Identification and Characterisation of Spore-Forming Bacteria in Bovine Raw Milk Collected from Four Dairy Farms in New Zealand
Abstract
:1. Introduction
2. Materials and Methods
2.1. Study Sites
2.2. Processing of Raw-Milk Samples
2.3. Bacterial Isolates
2.4. Isolation of Mesophilic Spore-Forming Bacteria from Raw-Milk Samples
2.5. Extraction of DNA
2.6. ERIC PCR Fingerprinting to Identify Unique DNA Patterns in Isolates for Further Analysis
2.7. Amplification and Sequencing of 16S rRNA Gene
2.8. Detection of Toxin Genes Using PCR
2.9. Toxin Production by Clostridium Perfringens
2.10. Screening for Spoilage Potential
3. Results
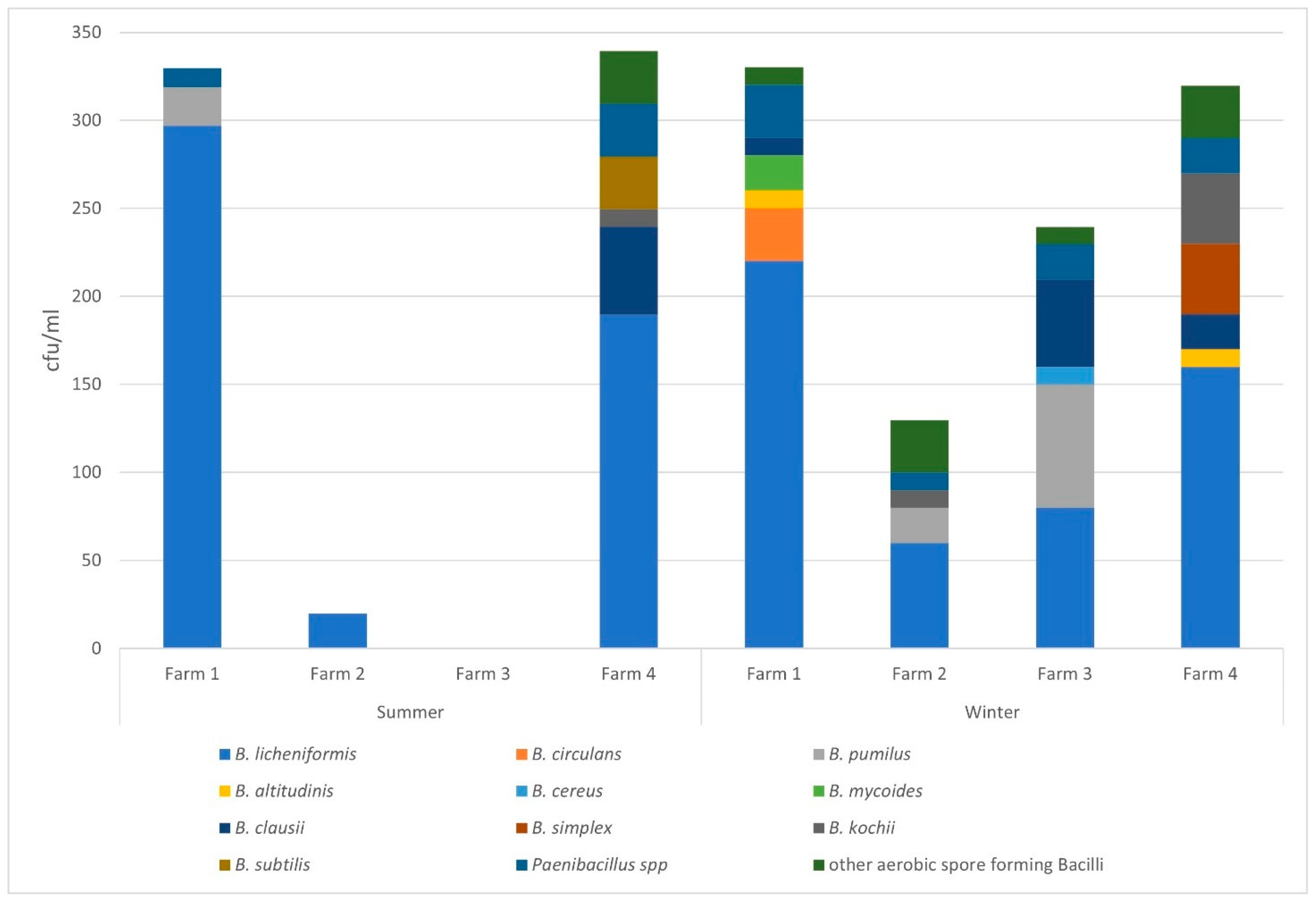
3.1. Enumeration of Aerobic Spore-Forming Bacteria Present in Raw Milk
3.2. ERIC Profiles and 16S rRNA Sequencing
3.3. Different Aerobic Spore-Forming Bacteria Present in Raw-Milk Samples
3.4. Different Anaerobic Spore-Forming Bacteria Present in Raw Milk
3.5. Detection of Toxin Genes in Bacillus spp. and C. perfringens
3.6. Spoilage Activity
4. Discussion
5. Conclusions
6. Future Directions
Supplementary Materials
Author Contributions
Funding
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- André, S.; Vallaeys, T.; Planchon, S. Spore-forming bacteria responsible for food spoilage. Res. Microbiol. 2017, 168, 379–387. [Google Scholar] [CrossRef]
- Haque, M.A.; Wang, F.; Chen, Y.; Hossen, F.; Islam, M.A.; Hossain, M.A.; Siddique, N.; He, C.; Ahmed, F. Bacillus spp. contamination: A novel risk originated from animal feed to human food chains in South-Eastern Bangladesh. Front. Microbiol. 2022, 12, 783103. [Google Scholar] [CrossRef]
- Chukwu, E.; Ogunsola, F.; Nwaokorie, F.; Coker, A. Characterization of Clostridium Species from Food Commodities and Faecal Specimens in Lagos State, Nigeria. West Afr. J. Med. 2015, 34, 167–173. [Google Scholar]
- Samaržija, D.; Zamberlin, Š.; Pogačić, T. Psychrotrophic bacteria and their negative effects on milk and dairy products quality. Mljekarstvo Časopis Unaprjeđenje Proizv. Prerade Mlijeka 2012, 62, 77–95. [Google Scholar]
- Christiansson, A.; Bertilsson, J.; Svensson, B. Bacillus cereus spores in raw milk: Factors affecting the contamination of milk during the grazing period. J. Dairy Sci. 1999, 82, 305–314. [Google Scholar] [CrossRef] [PubMed]
- Vissers, M.; Driehuis, F.; Te Giffel, M.; De Jong, P.; Lankveld, J. Minimizing the level of butyric acid bacteria spores in farm tank milk. J. Dairy Sci. 2007, 90, 3278–3285. [Google Scholar] [CrossRef] [PubMed]
- Klijn, N.; Herman, L.; Langeveld, L.; Vaerewijck, M.; Wagendorp, A.A.; Huemer, I.; Weerkamp, A.H. Genotypical and phenotypical characterization of Bacillus sporothermodurans strains, surviving UHT sterilisation. Int. Dairy J. 1997, 7, 421–428. [Google Scholar] [CrossRef]
- Griffiths, M.; Phillips, J.; West, I.; Muir, D. The effect of extended low-temperature storage of raw milk on the quality of pasteurized and UHT milk. Food Microbiol. 1988, 5, 75–87. [Google Scholar] [CrossRef]
- Hanson, M.; Wendorff, W.; Houck, K. Effect of heat treatment of milk on activation of Bacillus spores. J. Food Prot. 2005, 68, 1484–1486. [Google Scholar] [CrossRef] [PubMed]
- Rukke, E.O.; Sørhaug, T.; Stepaniak, L. HEAT TREATMENT OF MILK|Thermization of Milk. In Encyclopedia of Dairy Sciences (Second Edition); Fuquay, J.W., Ed.; Academic Press: Cambridge, MA, USA, 2011; pp. 693–698. [Google Scholar]
- Guinebretiere, M.; Girardin, H.; Dargaignaratz, C.; Carlin, F.; Nguyen-The, C. Contamination flows of Bacillus cereus and spore-forming aerobic bacteria in a cooked, pasteurized and chilled zucchini purée processing line. Int. J. Food Microbiol. 2003, 82, 223–232. [Google Scholar] [CrossRef]
- Peck, M. Clostridium botulinum and the safety of minimally heated, chilled foods: An emerging issue? J. Appl. Microbiol. 2006, 101, 556–570. [Google Scholar] [CrossRef] [PubMed]
- Ranieri, M.; Huck, J.; Sonnen, M.; Barbano, D.; Boor, K. High temperature, short time pasteurization temperatures inversely affect bacterial numbers during refrigerated storage of pasteurized fluid milk. J. Dairy Sci. 2009, 92, 4823–4832. [Google Scholar] [CrossRef]
