Evaluation of Holstein Cows with Tongue-Rolling: Plasma Metabolomics and Milk Proteomics
Abstract
1. Introduction
2. Materials and Methods
2.1. Animal Welfare Statement
2.2. Animal, Management, and Behavioral Observations
2.3. Sample Collection
2.4. Biochemical Testing of Plasma and Milk Samples
2.5. Untargeted Metabolomics Analysis
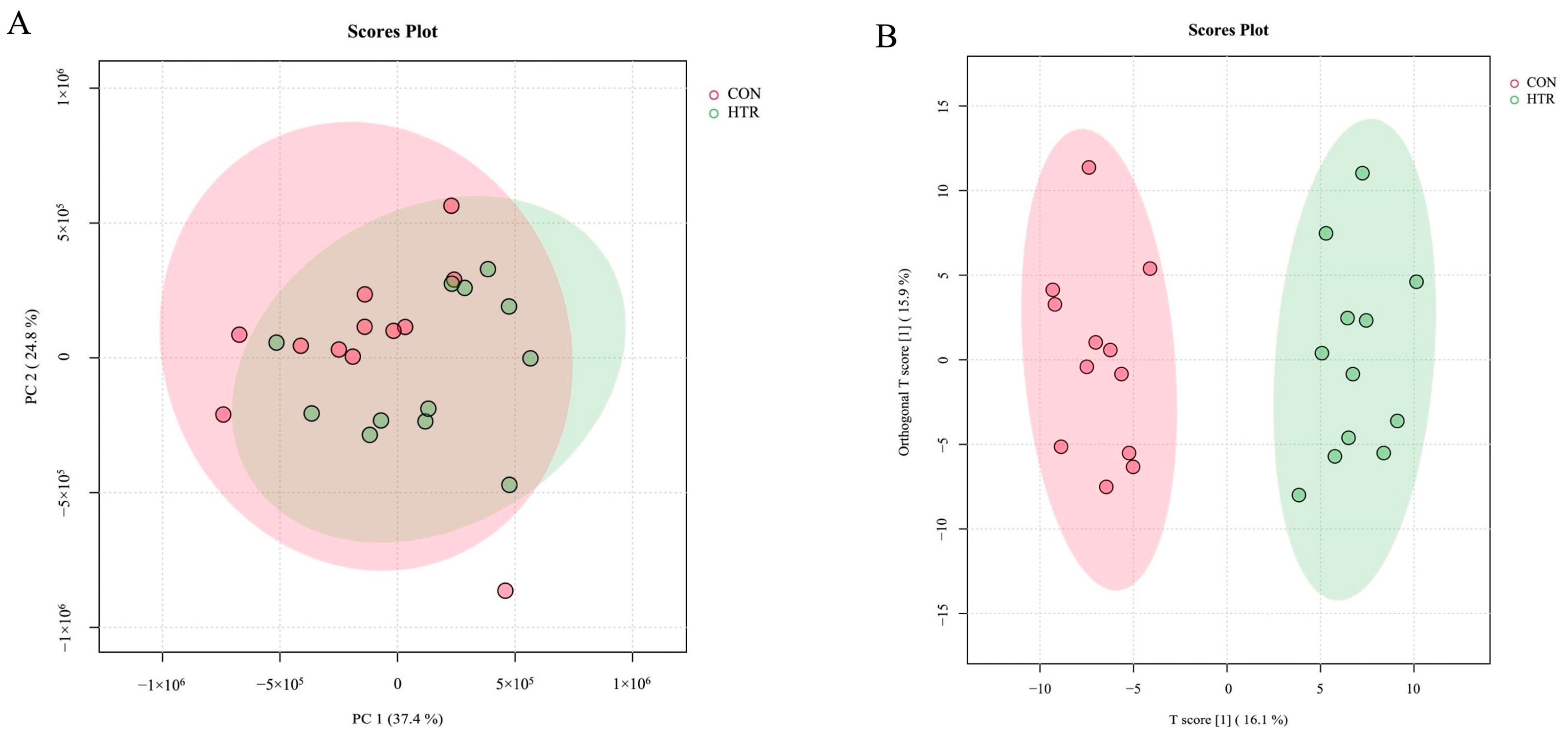
2.6. Statistical Analysis
2.6.1. Data-Independent Acquisition Proteomics Analysis
2.6.2. Biochemical Data Analysis
2.6.3. Metabolomics Data Analysis
2.6.4. Proteomics Data Analysis
3. Results
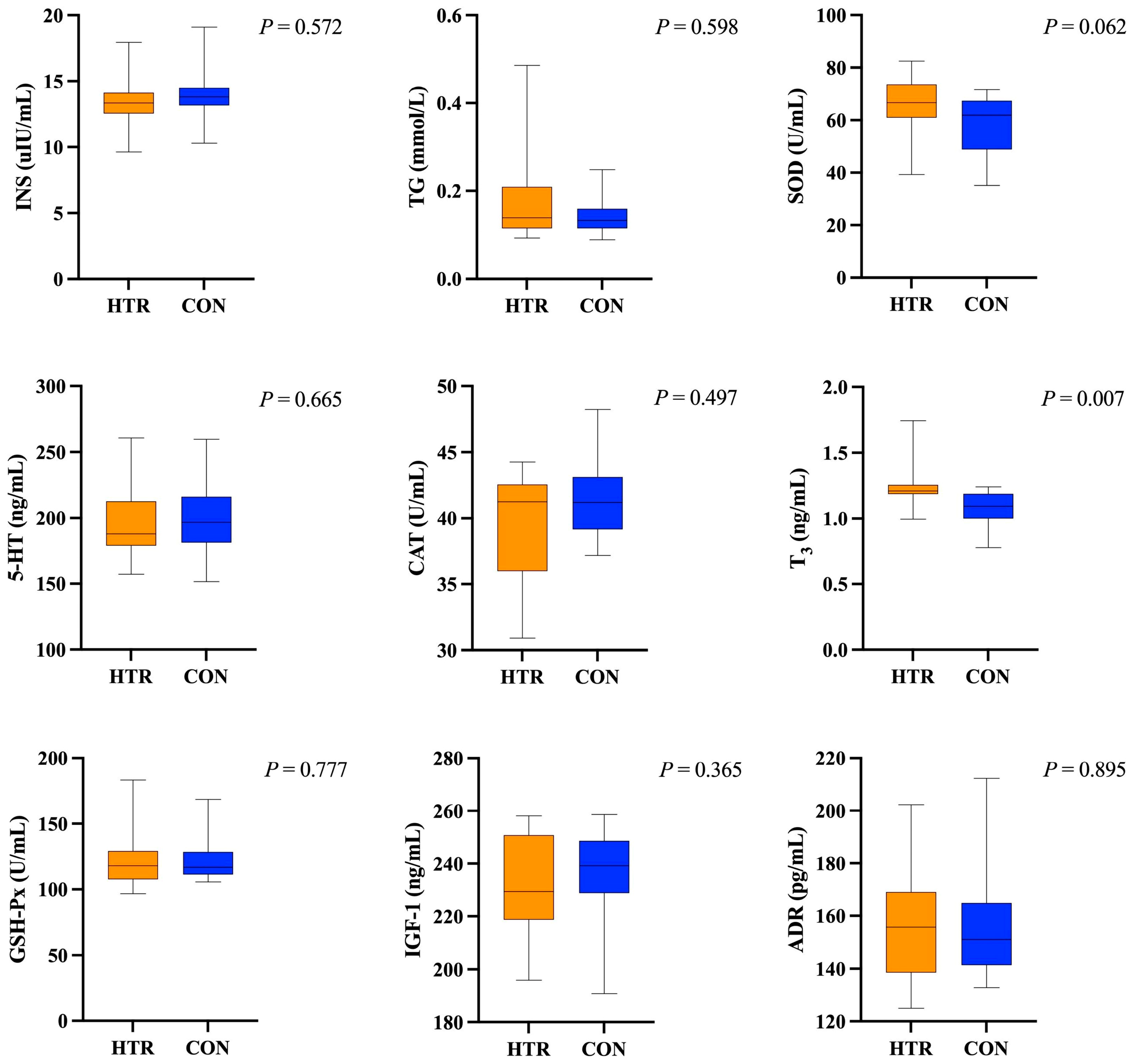
3.1. Comparative Analysis of Plasma and Milk Biochemical Indicators in HTR and CON Cows
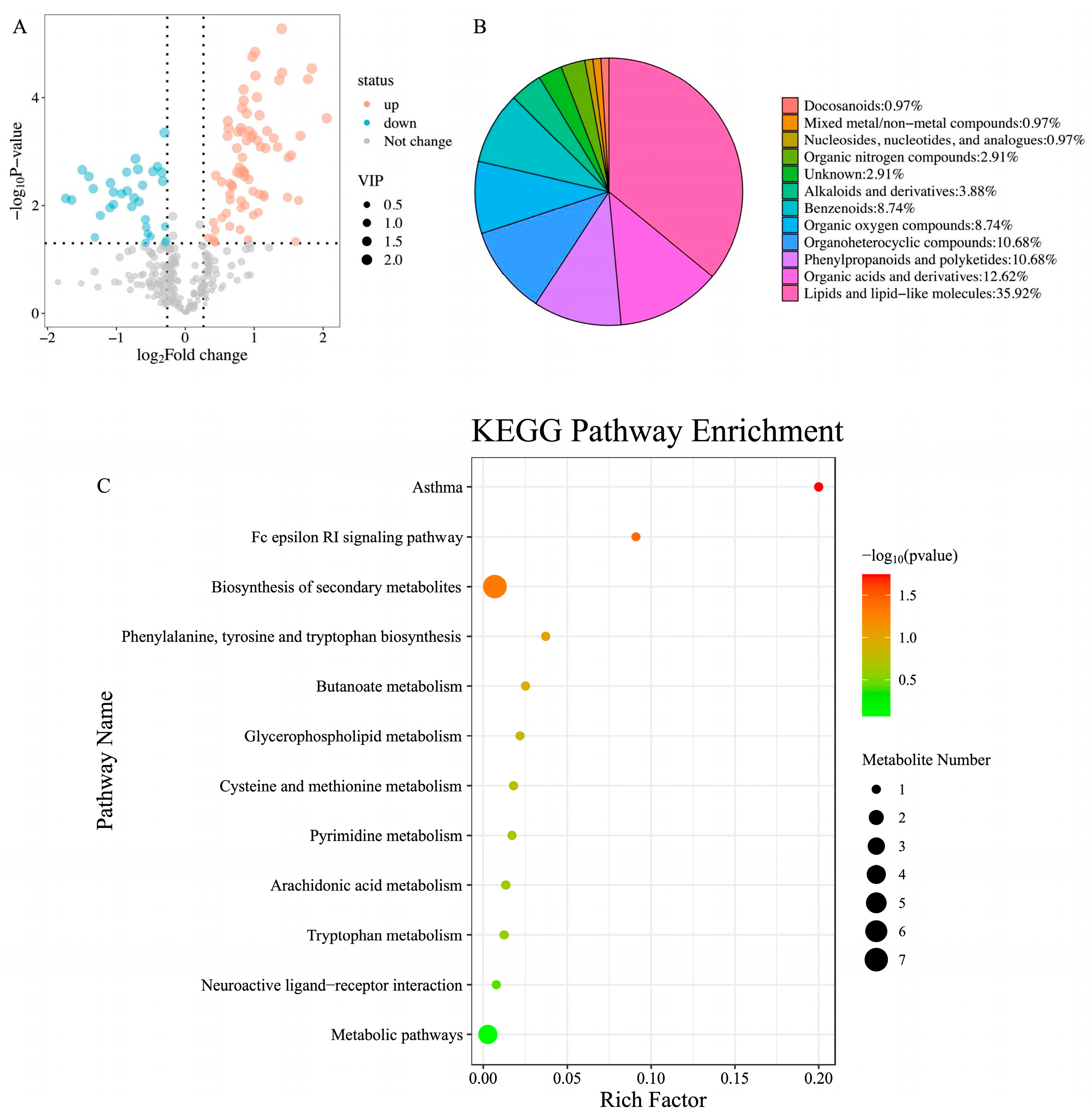
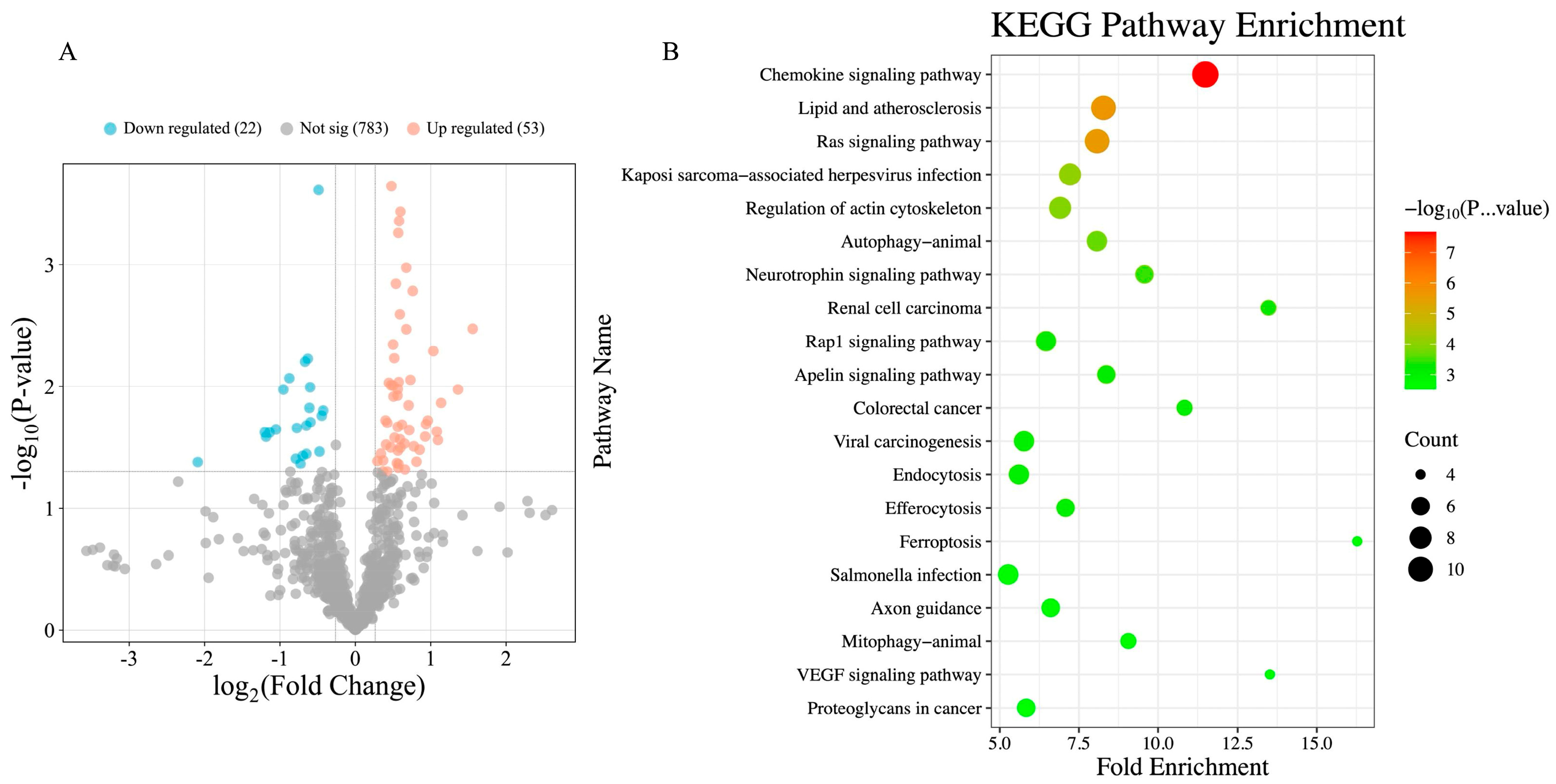
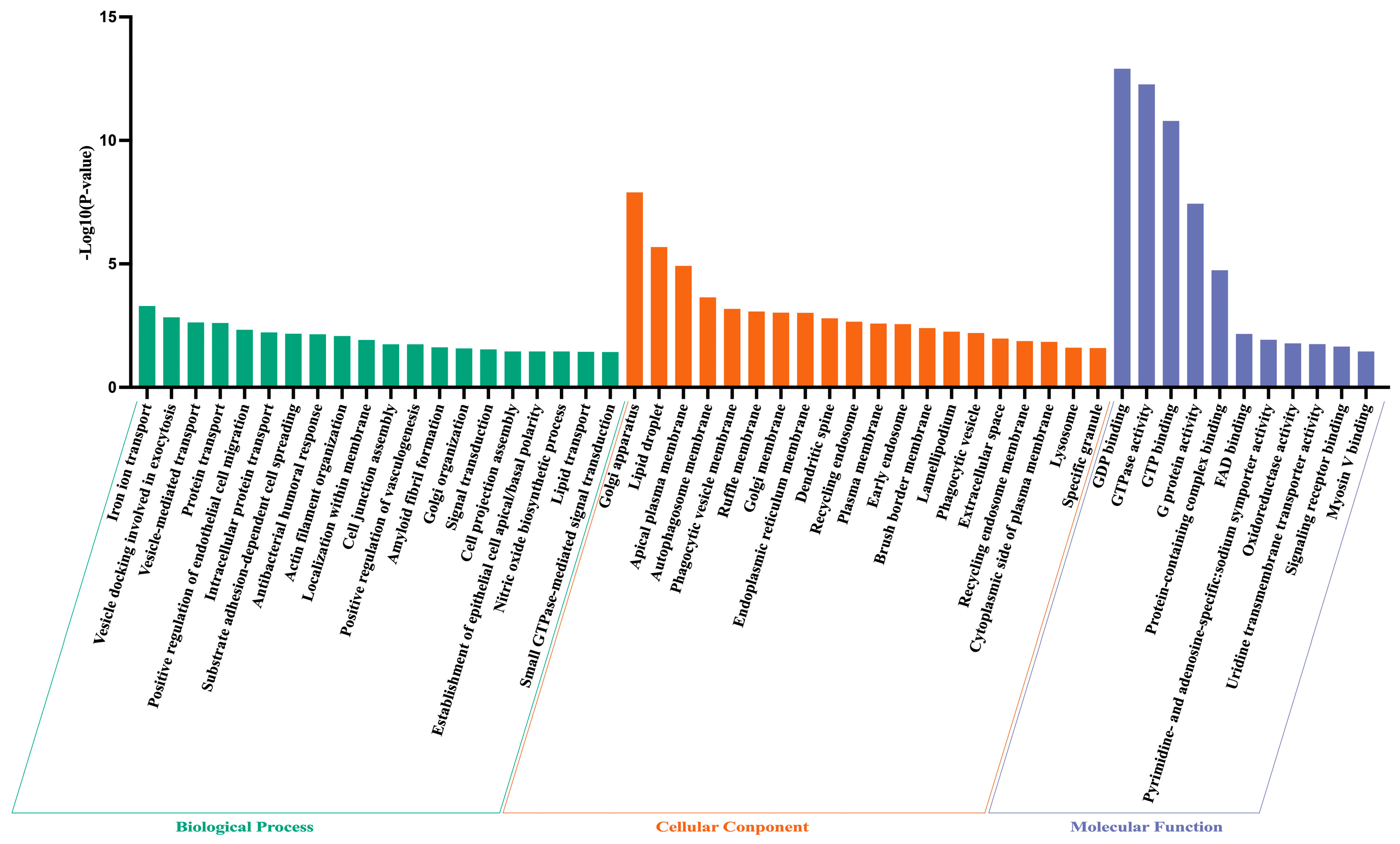
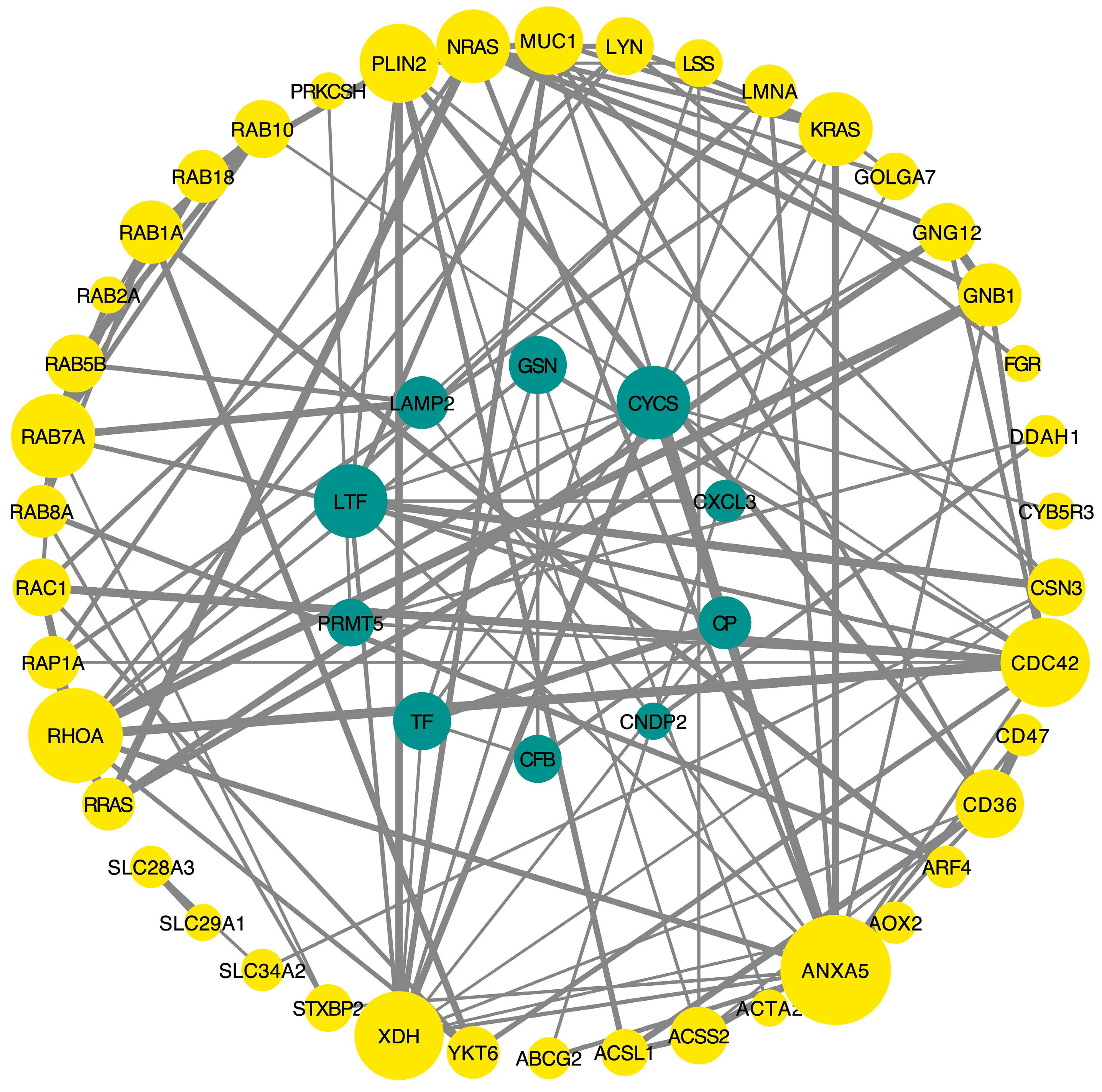
3.2. Metabolomics and Proteomics Profiling of HTR and CON Cows
4. Discussion
4.1. Enhanced Metabolic Activity in HTR Cows: Evidence from Plasma Biochemical Indicators and Milk Proteomics
4.1.1. Variations in Plasma Biochemical Indicators
4.1.2. Upregulation of GTP-Binding Protein Family Members Indicates Enhanced Cellular Activity in HTR Cows
4.2. Reduced Milk Quality in HTR Cows: Evidence from Milk Biochemical Indicators and Proteomics
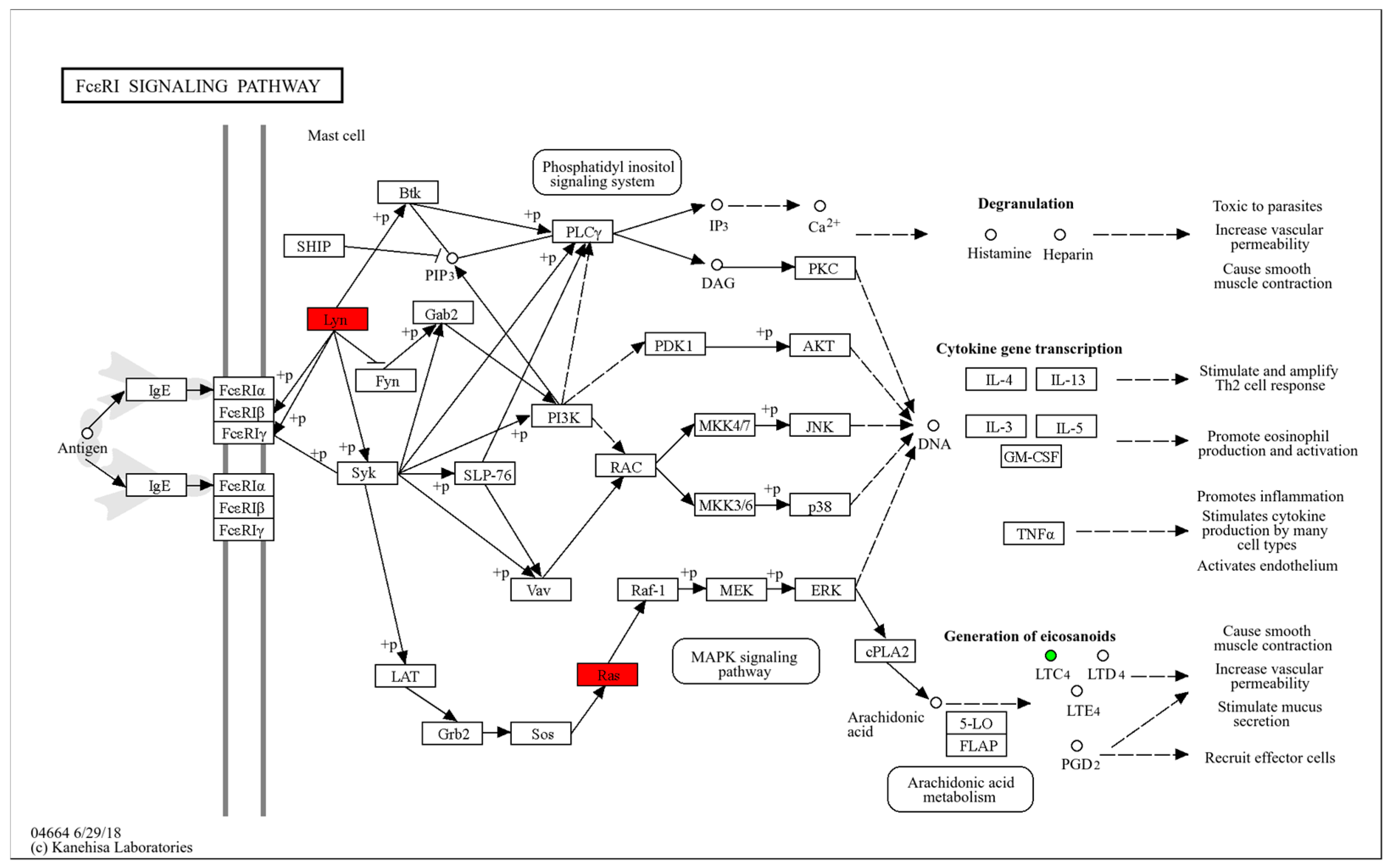
4.3. Metabolomics Reveals Impaired Immune Function in HTR Cows
4.4. Metabolomics and Proteomics Correlation Analysis
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
Abbreviations
| 5-HT | 5-hydroxytryptamine |
| ADR | Adrenaline |
| ALB | Albumin |
| BCS | Body condition score |
| BP | Biological processes |
| CAT | Catalase |
| CC | Cellular components |
| CON | Control |
| cUMP | 2′,3′-cyclic UMP |
| DA | Dopamine |
| DEP | Differentially expressed proteins |
| DHI | Dairy herd improvement |
| DIA | Data-independent acquisition |
| DIM | Days in milk |
| ELISA | Enzyme-linked immunosorbent assay |
| FC | Fold change |
| FFA | Free fatty acids |
| GLU | Glucose |
| GO | Gene ontology |
| GSH-Px | Glutathione peroxidase |
| HDL | High-density lipoprotein |
| HPT | Hypothalamic–pituitary–thyroid |
| HTR | High-frequency tongue-rolling |
| IgE | Immunoglobulin E |
| IGF-1 | Insulin-like growth factor-1 |
| INS | Insulin |
| KEGG | Kyoto encyclopedia of genes and genomes |
| LC-MS | Liquid chromatography–mass spectrometry |
| LC-NE | Locus coeruleus–norepinephrine axis |
| LDH | Lactate dehydrogenase |
| LDL | Low-density lipoprotein |
| LF | Lactoferrin |
| LTC4 | Leukotriene C4 |
| MF | Molecular functions |
| NNOB | Non-nutritive oral behavior |
| NOR | Norepinephrine |
| OPLS-DA | Orthogonal partial least squares–discriminant analysis |
| PABAK | Prevalence-adjusted bias-adjusted kappa |
| PCA | Principal components analysis |
| PPI | Protein–protein interactions |
| QC | Quality control |
| RH | Relative humidity |
| RIA | Radioimmunoassay |
| SOD | Superoxide dismutase |
| T3 | Triiodothyronine |
| T4 | Thyroxine |
| Ta | Ambient temperature |
| T-AOC | Total antioxidant capacity |
| TC | Total cholesterol |
| TG | Triglycerides |
| THI | Temperature–humidity index |
| TMR | Total mixed ration |
| TP | Total protein |
| TR | Tongue-rolling |
| UREA | Urea nitrogen |
| VIP | Variable importance in the projection |
| α-LA | α-lactalbumin |
| αs1-CN | αs1-casein |
| αs2-CN | αs2-casein |
| β-CN | β-casein |
| β-HB | β-hydroxybutyric acid |
| β-LG | β-lactoglobulin |
| κ-LG | κ-casein |
Appendix A
| Items | Value |
|---|---|
| Ingredients | Content, % |
| Alfalfa | 10.39 |
| Oat hay | 2.42 |
| Dandelion | 0.48 |
| Whole corn silage | 48.33 |
