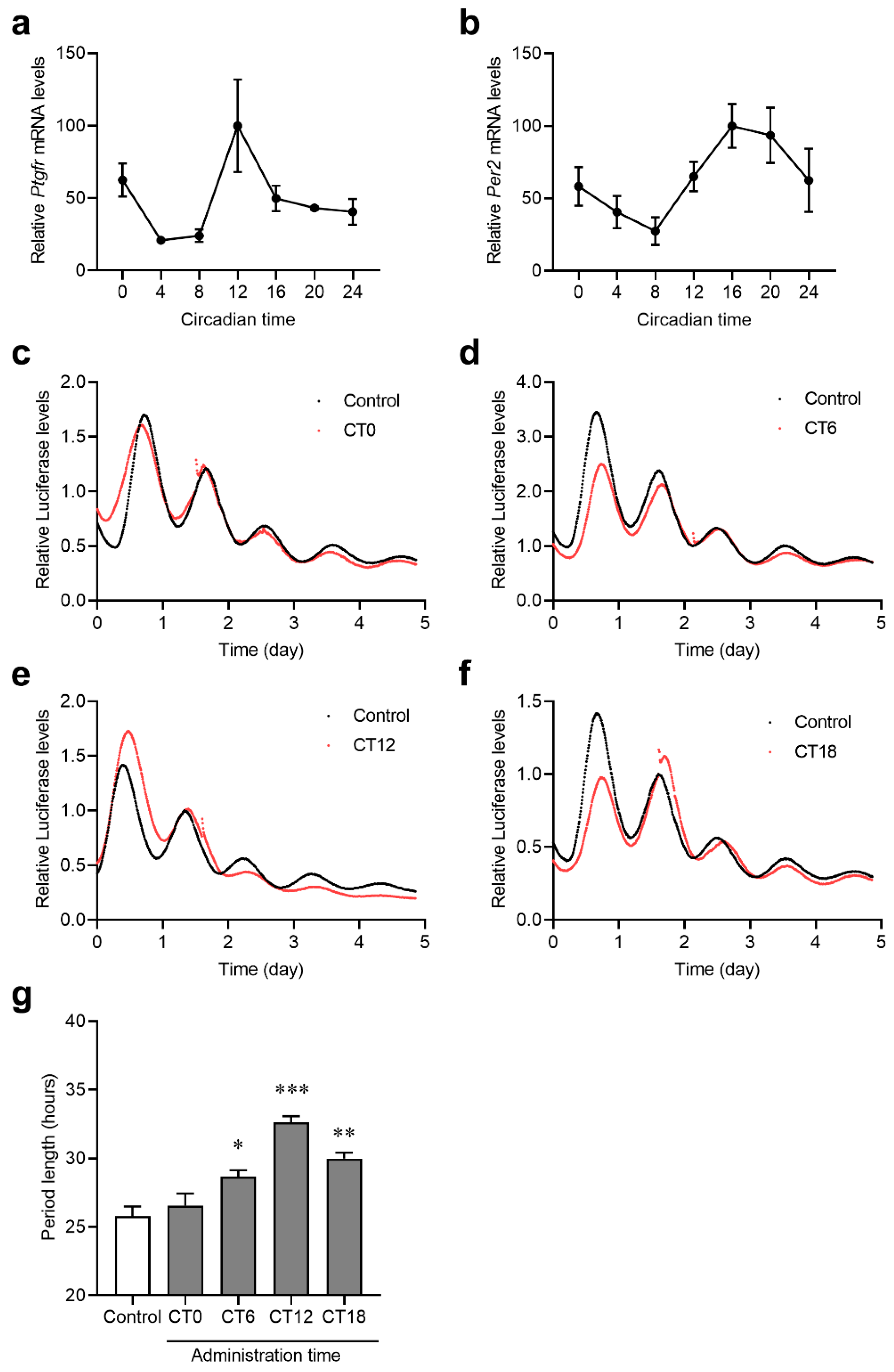
2.4. The Expression Rhythms of Ptgfr and the Secretion Rhythms of PGF2α in Mouse Retina
2.5. Effects of PTGFR Agonists on Per2 Expression Rhythm in Per2::luc C6 Cells
3. Discussion
4. Materials and Methods
4.1. Animal Experiments
4.2. Measurement of Locomotor Activity Rhythm
4.3. In Situ Hybridization
4.4. Quantitative RT-PCR Analysis
4.5. Quantification of PGF2α Using ELISA
4.6. Real-Time Monitoring of Circadian Bioluminescence
4.7. Statistical Analysis
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Langmesser, S.; Tallone, T.; Bordon, A.; Rusconi, S.; Albrecht, U. Interaction of circadian clock proteins PER2 and CRY with BMAL1 and CLOCK. BMC Mol. Biol. 2008, 9, 41. [Google Scholar] [CrossRef] [PubMed]
- Bass, J.; Takahashi, J.S. Circadian integration of metabolism and energetics. Science 2010, 330, 1349–1354. [Google Scholar] [CrossRef] [PubMed]
- Asher, G.; Schibler, U. Crosstalk between components of circadian and metabolic cycles in mammals. Cell Metab. 2011, 13, 125–137. [Google Scholar] [CrossRef]
- DeBruyne, J.P.; Weaver, D.R.; Reppert, S.M. CLOCK and NPAS2 have overlapping roles in the suprachiasmatic circadian clock. Nat. Neurosci. 2007, 10, 543–545. [Google Scholar] [CrossRef] [PubMed]
- Ueda, H.R.; Hayashi, S.; Chen, W.; Sano, M.; Machida, M.; Shigeyoshi, Y.; Iino, M.; Hashimoto, S. System-level identification of transcriptional circuits underlying mammalian circadian clocks. Nat. Genet. 2005, 37, 187–192. [Google Scholar] [CrossRef] [PubMed]
- Kwon, P.K.; Lee, K.H.; Kim, J.H.; Tae, S.; Ham, S.; Jeong, Y.H.; Kim, S.W.; Kang, B.; Kim, H.M.; Choi, J.H.; et al. hnRNP K Supports High-Amplitude D Site-Binding Protein mRNA (Dbp mRNA) Oscillation to Sustain Circadian Rhythms. Mol. Cell. Biol. 2020, 40, e00537-19. [Google Scholar] [CrossRef] [PubMed]
- Kanemitsu, T.; Tsurudome, Y.; Kusunose, N.; Oda, M.; Matsunaga, N.; Koyanagi, S.; Ohdo, S. Periodic variation in bile acids controls circadian changes in uric acid via regulation of xanthine oxidase by the orphan nuclear receptor PPARα. J. Biol. Chem. 2017, 292, 21397–21406. [Google Scholar] [CrossRef]
- Kim, W.; Shin, J.C.; Lee, K.H.; Kim, K.T. PTBP1 Positively Regulates the Translation of Circadian Clock Gene, Period1. Int. J. Mol. Sci. 2020, 21, 6921. [Google Scholar] [CrossRef]
- Mazzoccoli, G.; Colangelo, T.; Panza, A.; Rubino, R.; Tiberio, C.; Palumbo, O.; Carella, M.; Trombetta, D.; Gentile, A.; Tavano, F.; et al. Analysis of clock gene-miRNA correlation networks reveals candidate drivers in colorectal cancer. Oncotarget 2016, 7, 45444–45461. [Google Scholar] [CrossRef]