- Huck, J.; Hammond, B.; Murphy, S.; Woodcock, N.; Boor, K. Tracking spore-forming bacterial contaminants in fluid milk-processing systems. J. Dairy Sci. 2007, 90, 4872–4883. [Google Scholar] [CrossRef] [PubMed]
- Martinez, B.A.; Stratton, J.; Bianchini, A. Isolation and genetic identification of spore-forming bacteria associated with concentrated-milk processing in Nebraska. J. Dairy Sci. 2017, 100, 919–932. [Google Scholar] [CrossRef] [PubMed]
- Meer, R.; Baker, J.; Bodyfelt, F.; Griffiths, M. Psychrotrophic Bacillus spp. in fluid milk products: A review. J. Food Prot. 1991, 54, 969–979. [Google Scholar] [CrossRef] [PubMed]
- De Jonghe, V.; Coorevits, A.; De Block, J.; Van Coillie, E.; Grijspeerdt, K.; Herman, L.; De Vos, P.; Heyndrickx, M. Toxinogenic and spoilage potential of aerobic spore-formers isolated from raw milk. Int. J. Food Microbiol. 2010, 136, 318–325. [Google Scholar] [CrossRef]
- Fromm, H.I.; Boor, K. Characterization of pasteurized fluid milk shelf-life attributes. J. Food Sci. 2004, 69, M207–M214. [Google Scholar] [CrossRef]
- Ranieri, M.L.; Ivy, R.A.; Mitchell, W.R.; Call, E.; Masiello, S.N.; Wiedmann, M.; Boor, K.J. Real-time PCR detection of Paenibacillus spp. in raw milk to predict shelf life performance of pasteurized fluid milk products. Appl. Environ. Microbiol. 2012, 78, 5855–5863. [Google Scholar] [CrossRef]
- Pettersson, B.; Lembke, F.; Hammer, P.; Stackebrandt, E.; Priest, F.G. Bacillus sporothermodurans, a new species producing highly heat-resistant endospores. Int. J. Syst. Evol. Microbiol. 1996, 46, 759–764. [Google Scholar] [CrossRef]
- Scheldeman, P. Occurence and Resistance of Potentially Highly Heat Resistant Spore Forming Bacteria in Milk Products and at Dairy Farms. PhD Thesis, Ghent University, Gent, Belgium, 2004. [Google Scholar]
- Aouadhi, C.; Maaroufi, A.; Mejri, S. Incidence and characterisation of aerobic spore-forming bacteria originating from dairy milk in T unisia. Int. J. Dairy Technol. 2014, 67, 95–102. [Google Scholar] [CrossRef]
- Huck, J.R.; Woodcock, N.H.; Ralyea, R.D.; Boor, K.J. Molecular subtyping and characterization of psychrotolerant endospore-forming bacteria in two New York State fluid milk processing systems. J. Food Prot. 2007, 70, 2354–2364. [Google Scholar] [CrossRef] [PubMed]
- Feligini, M.; Brambati, E.; Panelli, S.; Ghitti, M.; Sacchi, R.; Capelli, E.; Bonacina, C. One-year investigation of Clostridium spp. occurrence in raw milk and curd of Grana Padano cheese by the automated ribosomal intergenic spacer analysis. Food Control 2014, 42, 71–77. [Google Scholar] [CrossRef]
- Cocolin, L.; Innocente, N.; Biasutti, M.; Comi, G. The late blowing in cheese: A new molecular approach based on PCR and DGGE to study the microbial ecology of the alteration process. Int. J. Food Microbiol. 2004, 90, 83–91. [Google Scholar] [CrossRef]
- Cremonesi, P.; Vanoni, L.; Silvetti, T.; Morandi, S.; Brasca, M. Identification of Clostridium beijerinckii, Cl. butyricum, Cl. sporogenes, Cl. tyrobutyricum isolated from silage, raw milk and hard cheese by a multiplex PCR assay. J. Dairy Res. 2012, 79, 318–323. [Google Scholar] [CrossRef]
- Reis, M.M.; Dixit, Y.; Carr, A.; Tu, C.; Palevich, F.; Gupta, T.; Reis, M.G. Hyperspectral imaging through vacuum packaging for monitoring cheese biochemical transformation caused by Clostridium metabolism. Food Res. Int. 2023, 169, 112866. [Google Scholar] [CrossRef]
- Klijn, N.; Nieuwenhof, F.; Hoolwerf, J.D.; Van Der Waals, C.; Weerkamp, A.H. Identification of Clostridium tyrobutyricum as the causative agent of late blowing in cheese by species-specific PCR amplification. Appl. Environ. Microbiol. 1995, 61, 2919–2924. [Google Scholar] [CrossRef] [PubMed]
- Vissers, M.; Driehuis, F.; Te Giffel, M.; De Jong, P.; Lankveld, J. Concentrations of butyric acid bacteria spores in silage and relationships with aerobic deterioration. J. Dairy Sci. 2007, 90, 928–936. [Google Scholar] [CrossRef]