| Cottonseed | 2.90 |
| Beet pulp | 2.42 |
| Ground corn | 7.49 |
| Pressed corn | 9.42 |
| Soybean meal | 8.70 |
| Rapeseed meal | 1.69 |
| DDGS 1 | 0.72 |
| Extruded soybean | 1.33 |
| Mineral and vitamin mix 2 | 3.70 |
| Nutrient composition | |
| DM, % of wet TMR | 62.40 |
| CP | 17.06 |
| EE | 3.32 |
| NDF | 35.75 |
| ADF | 18.20 |
| NEL/(MJ/kg) | 6.11 |
| Pathway Name | p-Value | Fold Enrichment | Z-Score |
|---|---|---|---|
| Chemokine signaling pathway | <0.001 | 11.487 | 2.714 |
| Lipid and atherosclerosis | <0.001 | 8.276 | 1.897 |
| Ras signaling pathway | <0.001 | 8.075 | 3.162 |
| Kaposi sarcoma-associated herpesvirus infection | <0.001 | 7.220 | 1.414 |
| Regulation of the actin cytoskeleton | <0.001 | 6.907 | 2.121 |
| Autophagy–animal | <0.001 | 8.070 | 1.890 |
| Neurotrophin signaling pathway | <0.001 | 9.574 | 2.449 |
| Renal cell carcinoma | <0.001 | 13.476 | 2.236 |
| Rap1 signaling pathway | <0.001 | 6.464 | 2.646 |
| Apelin signaling pathway | <0.001 | 8.368 | 2.449 |
| Colorectal cancer | 0.001 | 10.840 | 2.236 |
| Viral carcinogenesis | 0.001 | 5.769 | 1.890 |
| Endocytosis | 0.001 | 5.607 | 2.646 |
| Efferocytosis | 0.001 | 7.081 | 2.449 |
| Ferroptosis | 0.002 | 16.282 | −1.000 |
| Salmonella infection | 0.002 | 5.268 | 1.890 |
| Axon guidance | 0.002 | 6.612 | 2.449 |
| Mitophagy–animal | 0.002 | 9.066 | 2.236 |
| VEGF signaling pathway | 0.003 | 13.522 | 2.000 |
| Proteoglycans in cancer | 0.003 | 5.838 | 2.449 |
| MAPK signaling pathway | 0.003 | 4.654 | 2.646 |
| Relaxin signaling pathway | 0.004 | 7.555 | 2.236 |
| Fc epsilon RI signaling pathway | 0.005 | 11.237 | 2.000 |
| Phagosome | 0.009 | 6.007 | 1.342 |
| Adherens junction | 0.011 | 8.578 | 2.000 |
| B cell receptor signaling pathway | 0.011 | 8.487 | 2.000 |
| Tight junction | 0.012 | 5.540 | 2.236 |
| Fc gamma R-mediated phagocytosis | 0.012 | 8.141 | 1.000 |
| AGE-RAGE signaling pathway in diabetic complications | 0.014 | 7.822 | 2.000 |
| Pancreatic secretion | 0.014 | 7.746 | 2.000 |
| C-type lectin receptor signaling pathway | 0.015 | 7.526 | 2.000 |
| Pathways in cancer | 0.018 | 2.870 | 2.121 |
| Cholinergic synapse | 0.019 | 6.937 | 2.000 |
| Tuberculosis | 0.019 | 4.749 | 1.342 |
| Leukocyte transendothelial migration | 0.020 | 6.761 | 2.000 |
| Serotonergic synapse | 0.022 | 6.539 | 2.000 |
| Sphingolipid signaling pathway | 0.023 | 6.382 | 2.000 |
| AMPK signaling pathway | 0.024 | 6.332 | 2.000 |
| T cell receptor signaling pathway | 0.025 | 6.233 | 2.000 |
| Alcoholism | 0.027 | 4.280 | 2.236 |
| Apoptosis | 0.035 | 5.427 | 1.000 |
| Mineral absorption | 0.037 | 9.651 | −0.577 |
| Long-term depression | 0.038 | 9.498 | 1.732 |
| Phospholipase D signaling pathway | 0.040 | 5.180 | 2.000 |
| PI3K-Akt signaling pathway | 0.042 | 3.053 | 1.633 |
| Long-term potentiation | 0.046 | 8.548 | 1.732 |
| GO Type | GO Terms | p-Value | Fold Enrichment |
|---|---|---|---|
| BP | Iron ion transport | <0.001 | 25.228 |
| BP | Vesicle docking involved in exocytosis | 0.001 | 51.785 |
| BP | Vesicle-mediated transport | 0.002 | 8.816 |
| BP | Protein transport | 0.002 | 6.287 |
| BP | Positive regulation of endothelial cell migration | 0.005 | 28.938 |
| BP | Intracellular protein transport | 0.006 | 6.804 |
| BP | Substrate adhesion-dependent cell spreading | 0.007 | 23.998 |
| BP | Antibacterial humoral response | 0.007 | 23.426 |
| BP | Actin filament organization | 0.008 | 9.506 |
| BP | Localization within membrane | 0.012 | 163.984 |
| BP | Cell junction assembly | 0.018 | 109.323 |
| BP | Positive regulation of vasculogenesis | 0.018 | 109.323 |