- Welsh, D.K.; Takahashi, J.S.; Kay, S.A. Suprachiasmatic nucleus: Cell autonomy and network properties. Annu. Rev. Physiol. 2010, 72, 551–577. [Google Scholar] [CrossRef]
- Ashton, A.; Foster, R.G.; Jagannath, A. Photic Entrainment of the Circadian System. Int. J. Mol. Sci. 2022, 23, 729. [Google Scholar] [CrossRef] [PubMed]
- Le Minh, N.; Damiola, F.; Tronche, F.; Schütz, G.; Schibler, U. Glucocorticoid hormones inhibit food-induced phase-shifting of peripheral circadian oscillators. EMBO J. 2001, 20, 7128–7136. [Google Scholar] [CrossRef]
- Entzian, P.; Linnemann, K.; Schlaak, M.; Zabel, P. Obstructive sleep apnea syndrome and circadian rhythms of hormones and cytokines. Am. J. Respir. Crit. Care Med. 1996, 153, 1080–1086. [Google Scholar] [CrossRef] [PubMed]
- Narumiya, S.; Sugimoto, Y.; Ushikubi, F. Prostanoid receptors: Structures, properties, and functions. Physiol. Rev. 1999, 79, 1193–1226. [Google Scholar] [CrossRef] [PubMed]
- Kato, K.; Murai, I.; Asai, S.; Takahashi, Y.; Nagata, T.; Komuro, S.; Mizuno, S.; Iwasaki, A.; Ishikawa, K.; Arakawa, Y. Circadian rhythm of melatonin and prostaglandin in modulation of stress-induced gastric mucosal lesions in rats. Aliment. Pharmacol. Ther. 2002, 16, 29–34. [Google Scholar] [CrossRef]
- Tsuchiya, Y.; Minami, I.; Kadotani, H.; Nishida, E. Resetting of peripheral circadian clock by prostaglandin E2. EMBO Rep. 2005, 6, 256–261. [Google Scholar] [CrossRef] [PubMed]
- Zhao, Y.; Lei, Y.; Ning, H.; Zhang, Y.; Chen, G.; Wang, C.; Wan, Q.; Guo, S.; Liu, Q.; Xie, R.; et al. PGF2α facilitates pathological retinal angiogenesis by modulating endothelial FOS-driven ELR+ CXC chemokine expression. EMBO Mol. Med. 2023, 15, e16373. [Google Scholar] [CrossRef]
- Xu, C.; You, X.; Liu, W.; Sun, Q.; Ding, X.; Huang, Y.; Ni, X. Prostaglandin F2α regulates the expression of uterine activation proteins via multiple signalling pathways. Reproduction 2015, 149, 139–146. [Google Scholar] [CrossRef]
- Sharif, N.A.; Kelly, C.R.; Crider, J.Y.; Williams, G.W.; Xu, S.X. Ocular hypotensive FP prostaglandin (PG) analogs: PG receptor subtype binding affinities and selectivities, and agonist potencies at FP and other PG receptors in cultured cells. J. Ocul. Pharmacol. Ther. 2003, 19, 501–515. [Google Scholar] [CrossRef]
- van der Vinne, V.; Swoap, S.J.; Vajtay, T.J.; Weaver, D.R. Desynchrony between brain and peripheral clocks caused by CK1δ/ε disruption in GABA neurons does not lead to adverse metabolic outcomes. Proc. Natl. Acad. Sci. USA 2018, 115, E2437–E2446. [Google Scholar] [CrossRef]
- Dunlap, J.C. Molecular bases for circadian clocks. Cell 1999, 96, 271–290. [Google Scholar] [CrossRef] [PubMed]