- Zucali, M.; Bava, L.; Colombini, S.; Brasca, M.; Decimo, M.; Morandi, S.; Tamburini, A.; Crovetto, G.M. Management practices and forage quality affecting the contamination of milk with anaerobic spore-forming bacteria. J. Sci. Food Agric. 2015, 95, 1294–1302. [Google Scholar] [CrossRef] [PubMed]
- Turnbull, P.C.; Jackson, P.J.; Hill, K.K.; Keim, P.; Kolstø, A.B.; Beecher, D.J. Longstanding Taxonomic Enigmas within the ‘Bacillus cereus group’are on the Verge of being Resolved by Far-reaching Molecular Developments: Forecasts on the possible outcome by an ad hoc team. In Applications and Systematics of Bacillus and Relatives; John Wiley & Sons, Inc.: Hoboken, NJ, USA, 2002; pp. 23–36. [Google Scholar]
- Ghelardi, E.; Celandroni, F.; Salvetti, S.; Barsotti, C.; Baggiani, A.; Senesi, S. Identification and characterization of toxigenic Bacillus cereus isolates responsible for two food-poisoning outbreaks. FEMS Microbiol. Lett. 2002, 208, 129–134. [Google Scholar] [CrossRef]
- Mikkola, R.; Kolari, M.; Andersson, M.A.; Helin, J.; Salkinoja-Salonen, M.S. Toxic lactonic lipopeptide from food poisoning isolates of Bacillus licheniformis. Eur. J. Biochem. 2000, 267, 4068–4074. [Google Scholar] [CrossRef]
- Salkinoja-Salonen, M.S.; Vuorio, R.; Andersson, M.; Kampfer, P.; Andersson, M.; Honkanen-Buzalski, T.; Scoging, A. Toxigenic strains of Bacillus licheniformis related to food poisoning. Appl. Environ. Microbiol. 1999, 65, 4637–4645. [Google Scholar] [CrossRef]
- Suominen, I.; Andersson, M.A.; Andersson, M.C.; Hallaksela, A.-M.; Kämpfer, P.; Rainey, F.A.; Salkinoja-Salonen, M. Toxic Bacillus pumilus from indoor air, recycled paper pulp, Norway spruce, food poisoning outbreaks and clinical samples. Syst. Appl. Microbiol. 2001, 24, 267–276. [Google Scholar] [CrossRef]
- Mikkola, R.; Andersson, M.A.; Grigoriev, P.; Teplova, V.V.; Saris, N.-E.L.; Rainey, F.A.; Salkinoja-Salonen, M.S. Bacillus amyloliquefaciens strains isolated from moisture-damaged buildings produced surfactin and a substance toxic to mammalian cells. Arch. Microbiol. 2004, 181, 314–323. [Google Scholar]
- Lindsay, D.; Mosupye, F.; Brözel, V.; Von Holy, A. Cytotoxicity of alkaline-tolerant dairy-associated Bacillus spp. Lett. Appl. Microbiol. 2000, 30, 364–369. [Google Scholar] [CrossRef]
- From, C.; Hormazabal, V.; Granum, P.E. Food poisoning associated with pumilacidin-producing Bacillus pumilus in rice. Int. J. Food Microbiol. 2007, 115, 319–324. [Google Scholar] [CrossRef] [PubMed]
- Beattie, S.a.; Williams, A. Detection of toxigenic strains of Bacillus cereus and other Bacillus spp. with an improved cytotoxicity assay. Lett. Appl. Microbiol. 1999, 28, 221–225. [Google Scholar] [CrossRef]
- Taylor, J.M.; Sutherland, A.D.; Aidoo, K.E.; Logan, N.A. Heat-stable toxin production by strains of Bacillus cereus, Bacillus firmus, Bacillus megaterium, Bacillus simplex and Bacillus licheniformis. FEMS Microbiol. Lett. 2005, 242, 313–317. [Google Scholar] [CrossRef] [PubMed]
- Mcauley, C.M.; McMillan, K.; Moore, S.C.; Fegan, N.; Fox, E.M. Prevalence and characterization of foodborne pathogens from Australian dairy farm environments. J. Dairy Sci. 2014, 97, 7402–7412. [Google Scholar] [CrossRef]
- Camerini, S.; Marcocci, L.; Picarazzi, L.; Iorio, E.; Ruspantini, I.; Pietrangeli, P.; Crescenzi, M.; Franciosa, G. Type E botulinum neurotoxin-producing Clostridium butyricum strains are aerotolerant during vegetative growth. Msystems 2019, 4. [Google Scholar] [CrossRef] [PubMed]
- Griffiths, M.; Phillips, J. Incidence, source and some properties of psychrotrophic Bacillus spp found in raw and pasteurized milk. Int. J. Dairy Technol. 1990, 43, 62–66. [Google Scholar] [CrossRef]
- Gupta, T.B.; Brightwell, G. Farm level survey of spore-forming bacteria on four dairy farms in the Waikato region of New Zealand. Microbiologyopen 2017, 6, e00457. [Google Scholar] [CrossRef] [PubMed]
- Versalovic, J.; Koeuth, T.; Lupski, R. Distribution of repetitive DNA sequences in eubacteria and application to finerpriting of bacterial enomes. Nucleic Acids Res. 1991, 19, 6823–6831. [Google Scholar] [CrossRef] [PubMed]