| BP | Amyloid fibril formation | 0.024 | 81.992 |
| BP | Golgi organization | 0.026 | 11.713 |
| BP | Signal transduction | 0.029 | 3.422 |
| BP | Cell projection assembly | 0.035 | 54.661 |
| BP | Establishment of epithelial cell apical/basal polarity | 0.035 | 54.661 |
| BP | Nitric oxide biosynthetic process | 0.035 | 54.661 |
| BP | Lipid transport | 0.036 | 9.839 |
| BP | Small GTPase-mediated signal transduction | 0.037 | 9.742 |
| BP | Regulation of cell shape | 0.038 | 9.552 |
| BP | Rac protein signal transduction | 0.044 | 43.729 |
| BP | Protein localization to plasma membrane | 0.048 | 8.482 |
| BP | Protein stabilization | 0.048 | 8.409 |
| CC | Golgi apparatus | <0.001 | 7.169 |
| CC | Lipid droplet | <0.001 | 28.476 |
| CC | Apical plasma membrane | <0.001 | 10.200 |
| CC | Autophagosome membrane | <0.001 | 33.338 |
| CC | Phagocytic vesicle membrane | <0.001 | 23.167 |
| CC | Ruffle membrane | <0.001 | 21.357 |
| CC | Golgi membrane | <0.001 | 6.087 |
| CC | Endoplasmic reticulum membrane | <0.001 | 4.344 |
| CC | Dendritic spine | 0.002 | 17.086 |
| CC | Recycling endosome | 0.002 | 15.358 |
| CC | Plasma membrane | 0.003 | 1.669 |
| CC | Early endosome | 0.003 | 8.458 |
| CC | Brush border membrane | 0.004 | 31.065 |
| CC | Lamellipodium | 0.006 | 11.023 |
| CC | Phagocytic vesicle | 0.006 | 25.004 |
| CC | Extracellular space | 0.011 | 2.494 |
| CC | Recycling endosome membrane | 0.013 | 16.806 |
| CC | Cytoplasmic side of plasma membrane | 0.015 | 16.018 |
| CC | Lysosome | 0.025 | 6.328 |
| CC | Specific granule | 0.026 | 75.937 |
| CC | Extracellular exosome | 0.028 | 11.265 |
| CC | Cytoskeleton | 0.029 | 4.250 |
| CC | Late endosome | 0.034 | 10.150 |
| CC | Membrane raft | 0.042 | 9.072 |
| CC | Midbody | 0.045 | 8.762 |
| CC | Late endosome membrane | 0.045 | 8.762 |
| MF | GDP binding | <0.001 | 55.349 |
| MF | GTPase activity | <0.001 | 15.327 |
| MF | GTP binding | <0.001 | 11.889 |
| MF | G protein activity | <0.001 | 62.268 |
| MF | Protein-containing complex binding | <0.001 | 18.280 |
| MF | FAD binding | 0.007 | 23.721 |
| MF | Pyrimidine- and adenosine-specific: sodium symporter activity | 0.012 | 166.048 |
| MF | Oxidoreductase activity | 0.017 | 7.339 |
| MF | Uridine transmembrane transporter activity | 0.018 | 110.698 |
| MF | Signaling receptor binding | 0.022 | 6.576 |
| MF | Myosin V binding | 0.035 | 55.349 |
References
- Novak, J.; Bailoo, J.D.; Melotti, L.; Rommen, J.; Würbel, H. An Exploration Based Cognitive Bias Test for Mice: Effects of Handling Method and Stereotypic Behaviour. PLoS ONE 2015, 10, e0130718. [Google Scholar] [CrossRef]
- Ahola, M.K.; Vapalahti, K.; Lohi, H. Early Weaning Increases Aggression and Stereotypic Behaviour in Cats. Sci. Rep. 2017, 7, 10412. [Google Scholar] [CrossRef]
- Ridge, E.E.; Foster, M.J.; Daigle, C.L. Effect of Diet on Non-Nutritive Oral Behavior Performance in Cattle: A Systematic Review. Livest. Sci. 2020, 238, 104063. [Google Scholar] [CrossRef]
- Li, C.; Gu, X. Oral Stereotypic Behaviors in Farm Animals and Their Causes. Anim. Res. One Health 2024, 2, 337–351. [Google Scholar] [CrossRef]
- Kelly, K.R.; Harrison, M.L.; Size, D.D.; MacDonald, S.E. Individual Effects of Seasonal Changes, Visitor Density, and Concurrent Bear Behavior on Stereotypical Behaviors in Captive Polar Bears (Ursus Maritimus). J. Appl. Anim. Welf. Sci. 2015, 18, 17–31. [Google Scholar] [CrossRef] [PubMed]
- Boddicker, N.; Gabler, N.K.; Spurlock, M.E.; Nettleton, D.; Dekkers, J.C.M. Effects of Ad Libitum and Restricted Feed Intake on Growth Performance and Body Composition of Yorkshire Pigs Selected for Reduced Residual Feed Intake. J. Anim. Sci. 2011, 89, 40–51. [Google Scholar] [CrossRef] [PubMed]