- Matsuo, T.; Cynader, M.S. Localisation of prostaglandin F2 alpha and E2 binding sites in the human eye. Br. J. Ophthalmol. 1992, 76, 210–213. [Google Scholar] [CrossRef] [PubMed]
- De Zavalía, N.; Fernandez, D.C.; Sande, P.H.; Keller Sarmiento, M.I.; Golombek, D.A.; Rosenstein, R.E.; Silberman, D.M. Circadian variations of prostaglandin E2 and F2 alpha release in the golden hamster retina. J. Neurochem. 2010, 112, 972–979. [Google Scholar] [CrossRef] [PubMed]
- Kawaguchi, C.; Shintani, N.; Hayata-Takano, A.; Hatanaka, M.; Kuromi, A.; Nakamura, R.; Yamano, Y.; Shintani, Y.; Nagai, K.; Tsuchiya, S.; et al. Lipocalin-type prostaglandin D synthase regulates light-induced phase advance of the central circadian rhythm in mice. Commun. Biol. 2020, 3, 557. [Google Scholar] [CrossRef] [PubMed]
- Yan, J.; Chen, Y.; Zhu, Y.; Paquet-Durand, F. Programmed Non-Apoptotic Cell Death in Hereditary Retinal Degeneration: Crosstalk between cGMP-Dependent Pathways and PARthanatos? Int. J. Mol. Sci. 2021, 22, 10567. [Google Scholar] [CrossRef] [PubMed]
- Warren, E.J.; Allen, C.N.; Brown, R.L.; Robinson, D.W. The light-activated signaling pathway in SCN-projecting rat retinal ganglion cells. Eur. J. Neurosci. 2006, 23, 2477–2487. [Google Scholar] [CrossRef] [PubMed]
- Masana, M.I.; Benloucif, S.; Dubocovich, M.L. Light-induced c-fos mRNA expression in the suprachiasmatic nucleus and the retina of C3H/HeN mice. Brain Res. Mol. Brain Res. 1996, 42, 193–201. [Google Scholar] [CrossRef]
- Vitaterna, M.H.; Selby, C.P.; Todo, T.; Niwa, H.; Thompson, C.; Fruechte, E.M.; Hitomi, K.; Thresher, R.J.; Ishikawa, T.; Miyazaki, J.; et al. Differential regulation of mammalian period genes and circadian rhythmicity by cryptochromes 1 and 2. Proc. Natl. Acad. Sci. USA 1999, 96, 12114–12119. [Google Scholar] [CrossRef]
- Ukai-Tadenuma, M.; Yamada, R.G.; Xu, H.; Ripperger, J.A.; Liu, A.C.; Ueda, H.R. Delay in feedback repression by cryptochrome 1 is required for circadian clock function. Cell 2011, 144, 268–281. [Google Scholar] [CrossRef]
- Wang, Y.; Yan, S.; Xiao, B.; Zuo, S.; Zhang, Q.; Chen, G.; Yu, Y.; Chen, D.; Liu, Q.; Liu, Y.; et al. Prostaglandin F2α Facilitates Hepatic Glucose Production through CaMKIIγ/p38/FOXO1 Signaling Pathway in Fasting and Obesity. Diabetes 2018, 67, 1748–1760. [Google Scholar] [CrossRef]
- Fujimori, K. Prostaglandins as PPARγ Modulators in Adipogenesis. PPAR Res. 2012, 2012, 527607. [Google Scholar] [CrossRef] [PubMed]
- Tischkau, S.A.; Mitchell, J.W.; Tyan, S.H.; Buchanan, G.F.; Gillette, M.U. Ca2+/cAMP response element-binding protein (CREB)-dependent activation of Per1 is required for light-induced signaling in the suprachiasmatic nucleus circadian clock. J. Biol. Chem. 2003, 278, 718–723. [Google Scholar] [CrossRef] [PubMed]