- Weijtens, M.; Reinders, R.; Urlings, H.; Van der Plas, J. Campylobacter infections in fattening pigs; excretion pattern and genetic diversity. J. Appl. Microbiol. 1999, 86, 63–70. [Google Scholar] [CrossRef] [PubMed]
- Böddinghaus, B.; Wolters, J.; Heikens, W.; Böttger, E.C. Phylogenetic analysis and identification of different serovars of Mycobacterium intracellulare at the molecular level. FEMS Microbiol. Lett. 1990, 70, 197–203. [Google Scholar] [CrossRef]
- Cole, J.R.; Wang, Q.; Fish, J.A.; Chai, B.; McGarrell, D.M.; Sun, Y.; Brown, C.T.; Porras-Alfaro, A.; Kuske, C.R.; Tiedje, J.M. Ribosomal Database Project: Data and tools for high throughput rRNA analysis. Nucleic Acids Res. 2014, 42, D633–D642. [Google Scholar] [CrossRef]
- Nieminen, T.; Rintaluoma, N.; Andersson, M.; Taimisto, A.-M.; Ali-Vehmas, T.; Seppälä, A.; Priha, O.; Salkinoja-Salonen, M. Toxinogenic Bacillus pumilus and Bacillus licheniformis from mastitic milk. Vet. Microbiol. 2007, 124, 329–339. [Google Scholar] [CrossRef]
- Ehling-Schulz, M.; Fricker, M.; Scherer, S. Identification of emetic toxin producing Bacillus cereus strains by a novel molecular assay. FEMS Microbiol. Lett. 2004, 232, 189–195. [Google Scholar] [CrossRef]
- Yang, I.-C.; Shih, D.Y.-C.; Huang, T.-P.; Huang, Y.-P.; Wang, J.-Y.; Pan, T.-M. Establishment of a novel multiplex PCR assay and detection of toxigenic strains of the species in the Bacillus cereus group. J. Food Prot. 2005, 68, 2123–2130. [Google Scholar] [CrossRef]
- Baums, C.G.; Schotte, U.; Amtsberg, G.; Goethe, R. Diagnostic multiplex PCR for toxin genotyping of Clostridium perfringens isolates. Vet. Microbiol. 2004, 100, 11–16. [Google Scholar] [CrossRef]
- Coorevits, A.; De Jonghe, V.; Vandroemme, J.; Reekmans, R.; Heyrman, J.; Messens, W.; De Vos, P.; Heyndrickx, M. Comparative analysis of the diversity of aerobic spore-forming bacteria in raw milk from organic and conventional dairy farms. Syst. Appl. Microbiol. 2008, 31, 126–140. [Google Scholar] [CrossRef]
- Gopal, N.; Hill, C.; Ross, P.R.; Beresford, T.P.; Fenelon, M.A.; Cotter, P.D. The prevalence and control of Bacillus and related spore-forming bacteria in the dairy industry. Front. Microbiol. 2015, 6, 1418. [Google Scholar] [CrossRef]
- Chen, L.; Coolbear, T.; Daniel, R.M. Characteristics of proteinases and lipases produced by seven Bacillus sp. isolated from milk powder production lines. Int. Dairy J. 2004, 14, 495–504. [Google Scholar] [CrossRef]
- Ternström, A.; Lindberg, A.M.; Molin, G. Classification of the spoilage flora of raw and pasteurized bovine milk, with special reference to Pseudomonas and Bacillus. J. Appl. Bacteriol. 1993, 75, 25–34. [Google Scholar] [CrossRef]
- Dračková, J.M.; Vorlová, L. Influence of Bacillus spp. enzymes on ultra high temperature-treated milk proteins. Acta Vet. Brno 2004, 73, 393–400. [Google Scholar] [CrossRef]
- Martin, N.; Ranieri, M.; Murphy, S.; Ralyea, R.; Wiedmann, M.; Boor, K. Results from raw milk microbiological tests do not predict the shelf-life performance of commercially pasteurized fluid milk. J. Dairy Sci. 2011, 94, 1211–1222. [Google Scholar] [CrossRef]
- Buehner, K.P.; Anand, S.; Garcia, A. Prevalence of thermoduric bacteria and spores on 10 Midwest dairy farms. J. Dairy Sci. 2014, 97, 6777–6784. [Google Scholar] [CrossRef] [PubMed]
- Crielly, E.; Logan, N.; Anderton, A. Studies on the Bacillus flora of milk and milk products. J. Appl. Microbiol. 1994, 77, 256–263. [Google Scholar] [CrossRef] [PubMed]
- Shehata, A.; Magdoub, M.; Sultan, N.E.; El-Samragy, Y. Aerobic mesophilic and psychrotrophic sporeforming bacteria in buffalo milk. J. Dairy Sci. 1983, 66, 1228–1231. [Google Scholar] [CrossRef]
- Scheldeman, P.; Herman, L.; Foster, S.; Heyndrickx, M. Bacillus sporothermodurans and other highly heat-resistant spore formers in milk. J. Appl. Microbiol. 2006, 101, 542–555. [Google Scholar] [CrossRef]
- Ronimus, R.S.; Parker, L.E.; Turner, N.; Poudel, S.; Rückert, A.; Morgan, H.W. A RAPD-based comparison of thermophilic bacilli from milk powders. Int. J. Food Microbiol. 2003, 85, 45–61. [Google Scholar] [CrossRef]
- Kalogridou-Vassiliadou, D. Biochemical activities of Bacillus species isolated from flat sour evaporated milk. J. Dairy Sci. 1992, 75, 2681–2686. [Google Scholar] [CrossRef]