- Riber, A.B.; Tahamtani, F.M.; Steenfeldt, S. Effects of Qualitative Feed Restriction in Broiler Breeder Pullets on Behaviour in the Home Environment. Appl. Anim. Behav. Sci. 2021, 235, 105225. [Google Scholar] [CrossRef]
- Li, C.; Chen, X.; Fang, T.; Gu, X. Do Cows with Stereotypic Tongue-Rolling Behaviour Cope Better with Their Environment? Front. Vet. Sci. 2024, 11, 1404539. [Google Scholar] [CrossRef]
- Redbo, I. Relations between Oral Stereotypies, Open-Field Behavior, and Pituitary–Adrenal System in Growing Dairy Cattle. Physiol. Behav. 1998, 64, 273–278. [Google Scholar] [CrossRef]
- Robbins, J.A.; McCandless, K.; Weary, D.M.; Paros, M. Breed, Parity, and Days in Milk Affect Risk of Tongue Rolling in Dairy Cows. JDS Commun. 2023, 4, 210–213. [Google Scholar] [CrossRef]
- Redbo, I.; Emanuelson, M.; Lundberg, K.; Oredsson, N. Feeding Level and Oral Stereotypies in Dairy Cows. Anim. Sci. 1996, 62, 199–206. [Google Scholar] [CrossRef]
- Redbo, I.; Nordblad, A. Stereotypies in Heifers Are Affected by Feeding Regime. Appl. Anim. Behav. Sci. 1997, 53, 193–202. [Google Scholar] [CrossRef]
- Lindström, T.; Redbo, I. Effect of Feeding Duration and Rumen Fill on Behaviour in Dairy Cows. Appl. Anim. Behav. Sci. 2000, 70, 83–97. [Google Scholar] [CrossRef]
- Downey, B.C.; Tucker, C.B. Early Life Access to Hay Does Not Affect Later Life Oral Behavior in Feed-Restricted Heifers. J. Dairy. Sci. 2023, 106, 5672–5686. [Google Scholar] [CrossRef]
- Horvath, K.C.; Miller-Cushon, E.K. Evaluating Effects of Providing Hay on Behavioral Development and Performance of Group-Housed Dairy Calves. J. Dairy Sci. 2019, 102, 10411–10422. [Google Scholar] [CrossRef]
- Sun, F.; Zhao, Q.; Chen, X.; Zhao, G.; Gu, X. Physiological Indicators and Production Performance of Dairy Cows With Tongue Rolling Stereotyped Behavior. Front. Vet. Sci. 2022, 9, 840726. [Google Scholar] [CrossRef]
- Mahmoud, M.M.; Saeed, H.E.; Abdel-Hamied, E. Clinical, Hemato-Biochemical and Oxidative Stress Biomarkers Alterations in Dairy Heifers with Tongue-Rolling. Egypt. J. Vet. Sci. 2024, 1–8. [Google Scholar] [CrossRef]
- Welch, J.G.; Smith, A.M. Forage Quality and Rumination Time in Cattle. J. Dairy Sci. 1970, 53, 797–800. [Google Scholar] [CrossRef]
- Castells, L.; Bach, A.; Araujo, G.; Montoro, C.; Terré, M. Effect of Different Forage Sources on Performance and Feeding Behavior of Holstein Calves. J. Dairy Sci. 2012, 95, 286–293. [Google Scholar] [CrossRef]
- Devant, M.; Penner, G.B.; Marti, S.; Quintana, B.; Fábregas, F.; Bach, A.; Arís, A. Behavior and Inflammation of the Rumen and Cecum in Holstein Bulls Fed High-Concentrate Diets with Different Concentrate Presentation Forms with or without Straw Supplementation1. J. Anim. Sci. 2016, 94, 3902–3917. [Google Scholar] [CrossRef]
- Horvath, K.C.; Miller-Cushon, E.K. The Effect of Milk-Feeding Method and Hay Provision on the Development of Feeding Behavior and Non-Nutritive Oral Behavior of Dairy Calves. J. Dairy Sci. 2017, 100, 3949–3957. [Google Scholar] [CrossRef]
- Mirzaei, M.; Khorvash, M.; Ghorbani, G.R.; Kazemi-Bonchenari, M.; Ghaffari, M.H. Growth Performance, Feeding Behavior, and Selected Blood Metabolites of Holstein Dairy Calves Fed Restricted Amounts of Milk: No Interactions between Sources of Finely Ground Grain and Forage Provision. J. Dairy Sci. 2017, 100, 1086–1094. [Google Scholar] [CrossRef]
- Fabrile, M.P.; Ghidini, S.; Conter, M.; Varrà, M.O.; Ianieri, A.; Zanardi, E. Filling Gaps in Animal Welfare Assessment through Metabolomics. Front. Vet. Sci. 2023, 10, 1129741. [Google Scholar] [CrossRef]