- Terazono, H.; Mutoh, T.; Yamaguchi, S.; Kobayashi, M.; Akiyama, M.; Udo, R.; Ohdo, S.; Okamura, H.; Shibata, S. Adrenergic regulation of clock gene expression in mouse liver. Proc. Natl. Acad. Sci. USA 2003, 100, 6795–6800. [Google Scholar] [CrossRef] [PubMed]
- Kholodenko, I.V.; Yarygin, K.N. Cellular Mechanisms of Liver Regeneration and Cell-Based Therapies of Liver Diseases. Biomed. Res. Int. 2017, 2017, 8910821. [Google Scholar] [CrossRef] [PubMed]
- Greco, C.M.; Koronowski, K.B.; Smith, J.G.; Shi, J.; Kunderfranco, P.; Carriero, R.; Chen, S.; Samad, M.; Welz, P.S.; Zinna, V.M.; et al. Integration of feeding behavior by the liver circadian clock reveals network dependency of metabolic rhythms. Sci. Adv. 2021, 7, eabi7828. [Google Scholar] [CrossRef] [PubMed]
- Asano, F.; Kim, S.J.; Fujiyama, T.; Miyoshi, C.; Hotta-Hirashima, N.; Asama, N.; Iwasaki, K.; Kakizaki, M.; Mizuno, S.; Mieda, M.; et al. SIK3-HDAC4 in the suprachiasmatic nucleus regulates the timing of arousal at the dark onset and circadian period in mice. Proc. Natl. Acad. Sci. USA 2023, 120, e2218209120. [Google Scholar] [CrossRef] [PubMed]
- Winkler, N.S.; Fautsch, M.P. Effects of prostaglandin analogues on aqueous humor outflow pathways. J. Ocul. Pharmacol. Ther. 2014, 30, 102–109. [Google Scholar] [CrossRef]
- Cuppoletti, J.; Malinowska, D.H.; Tewari, K.P.; Chakrabarti, J.; Ueno, R. Cellular and molecular effects of unoprostone as a BK channel activator. Biochim. Biophys. Acta 2007, 1768, 1083–1092. [Google Scholar] [CrossRef]
- Foster, R.G.; Hughes, S.; Peirson, S.N. Circadian Photoentrainment in Mice and Humans. Biology 2020, 9, 180. [Google Scholar] [CrossRef]
- Csoma, B.; Bikov, A. The Role of the Circadian Rhythm in Dyslipidaemia and Vascular Inflammation Leading to Atherosclerosis. Int. J. Mol. Sci. 2023, 24, 14145. [Google Scholar] [CrossRef]
- Škrlec, I.; Talapko, J.; Džijan, S.; Cesar, V.; Lazić, N.; Lepeduš, H. The Association between Circadian Clock Gene Polymorphisms and Metabolic Syndrome: A Systematic Review and Meta-Analysis. Biology 2021, 11, 20. [Google Scholar] [CrossRef] [PubMed]
- Early, J.O.; Menon, D.; Wyse, C.A.; Cervantes-Silva, M.P.; Zaslona, Z.; Carroll, R.G.; Palsson-McDermott, E.M.; Angiari, S.; Ryan, D.G.; Corcoran, S.E.; et al. Circadian clock protein BMAL1 regulates IL-1β in macrophages via NRF2. Proc. Natl. Acad. Sci. USA 2018, 115, E8460–E8468. [Google Scholar] [CrossRef] [PubMed]
- Estarlich, M.; Tolsa, C.; Trapero, I.; Buigues, C. Circadian Variations and Associated Factors in Patients with Ischaemic Heart Disease. Int. J. Environ. Res. Public Health 2022, 19, 15628. [Google Scholar] [CrossRef] [PubMed]