- Reginensi, S.M.; González, M.J.; Olivera, J.A.; Sosa, M.; Juliano, P.; Bermúdez, J. RAPD-based screening for spore-forming bacterial populations in Uruguayan commercial powdered milk. Int. J. Food Microbiol. 2011, 148, 36–41. [Google Scholar] [CrossRef]
- Mehta, D.; Metzger, L.; Hassan, A.; Nelson, B.; Patel, H. The ability of spore formers to degrade milk proteins, fat, phospholipids, common stabilizers, and exopolysaccharides. J. Dairy Sci. 2019, 102, 10799–10813. [Google Scholar] [CrossRef]
- Rowan, N.J.; Deans, K.; Anderson, J.G.; Gemmell, C.G.; Hunter, I.S.; Chaithong, T. Putative virulence factor expression by clinical and food isolates of Bacillus spp. after growth in reconstituted infant milk formulae. Appl. Environ. Microbiol. 2001, 67, 3873–3881. [Google Scholar] [CrossRef]
- Harwood, C.R.; Mouillon, J.-M.; Pohl, S.; Arnau, J. Secondary metabolite production and the safety of industrially important members of the Bacillus subtilis group. FEMS Microbiol. Rev. 2018, 42, 721–738. [Google Scholar] [CrossRef]
- Coronel-León, J.; de Grau, G.; Grau-Campistany, A.; Farfan, M.; Rabanal, F.; Manresa, A.; Marqués, A.M. Biosurfactant production by AL 1.1, a Bacillus licheniformis strain isolated from Antarctica: Production, chemical characterization and properties. Ann. Microbiol. 2015, 65, 2065–2078. [Google Scholar] [CrossRef]
- Sutherland, A.; Murdoch, R. Seasonal occurrence of psychrotrophic Bacillus species in raw milk, and studies on the interactions with mesophilic Bacillus sp. Int. J. Food Microbiol. 1994, 21, 279–292. [Google Scholar] [CrossRef] [PubMed]
- Tatzel, R.; Ludwig, W.; Schleifer, K.H.; Wallnöfer, P.R. Identification of Bacillus strains isolated from milk and cream with classical and nucleic acid hybridization methods. J. Dairy Res. 1994, 61, 529–535. [Google Scholar] [CrossRef] [PubMed]
- Lukasova, J.; Vyhnalkova, J.; Pacova, Z. Bacillus species in raw milk and in the farm environment. Milchwissenschaft 2001, 56, 609–611. [Google Scholar]
- Lan, X.; Wang, J.; Zheng, N.; Zhao, S.; Li, S.; Li, F. Prevalence and risk factors for Bacillus cereus in raw milk in Inner Mongolia, Northern China. Int. J. Dairy Technol. 2018, 71, 269–273. [Google Scholar] [CrossRef]
- Fei, P.; Yuan, X.; Zhao, S.; Yang, T.; Xiang, J.; Chen, X.; Zhou, L.; Ji, M. Prevalence and Genetic Diversity of Bacillus cereus Isolated from Raw Milk and Cattle Farm Environments. Curr. Microbiol. 2019, 76, 1355–1360. [Google Scholar] [CrossRef] [PubMed]
- Zervas, A.; Aggerbeck, M.R.; Allaga, H.; Güzel, M.; Hendriks, M.; Jonuškienė, I.; Kedves, O.; Kupeli, A.; Lamovšek, J.; Mülner, P. Identification and characterization of 33 Bacillus cereus sensu lato isolates from agricultural fields from eleven widely distributed countries by whole genome sequencing. Microorganisms 2020, 8, 2028. [Google Scholar] [CrossRef]
- Borge, G.I.A.; Skeie, M.; Sørhaug, T.; Langsrud, T.; Granum, P.E. Growth and toxin profiles of Bacillus cereus isolated from different food sources. Int. J. Food Microbiol. 2001, 69, 237–246. [Google Scholar] [CrossRef]
- Catania, A.M.; Civera, T.; Di Ciccio, P.A.; Grassi, M.A.; Morra, P.; Dalmasso, A. Characterization of vegetative Bacillus cereus and Bacillus subtilis strains isolated from processed cheese products in an Italian dairy plant. Foods 2021, 10, 2876. [Google Scholar] [CrossRef]
- Svensson, B.; Monthan, A.; Shaheen, R.; Andersson, M.A.; Salkinoja-Salonen, M.; Christiansson, A. Occurrence of emetic toxin producing Bacillus cereus in the dairy production chain. Int. Dairy J. 2006, 16, 740–749. [Google Scholar] [CrossRef]
- Lindback, T.; Granum, P. Detection and Purification of Bacillus cereus Enterotoxins Food-Borne Pathogens: Methods and Protocols. In Methods in Biotechnology; Humana: Totowa, NJ, USA, 2006; pp. 15–26. [Google Scholar]
- Kotiranta, A.; Lounatmaa, K.; Haapasalo, M. Epidemiology and pathogenesis of Bacillus cereus infections. Microbes Infect. 2000, 2, 189–198. [Google Scholar] [CrossRef] [PubMed]