- Liu, B.; Zhao, R.; Wu, T.; Ma, Y.; Gao, Y.; Wu, Y.; Hao, B.; Yin, J.; Li, Y. Transcriptomes Reveal microRNAs and mRNAs in Different Photoperiods Influencing Cashmere Growth in Goat. PLoS ONE 2023, 18, e0282772. [Google Scholar] [CrossRef]
- Zeng, M.; Wang, X.; Qiu, Y.; Sun, X.; Qiu, H.; Ma, X.; Lv, Q.; Gao, J.; Wang, C.; Chang, Q. Metabolomic and Systematic Biochemical Analysis of Sheep Infected with Fasciola Hepatica. Vet. Parasitol. 2023, 313, 109852. [Google Scholar] [CrossRef]
- Brinker, T.; Bijma, P.; Vereijken, A.; Ellen, E.D. The Genetic Architecture of Socially-Affected Traits: A GWAS for Direct and Indirect Genetic Effects on Survival Time in Laying Hens Showing Cannibalism. Genet. Sel. Evol. 2018, 50, 38. [Google Scholar] [CrossRef]
- Wilson, K.; Zanella, R.; Ventura, C.; Johansen, H.L.; Framstad, T.; Janczak, A.; Zanella, A.J.; Neibergs, H.L. Identification of Chromosomal Locations Associated with Tail Biting and Being a Victim of Tail-Biting Behaviour in the Domestic Pig (Sus Scrofa Domesticus). J. Appl. Genet. 2012, 53, 449–456. [Google Scholar] [CrossRef]
- Lehner, P.N. Sampling Methods in Behavior Research. Poult. Sci. 1992, 71, 643–649. [Google Scholar] [CrossRef]
- Wiśniewski, J.R. Quantitative Evaluation of Filter Aided Sample Preparation (FASP) and Multienzyme Digestion FASP Protocols. Anal. Chem. 2016, 88, 5438–5443. [Google Scholar] [CrossRef]
- Pang, Z.; Lu, Y.; Zhou, G.; Hui, F.; Xu, L.; Viau, C.; Spigelman, A.F.; MacDonald, P.E.; Wishart, D.S.; Li, S.; et al. MetaboAnalyst 6.0: Towards a Unified Platform for Metabolomics Data Processing, Analysis and Interpretation. Nucleic Acids Res. 2024, 52, W398–W406. [Google Scholar] [CrossRef]
- López-Ibáñez, J.; Pazos, F.; Chagoyen, M. MBROLE 2.0-Functional Enrichment of Chemical Compounds. Nucleic Acids Res. 2016, 44, W201–W204. [Google Scholar] [CrossRef]
- Tang, D.; Chen, M.; Huang, X.; Zhang, G.; Zeng, L.; Zhang, G.; Wu, S.; Wang, Y. SRplot: A Free Online Platform for Data Visualization and Graphing. PLoS ONE 2023, 18, e0294236. [Google Scholar] [CrossRef]
- Lyu, F.; Han, F.; Ge, C.; Mao, W.; Chen, L.; Hu, H.; Chen, G.; Lang, Q.; Fang, C. OmicStudio: A Composable Bioinformatics Cloud Platform with Real-Time Feedback That Can Generate High-Quality Graphs for Publication. Imeta 2023, 2, e85. [Google Scholar] [CrossRef]
- Sherman, B.T.; Hao, M.; Qiu, J.; Jiao, X.; Baseler, M.W.; Lane, H.C.; Imamichi, T.; Chang, W. DAVID: A Web Server for Functional Enrichment Analysis and Functional Annotation of Gene Lists (2021 Update). Nucleic Acids Res. 2022, 50, W216–W221. [Google Scholar] [CrossRef]
- Szklarczyk, D.; Kirsch, R.; Koutrouli, M.; Nastou, K.; Mehryary, F.; Hachilif, R.; Gable, A.L.; Fang, T.; Doncheva, N.T.; Pyysalo, S.; et al. The STRING Database in 2023: Protein-Protein Association Networks and Functional Enrichment Analyses for Any Sequenced Genome of Interest. Nucleic Acids Res. 2023, 51, D638–D646. [Google Scholar] [CrossRef]
- Busato, A.; Faissler, D.; Küpfer, U.; Blum, J.W. Body Condition Scores in Dairy Cows: Associations with Metabolic and Endocrine Changes in Healthy Dairy Cows. J. Vet. Med. Ser. A 2002, 49, 455–460. [Google Scholar] [CrossRef]
- Rowntree, J.E.; Hill, G.M.; Hawkins, D.R.; Link, J.E.; Rincker, M.J.; Bednar, G.W.; Kreft, R.A. Effect of Se on Selenoprotein Activity and Thyroid Hormone Metabolism in Beef and Dairy Cows and Calves. J. Anim. Sci. 2004, 82, 2995–3005. [Google Scholar] [CrossRef]
- Mendoza, A.; Hollenberg, A.N. New Insights into Thyroid Hormone Action. Pharmacol. Ther. 2017, 173, 135–145. [Google Scholar] [CrossRef]