- Liu, Y.; Zhang, Y.; Li, T.; Han, J.; Wang, Y. The tight junction protein TJP1 regulates the feeding-modulated hepatic circadian clock. Nat. Commun. 2020, 11, 589. [Google Scholar] [CrossRef] [PubMed]
- Pignatelli, J.; de Sevilla, M.E.; Sperber, J.; Horrillo, D.; Medina-Gomez, G.; Aleman, I.T. Insulin-like Growth Factor I Couples Metabolism with Circadian Activity through Hypothalamic Orexin Neurons. Int. J. Mol. Sci. 2022, 23, 4679. [Google Scholar] [CrossRef] [PubMed]
- Watanabe, T.; Suzuki, T.; Ishikawa, A.; Yokota, Y.; Ueda, H.R.; Yamada, R.G.; Tei, H.; Imai, S.; Tomida, S.; Kobayashi, J.; et al. Genetic and Molecular Analysis of Wild-Derived Arrhythmic Mice. PLoS ONE 2009, 4, e4301. [Google Scholar] [CrossRef] [PubMed]
- Kusunose, N.; Matsunaga, N.; Kimoto, K.; Akamine, T.; Hamamura, K.; Koyanagi, S.; Ohdo, S.; Kubota, T. Mitomycin C modulates the circadian oscillation of clock gene period 2 expression through attenuating the glucocorticoid signaling in mouse fibroblasts. Biochem. Biophys. Res. Commun. 2015, 467, 157–163. [Google Scholar] [CrossRef]
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Tsurudome, Y.; Yoshida, Y.; Hamamura, K.; Ogino, T.; Yasukochi, S.; Yasuo, S.; Iwamoto, A.; Yoshihara, T.; Inazumi, T.; Tsuchiya, S.; et al. Prostaglandin F2α Affects the Cycle of Clock Gene Expression and Mouse Behavior. Int. J. Mol. Sci. 2024, 25, 1841. https://doi.org/10.3390/ijms25031841
Tsurudome Y, Yoshida Y, Hamamura K, Ogino T, Yasukochi S, Yasuo S, Iwamoto A, Yoshihara T, Inazumi T, Tsuchiya S, et al. Prostaglandin F2α Affects the Cycle of Clock Gene Expression and Mouse Behavior. International Journal of Molecular Sciences. 2024; 25(3):1841. https://doi.org/10.3390/ijms25031841
Chicago/Turabian StyleTsurudome, Yuya, Yuya Yoshida, Kengo Hamamura, Takashi Ogino, Sai Yasukochi, Shinobu Yasuo, Ayaka Iwamoto, Tatsuya Yoshihara, Tomoaki Inazumi, Soken Tsuchiya, and et al. 2024. "Prostaglandin F2α Affects the Cycle of Clock Gene Expression and Mouse Behavior" International Journal of Molecular Sciences 25, no. 3: 1841. https://doi.org/10.3390/ijms25031841
APA StyleTsurudome, Y., Yoshida, Y., Hamamura, K., Ogino, T., Yasukochi, S., Yasuo, S., Iwamoto, A., Yoshihara, T., Inazumi, T., Tsuchiya, S., Takeo, T., Nakagata, N., Higuchi, S., Sugimoto, Y., Tsuruta, A., Koyanagi, S., Matsunaga, N., & Ohdo, S. (2024). Prostaglandin F2α Affects the Cycle of Clock Gene Expression and Mouse Behavior. International Journal of Molecular Sciences, 25(3), 1841. https://doi.org/10.3390/ijms25031841