- Asano, S.-I.; Nukumizu, Y.; Bando, H.; Iizuka, T.; Yamamoto, T. Cloning of novel enterotoxin genes from Bacillus cereus and Bacillus thuringiensis. Appl. Environ. Microbiol. 1997, 63, 1054–1057. [Google Scholar] [CrossRef]
- Tran, S.-L.; Guillemet, E.; Gohar, M.; Lereclus, D.; Ramarao, N. CwpFM (EntFM) is a Bacillus cereus potential cell wall peptidase implicated in adhesion, biofilm formation, and virulence. J. Bacteriol. 2010, 192, 2638–2642. [Google Scholar] [CrossRef] [PubMed]
- Granum, P.E.; Lund, T. Bacillus cereus and its food poisoning toxins. FEMS Microbiol. Lett. 1997, 157, 223–228. [Google Scholar] [CrossRef]
- Ngamwongsatit, P.; Buasri, W.; Pianariyanon, P.; Pulsrikarn, C.; Ohba, M.; Assavanig, A.; Panbangred, W. Broad distribution of enterotoxin genes (hblCDA, nheABC, cytK, and entFM) among Bacillus thuringiensis and Bacillus cereus as shown by novel primers. Int. J. Food Microbiol. 2008, 121, 352–356. [Google Scholar] [CrossRef] [PubMed]
- Carter, L.; Chase, H.R.; Gieseker, C.M.; Hasbrouck, N.R.; Stine, C.B.; Khan, A.; Ewing-Peeples, L.J.; Tall, B.D.; Gopinath, G.R. Analysis of enterotoxigenic Bacillus cereus strains from dried foods using whole genome sequencing, multi-locus sequence analysis and toxin gene prevalence and distribution using endpoint PCR analysis. Int. J. Food Microbiol. 2018, 284, 31–39. [Google Scholar] [CrossRef]
- Tirloni, E.; Bernardi, C.; Ghelardi, E.; Celandroni, F.; Andrighetto, C.; Rota, N.; Stella, S. Biopreservation as a potential hurdle for Bacillus cereus growth in fresh cheese. J. Dairy Sci. 2020, 103, 150–160. [Google Scholar] [CrossRef]
- Hwang, J.-Y.; Park, J.-H. Characteristics of enterotoxin distribution, hemolysis, lecithinase, and starch hydrolysis of Bacillus cereus isolated from infant formulas and ready-to-eat foods. J. Dairy Sci. 2015, 98, 1652–1660. [Google Scholar] [CrossRef] [PubMed]
- Guinebretière, M.H.; Thompson, F.L.; Sorokin, A.; Normand, P.; Dawyndt, P.; Ehling-Schulz, M.; Svensson, B.; Sanchis, V.; Nguyen-The, C.; Heyndrickx, M. Ecological diversification in the Bacillus cereus group. Environ. Microbiol. 2008, 10, 851–865. [Google Scholar] [CrossRef] [PubMed]
- Di Pinto, A.; Bonerba, E.; Bozzo, G.; Ceci, E.; Terio, V.; Tantillo, G. Occurence of potentially enterotoxigenic Bacillus cereus in infant milk powder. Eur. Food Res. Technol. 2013, 237, 275–279. [Google Scholar] [CrossRef]
- Bağcıoğlu, M.; Fricker, M.; Johler, S.; Ehling-Schulz, M. Detection and identification of Bacillus cereus, Bacillus cytotoxicus, Bacillus thuringiensis, Bacillus mycoides and Bacillus weihenstephanensis via machine learning based FTIR spectroscopy. Front. Microbiol. 2019, 10, 902. [Google Scholar] [CrossRef]
- Ibrahim, A.S.; Hafiz, N.M.; Saad, M. Prevalence of Bacillus cereus in dairy powders focusing on its toxigenic genes and antimicrobial resistance. Arch. Microbiol. 2022, 204, 339. [Google Scholar] [CrossRef]
- Heidt, J.; Papaloukas, N.; Timmerman, C. A rare bloodstream infection: Bacillus mycoides. Neth J. Med. 2019, 77, 227. [Google Scholar]
- Lindback, T.; Fagerlund, A.; Rødland, M.S.; Granum, P.E. Characterization of the Bacillus cereus Nhe enterotoxin. Microbiology 2004, 150, 3959–3967. [Google Scholar] [CrossRef]
- Trmčić, A.; Martin, N.; Boor, K.; Wiedmann, M. A standard bacterial isolate set for research on contemporary dairy spoilage. J. Dairy Sci. 2015, 98, 5806–5817. [Google Scholar] [CrossRef]
- Heyndrickx, M.; Scheldeman, P. Bacilli Associated with Spoilage in Dairy Products and Other Food. In Applications and Systematics of Bacillus and Relatives; Blackwell Science Ltd.: Malden, MA, USA, 2002; pp. 64–82. [Google Scholar]
- Scheldeman, P.; Goossens, K.; Rodriguez-Diaz, M.; Pil, A.; Goris, J.; Herman, L.; De Vos, P.; Logan, N.A.; Heyndrickx, M. Paenibacillus lactis sp. nov., isolated from raw and heat-treated milk. Int. J. Syst. Evol. Microbiol. 2004, 54, 885–891. [Google Scholar] [CrossRef]
- Sattin, E.; Andreani, N.; Carraro, L.; Fasolato, L.; Balzan, S.; Novelli, E.; Squartini, A.; Telatin, A.; Simionati, B.; Cardazzo, B. Microbial dynamics during shelf-life of industrial Ricotta cheese and identification of a Bacillus strain as a cause of a pink discolouration. Food Microbiol. 2016, 57, 8–15. [Google Scholar] [CrossRef]