- Wennerberg, K.; Rossman, K.L.; Der, C.J. The Ras Superfamily at a Glance. J. Cell Sci. 2005, 118, 843–846. [Google Scholar] [CrossRef]
- Eckenstaler, R.; Hauke, M.; Benndorf, R.A. A Current Overview of RhoA, RhoB, and RhoC Functions in Vascular Biology and Pathology. Biochem. Pharmacol. 2022, 206, 115321. [Google Scholar] [CrossRef]
- Langemeyer, L.; Fröhlich, F.; Ungermann, C. Rab GTPase Function in Endosome and Lysosome Biogenesis. Trends Cell Biol. 2018, 28, 957–970. [Google Scholar] [CrossRef]
- Hatoyama, Y.; Homma, Y.; Hiragi, S.; Fukuda, M. Establishment and Analysis of Conditional Rab1- and Rab5-Knockout Cells Using the Auxin-Inducible Degron System. J. Cell Sci. 2021, 134, jcs259184. [Google Scholar] [CrossRef] [PubMed]
- Ding, X.; Jiang, X.; Tian, R.; Zhao, P.; Li, L.; Wang, X.; Chen, S.; Zhu, Y.; Mei, M.; Bao, S.; et al. RAB2 Regulates the Formation of Autophagosome and Autolysosome in Mammalian Cells. Autophagy 2019, 15, 1774–1786. [Google Scholar] [CrossRef]
- Kuchitsu, Y.; Fukuda, M. Revisiting Rab7 Functions in Mammalian Autophagy: Rab7 Knockout Studies. Cells 2018, 7, 215. [Google Scholar] [CrossRef]
- Kern, A.; Dikic, I.; Behl, C. The Integration of Autophagy and Cellular Trafficking Pathways via RAB GAPs. Autophagy 2015, 11, 2393–2397. [Google Scholar] [CrossRef] [PubMed]
- English, A.R.; Voeltz, G.K. Rab10 GTPase Regulates ER Dynamics and Morphology. Nat. Cell Biol. 2013, 15, 169–178. [Google Scholar] [CrossRef]
- Salloum, S.; Wang, H.; Ferguson, C.; Parton, R.G.; Tai, A.W. Rab18 Binds to Hepatitis C Virus NS5A and Promotes Interaction between Sites of Viral Replication and Lipid Droplets. PLoS Pathog. 2013, 9, e1003513. [Google Scholar] [CrossRef] [PubMed]
- Xu, D.; Li, Y.; Wu, L.; Li, Y.; Zhao, D.; Yu, J.; Huang, T.; Ferguson, C.; Parton, R.G.; Yang, H.; et al. Rab18 Promotes Lipid Droplet (LD) Growth by Tethering the ER to LDs through SNARE and NRZ Interactions. J. Cell Biol. 2018, 217, 975–995. [Google Scholar] [CrossRef]
- Jansen, S.; Gosens, R.; Wieland, T.; Schmidt, M. Paving the Rho in Cancer Metastasis: Rho GTPases and Beyond. Pharmacol. Ther. 2018, 183, 1–21. [Google Scholar] [CrossRef]
- Llavero, F.; Arrazola Sastre, A.; Luque Montoro, M.; Martín, M.A.; Arenas, J.; Lucia, A.; Zugaza, J.L. Small GTPases of the Ras Superfamily and Glycogen Phosphorylase Regulation in T Cells. Small GTPases 2021, 12, 106–113. [Google Scholar] [CrossRef]
- Samovski, D.; Sun, J.; Pietka, T.; Gross, R.W.; Eckel, R.H.; Su, X.; Stahl, P.D.; Abumrad, N.A. Regulation of AMPK Activation by CD36 Links Fatty Acid Uptake to β-Oxidation. Diabetes 2015, 64, 353–359. [Google Scholar] [CrossRef]
- Zhao, L.; Zhang, C.; Luo, X.; Wang, P.; Zhou, W.; Zhong, S.; Xie, Y.; Jiang, Y.; Yang, P.; Tang, R.; et al. CD36 Palmitoylation Disrupts Free Fatty Acid Metabolism and Promotes Tissue Inflammation in Non-Alcoholic Steatohepatitis. J. Hepatol. 2018, 69, 705–717. [Google Scholar] [CrossRef]
- Li, Y.; Huang, X.; Yang, G.; Xu, K.; Yin, Y.; Brecchia, G.; Yin, J. CD36 Favours Fat Sensing and Transport to Govern Lipid Metabolism. Prog. Lipid Res. 2022, 88, 101193. [Google Scholar] [CrossRef]
- Logtenberg, M.E.W.; Scheeren, F.A.; Schumacher, T.N. The CD47-SIRPα Immune Checkpoint. Immunity 2020, 52, 742–752. [Google Scholar] [CrossRef] [PubMed]
- Tanase, C.; Enciu, A.M.; Codrici, E.; Popescu, I.D.; Dudau, M.; Dobri, A.M.; Pop, S.; Mihai, S.; Gheorghișan-Gălățeanu, A.-A.; Hinescu, M.E. Fatty Acids, CD36, Thrombospondin-1, and CD47 in Glioblastoma: Together and/or Separately? Int. J. Mol. Sci. 2022, 23, 604. [Google Scholar] [CrossRef]
- Zhao, L.L.; Wang, X.L.; Tian, Q.; Mao, X.Y. Effect of Casein to Whey Protein Ratios on the Protein Interactions and Coagulation Properties of Low-Fat Yogurt. J. Dairy Sci. 2016, 99, 7768–7775. [Google Scholar] [CrossRef] [PubMed]
- Runthala, A.; Mbye, M.; Ayyash, M.; Xu, Y.; Kamal-Eldin, A. Caseins: Versatility of Their Micellar Organization in Relation to the Functional and Nutritional Properties of Milk. Molecules 2023, 28, 2023. [Google Scholar] [CrossRef]
- Roy, D.; Ye, A.; Moughan, P.J.; Singh, H. Composition, Structure, and Digestive Dynamics of Milk From Different Species—A Review. Front. Nutr. 2020, 7, 577759. [Google Scholar] [CrossRef]
- De Kruif, C.G.; Huppertz, T.; Urban, V.S.; Petukhov, A.V. Casein Micelles and Their Internal Structure. Adv. Colloid Interface Sci. 2012, 171–172, 36–52. [Google Scholar] [CrossRef] [PubMed]
- Dalgleish, D.G.; Corredig, M. The Structure of the Casein Micelle of Milk and Its Changes During Processing. Annu. Rev. Food Sci. Technol. 2012, 3, 449–467. [Google Scholar] [CrossRef]
- De Kruif, C.G.; Zhulina, E.B. κ-Casein as a Polyelectrolyte Brush on the Surface of Casein Micelles. Colloids Surf. A Physicochem. Eng. Asp. 1996, 117, 151–159. [Google Scholar] [CrossRef]
- Ragland, S.A.; Criss, A.K. From Bacterial Killing to Immune Modulation: Recent Insights into the Functions of Lysozyme. PLoS Pathog. 2017, 13, e1006512. [Google Scholar] [CrossRef]
- Bonaccorsi Di Patti, M.C.; Cutone, A.; Polticelli, F.; Rosa, L.; Lepanto, M.S.; Valenti, P.; Musci, G. The Ferroportin-Ceruloplasmin System and the Mammalian Iron Homeostasis Machine: Regulatory Pathways and the Role of Lactoferrin. Biometals 2018, 31, 399–414. [Google Scholar] [CrossRef]
- Cutone, A.; Lepanto, M.S.; Rosa, L.; Scotti, M.J.; Rossi, A.; Ranucci, S.; De Fino, I.; Bragonzi, A.; Valenti, P.; Musci, G.; et al. Aerosolized Bovine Lactoferrin Counteracts Infection, Inflammation and Iron Dysbalance in A Cystic Fibrosis Mouse Model of Pseudomonas Aeruginosa Chronic Lung Infection. Int. J. Mol. Sci. 2019, 20, 2128. [Google Scholar] [CrossRef]
- Vega-Bautista, A.; de la Garza, M.; Carrero, J.C.; Campos-Rodríguez, R.; Godínez-Victoria, M.; Drago-Serrano, M.E. The Impact of Lactoferrin on the Growth of Intestinal Inhabitant Bacteria. Int. J. Mol. Sci. 2019, 20, 4707. [Google Scholar] [CrossRef]
- Fan, L.L.; Yao, Q.Q.; Wu, H.M.; Wen, F.; Wang, J.Q.; Li, H.Y.; Zheng, N. Protective Effects of Recombinant Lactoferrin with Different Iron Saturations on Enteritis Injury in Young Mice. J. Dairy. Sci. 2022, 105, 4791–4803. [Google Scholar] [CrossRef]
- Bartnikas, T.B. Known and Potential Roles of Transferrin in Iron Biology. Biometals 2012, 25, 677–686. [Google Scholar] [CrossRef]
- Wattez, J.S.; Delmont, A.; Bouvet, M.; Beseme, O.; Goers, S.; Delahaye, F.; Laborie, C.; Lesage, J.; Foligné, B.; Breton, C.; et al. Maternal Perinatal Undernutrition Modifies Lactose and Serotranferrin in Milk: Relevance to the Programming of Metabolic Diseases? Am. J. Physiol. Endocrinol. Metab. 2015, 308, E393–E401. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Bonay, P.; Durán-Chica, I.; Fresno, M.; Alarcón, B.; Alcina, A. Antiparasitic Effects of the Intra-Golgi Transport Inhibitor Megalomicin. Antimicrob. Agents Chemother. 1998, 42, 2668–2673. [Google Scholar] [CrossRef] [PubMed]
- Volchegursky, Y.; Hu, Z.; Katz, L.; McDaniel, R. Biosynthesis of the Anti-Parasitic Agent Megalomicin: Transformation of Erythromycin to Megalomicin in Saccharopolyspora Erythraea. Mol. Microbiol. 2000, 37, 752–762. [Google Scholar] [CrossRef] [PubMed]