- Ivy, R.A.; Ranieri, M.L.; Martin, N.H.; den Bakker, H.C.; Xavier, B.M.; Wiedmann, M.; Boor, K.J. Identification and characterization of psychrotolerant sporeformers associated with fluid milk production and processing. Appl. Environ. Microbiol. 2012, 78, 1853–1864. [Google Scholar] [CrossRef]
- Ribeiro-Júnior, J.; Tamanini, R.; Alfieri, A.; Beloti, V. Effect of milk bactofugation on the counts and diversity of thermoduric bacteria. J. Dairy Sci. 2020, 103, 8782–8790. [Google Scholar] [CrossRef]
- Goldsztejn, M.; Grenda, T.; Kozieł, N.; Sapała, M.; Mazur, M.; Sieradzki, Z.; Król, B.; Kwiatek, K. Potential determinants of spp. occurrence in Polish silage. J. Vet. Res. 2020, 64, 549–555. [Google Scholar] [CrossRef] [PubMed]
- Le Bourhis, A.-G.; Saunier, K.; Doré, J.; Carlier, J.-P.; Chamba, J.-F.; Popoff, M.-R.; Tholozan, J.-L. Development and validation of PCR primers to assess the diversity of Clostridium spp. in cheese by temporal temperature gradient gel electrophoresis. Appl. Environ. Microbiol. 2005, 71, 29–38. [Google Scholar] [CrossRef] [PubMed]
- Driehuis, F.; Hoolwerf, J.; Rademaker, J.L. Concurrence of spores of Clostridium tyrobutyricum, Clostridium beijerinckii and Paenibacillus polymyxa in silage, dairy cow faeces and raw milk. Int. Dairy J. 2016, 63, 70–77. [Google Scholar] [CrossRef]
- Julien, M.-C.; Dion, P.; Lafreniere, C.; Antoun, H.; Drouin, P. Sources of clostridia in raw milk on farms. Appl. Environ. Microbiol. 2008, 74, 6348–6357. [Google Scholar] [CrossRef]
- Grass, J.E.; Gould, L.H.; Mahon, B.E. Epidemiology of foodborne disease outbreaks caused by Clostridium perfringens, United States, 1998–2010. Foodborne Pathog. Dis. 2013, 10, 131–136. [Google Scholar] [CrossRef] [PubMed]
- Osman, K.; El-Enbaawy, M.; Ezzeldeen, N.; Hussein, H. Mastitis in dairy buffalo and cattle in Egypt due to Clostridium perfringens: Prevalence, incidence, risk factors and costs. Rev. Sci. Et Tech. 2009, 28, 975. [Google Scholar] [CrossRef] [PubMed]
- El-Bassiony, T. Occurrence of Clostridium perfringens in milk and dairy products. J. Food Prot. 1980, 43, 536–537. [Google Scholar] [CrossRef]
- Hernández, M.; López-Enríquez, L.; Rodríguez-Lázaro, D. Quantitative Detection of Clostridium perfringens by Real-Time PCR in Raw Milk. Food Anal. Methods 2017, 10, 1139–1147. [Google Scholar] [CrossRef]
- Barash, J.R.; Hsia, J.K.; Arnon, S.S. Presence of soil-dwelling clostridia in commercial powdered infant formulas. J. Pediatr. 2010, 156, 402–408. [Google Scholar] [CrossRef]
- Turchi, B.; Pero, S.; Torracca, B.; Fratini, F.; Mancini, S.; Galiero, A.; Pedonese, F.; Nuvoloni, R.; Cerri, D. Occurrence of Clostridium spp. in ewe’s milk: Enumeration and identification of isolates. Dairy Sci. Technol. 2016, 96, 693–701. [Google Scholar] [CrossRef]
- Uzal, F.; Vidal, J.; McClane, B.; Gurjar, A. Clostridium perfringens toxins involved in mammalian veterinary diseases. Open Toxinol. J. 2010, 2, 24. [Google Scholar] [CrossRef]
- Canard, B.; Cole, S.T. Genome organization of the anaerobic pathogen Clostridium perfringens. Proc. Natl. Acad. Sci. USA 1989, 86, 6676–6680. [Google Scholar] [CrossRef]
- Uzal, F.A.; Freedman, J.C.; Shrestha, A.; Theoret, J.R.; Garcia, J.; Awad, M.M.; Adams, V.; Moore, R.J.; Rood, J.I.; McClane, B.A. Towards an understanding of the role of Clostridium perfringens toxins in human and animal disease. Future Microbiol. 2014, 9, 361–377. [Google Scholar] [CrossRef] [PubMed]
- Immerseel, F.V.; Buck, J.D.; Pasmans, F.; Huyghebaert, G.; Haesebrouck, F.; Ducatelle, R. Clostridium perfringens in poultry: An emerging threat for animal and public health. Avian Pathol. 2004, 33, 537–549. [Google Scholar] [CrossRef]
- Songer, J.G.; Miskimmins, D.W. Clostridium perfringens type E enteritis in calves: Two cases and a brief review of the literature. Anaerobe 2004, 10, 239–242. [Google Scholar] [CrossRef] [PubMed]
- Songer, J.G. Clostridial enteric diseases of domestic animals. Clin. Microbiol. Rev. 1996, 9, 216–234. [Google Scholar] [CrossRef] [PubMed]