- Tal, N.; Morehouse, B.R.; Millman, A.; Stokar-Avihail, A.; Avraham, C.; Fedorenko, T.; Yirmiya, E.; Herbst, E.; Brandis, A.; Mehlman, T.; et al. Cyclic CMP and Cyclic UMP Mediate Bacterial Immunity against Phages. Cell 2021, 184, 5728–5739.e16. [Google Scholar] [CrossRef]
- Harizi, H.; Gualde, N. The Impact of Eicosanoids on the Crosstalk between Innate and Adaptive Immunity: The Key Roles of Dendritic Cells. Tissue Antigens 2005, 65, 507–514. [Google Scholar] [CrossRef] [PubMed]
- Rinaldo-Matthis, A.; Haeggström, J.Z. Structures and Mechanisms of Enzymes in the Leukotriene Cascade. Biochimie 2010, 92, 676–681. [Google Scholar] [CrossRef] [PubMed]
- Turner, H.; Kinet, J.-P. Signalling through the High-Affinity IgE Receptor FcεRI. Nature 1999, 402, 24–30. [Google Scholar] [CrossRef] [PubMed]
- Oettgen, H.C.; Burton, O.T. IgE Receptor Signaling in Food Allergy Pathogenesis. Curr. Opin. Immunol. 2015, 36, 109–114. [Google Scholar] [CrossRef]
- Nagata, Y.; Suzuki, R. FcεRI: A Master Regulator of Mast Cell Functions. Cells 2022, 11, 622. [Google Scholar] [CrossRef]
| Item | HTR 1 | CON 2 | p-Value |
|---|---|---|---|
| DA (pg/mL) | 120.07 ± 11.56 | 121.06 ± 11.16 | 0.809 |
| NOR (pg/mL) | 423.51 ± 53.99 | 429.28 ± 46.63 | 0.746 |
| TC (mmol/L) | 5.24 ± 0.97 | 5.40 ± 1.06 | 0.650 |
| HDL (mmol/L) | 2.99 ± 0.48 | 3.05 ± 0.52 | 0.745 |
| LDL (mmol/L) | 1.35 ± 0.36 | 1.44 ± 0.41 | 0.474 |
| GLU (mmol/L) | 3.10 ± 0.41 | 3.35 ± 0.35 | 0.074 |
| FFA (mmol/L) | 0.28 ± 0.06 | 0.24 ± 0.07 | 0.123 |
| β-HB (mmol/L) | 0.43 ± 0.12 | 0.47 ± 0.12 | 0.344 |
| T4 (ng/mL) | 58.34 ± 3.80 | 58.82 ± 5.39 | 0.773 |
| ALB (g/L) | 31.85 ± 4.29 | 30.37 ± 4.07 | 0.327 |
| TP (g/L) | 68.91 ± 7.74 | 65.98 ± 7.69 | 0.297 |
| UREA (mmol/L) | 4.33 ± 0.59 | 4.61 ± 0.81 | 0.277 |
| LDH (U/L) | 1141.03 ± 223.94 | 1071.17 ± 195.14 | 0.360 |
| T-AOC (U/mL) | 9.12 ± 1.67 | 9.66 ± 1.05 | 0.288 |
| Item | HTR 1 | CON 2 | p-Value |
|---|---|---|---|
| Milk yield (kg/d) | 33.56 ± 9.86 | 31.23 ± 7.94 | 0.449 |
| Milk protein percentage (%) | 3.37 ± 0.15 | 3.71 ± 0.17 | <0.001 |
| Milk fat percentage (%) | 3.80 ± 0.68 | 3.92 ± 0.47 | 0.587 |
| Milk lactose percentage (%) | 5.28 ± 0.90 | 5.23 ± 0.10 | 0.155 |
| Milk true protein percentage (%) | 3.37 ± 0.15 | 3.69 ± 0.17 | <0.001 |
| Milk crude protein percentage (%) | 3.57 ± 0.14 | 3.90 ± 0.17 | <0.001 |
| αs1-CN (μg/mL) | 54.91 ± 1.50 | 56.25 ± 1.50 | 0.016 |
| αs2-CN (μg/mL) | 40.54 ± 1.67 | 41.13 ± 2.07 | 0.381 |
| β-CN (μg/mL) | 99.31 ± 2.42 | 103.73 ± 2.71 | <0.001 |
| κ-CN (μg/mL) | 23.46 ± 0.86 | 24.25 ± 0.91 | 0.020 |
| α-LA (μg/mL) | 26.09 ± 1.05 | 26.44 ± 0.62 | 0.282 |
| β-LG (μg/mL) | 334.06 ± 11.54 | 341.09 ± 9.22 | 0.070 |
| LF (μg/mL) | 789.57 ± 31.37 | 826.24 ± 32.68 | 0.002 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Li, C.; Chen, X.; Fang, T.; Gao, J.; Zhao, G.; Gu, X. Evaluation of Holstein Cows with Tongue-Rolling: Plasma Metabolomics and Milk Proteomics. Dairy 2025, 6, 53. https://doi.org/10.3390/dairy6050053
Li C, Chen X, Fang T, Gao J, Zhao G, Gu X. Evaluation of Holstein Cows with Tongue-Rolling: Plasma Metabolomics and Milk Proteomics. Dairy. 2025; 6(5):53. https://doi.org/10.3390/dairy6050053
Chicago/Turabian StyleLi, Chenyang, Xiaoyang Chen, Tingting Fang, Jie Gao, Guangyong Zhao, and Xianhong Gu. 2025. "Evaluation of Holstein Cows with Tongue-Rolling: Plasma Metabolomics and Milk Proteomics" Dairy 6, no. 5: 53. https://doi.org/10.3390/dairy6050053
APA StyleLi, C., Chen, X., Fang, T., Gao, J., Zhao, G., & Gu, X. (2025). Evaluation of Holstein Cows with Tongue-Rolling: Plasma Metabolomics and Milk Proteomics. Dairy, 6(5), 53. https://doi.org/10.3390/dairy6050053