- Greco, G.; Madio, A.; Buonavoglia, D.; Totaro, M.; Corrente, M.; Martella, V.; Buonavoglia, C. Clostridium perfringens toxin-types in lambs and kids affected with gastroenteric pathologies in Italy. Vet. J. 2005, 170, 346–350. [Google Scholar] [CrossRef] [PubMed]
- Dos Santos, R.A.N.; Abdel-Nour, J.; McAuley, C.; Moore, S.C.; Fegan, N.; Fox, E.M. Clostridium perfringens associated with dairy farm systems show diverse genotypes. Int. J. Food Microbiol. 2022, 382, 109933. [Google Scholar] [CrossRef] [PubMed]
- Bendary, M.M.; Abd El-Hamid, M.I.; El-Tarabili, R.M.; Hefny, A.A.; Algendy, R.M.; Elzohairy, N.A.; Ghoneim, M.M.; Al-Sanea, M.M.; Nahari, M.H.; Moustafa, W.H. Clostridium perfringens Associated with Foodborne Infections of Animal Origins: Insights into Prevalence, Antimicrobial Resistance, Toxin Genes Profiles, and Toxinotypes. Biology 2022, 11, 551. [Google Scholar] [CrossRef]
- Sallam, A.; Steinbüchel, A. Clostridium sulfidigenes sp. nov., a mesophilic, proteolytic, thiosulfate-and sulfur-reducing bacterium isolated from pond sediment. Int. J. Syst. Evol. Microbiol. 2009, 59, 1661–1665. [Google Scholar] [CrossRef]
- Gong, R.; Ye, X.; Wang, S.; Ren, Z. Isolation, identification, and biological characteristics of Clostridium sartagoforme from rabbit. PLoS ONE 2021, 16, e0259715. [Google Scholar] [CrossRef] [PubMed]
- Doyle, C.J.; O’Toole, P.W.; Cotter, P.D. Genomic characterization of sulphite reducing bacteria isolated from the dairy production chain. Front. Microbiol. 2018, 9, 1507. [Google Scholar] [CrossRef] [PubMed]
| Primers and Target Genes | Sequence (5′-3′) | Product Size bp | Reference |
|---|---|---|---|
| LicA-F | GTGCCTGATGTAACGAATG | 735 | [49] |
| LicA-R | CACTTCCTGCCATATACC | ||
| LicB-F | TGATCAGCCGGCCGTTGTCT | 904 | [49] |
| LicB-R | GGCGAATTGTCCGATCATGTCC | ||
| LicC-F | GCCTATCTGCCGATTGAC | 1195 | [49] |
| LicC-R | TATATGCATCCGGCACCA | ||
| cesB-F | GACAAGAGAAATTTCTACGAGCAAGTACAAT | 635 | [50] |
| cesB-R | GCAGCCTTCCAATTACTCCTTCTGCCACAGT |
| Primers and Target Genes | Sequence (5′-3′) | Product Size bp | Reference |
|---|---|---|---|
| hblA-F | ATT AAT ACA GGG GAT GGA GAA ACT T | 237 | [51] |
| hblA-R | TGA TCC TAA TAC TTC TAG ACG CTT | ||
| hblC-F | CCT ATC AAT ACT CTC GCA ACA CCA AT | 386 | [51] |
| hblC-R | TTT TCT TGA TTC GTC ATA GCC ATT TCT | ||
| hblD-F | AGA TGC TAC AAG ACT TCA AAG GGA AAC TAT | 436 | [51] |
| hblD-R | TGA TTA GCA CGA TCT GCT TTC ATA CTT | ||
| nheA-F | ATT ACA GGG TTA TTG GTT ACA GCA GT | 475 | [51] |
| nheA-R | AAT CTT GCT CCA TAC TCT CTT GGA TGC T | ||
| nheB-F | GTG CAG CTG TAG GCG GT | 328 | [51] |
| nheB-R | ATG TTT TTC CAG CTA TCT TTC GCA AT | ||
| nheC-F | GCG GAT ATT GTA AAG AAT CAA AAT GAG GT | 557 | [51] |
| nheC-R | TTT CCA GCT ATC TTT CGC TGT ATG TAA AT | ||
| entFM-F | CAA AGA CTT CGT AAC AAA AGG TGG T | 290 | [51] |
| entFM-R | TGT TTA CTC CGC CTT TTA CAA ACT T | ||
| cytK1-F | ATC GGG CAA AAT GCA AAA ACA CAT | 800 | [51] |
| ctyK1-R | ACC CAG TTT GCA GTT CCG AAT GT | ||
| cesB-F | GACAAGAGAAATTTCTACGAGCAAGTACAAT | 635 | [50] |
| cesB-R | GCAGCCTTCCAATTACTCCTTCTGCCACAGT |
| Primers and Target Genes | Sequence (5′-3′) | Product Size bp |
|---|---|---|
| CPA-F | GTTGATAGCGCAGGACATGTTAAG | 402 |
| CPA-R | CATGTAGTCATCTGTTCCAGCATC | |
| CPB-F | ACTATACAGACAGATCATTCAACC | 236 |
| CPB-R | TTAGGAGCAGTTAGAACTACAGAC | |
| CPE-F | ACTGCAACTACTACTCATACTGTG | 541 |
| CPE-R | CTGGTGCCTTAATAGAAAGACTCC | |
| CPI-F | GCGATGAAAAGCCTACACCACTAC | 317 |
| CPI-R | GGTATATCCTCCACGCATATAGTC |
| Isolates | LicA | LicB | LicC | cesB |
|---|---|---|---|---|
| Farm 1, Summer 1 | − | − | + | − |
| Farm 1, Summer 5 | + | + | + | − |
| Farm 1, Summer 7 | + | − | + | − |
| Farm 1, Summer 8 | + | − | + | − |
| Farm 1, Summer 9 | − | + | + | − |
| Farm 1, Summer 11 | + | − | + | − |
| Farm 1, Summer 12 | + | − | − | − |
| Farm 1, Summer 13 | + | − | − | − |
| Farm 1, Summer 2 (B. pumilus) | − | − | − | − |
| B. licheniformis +ve control | + | + | + | − |
| B. pumilus -ve control | − | − | − | − |
| Farm 1, Winter 1 | + | + | + | − |
| Farm 1, Winter 3 | + | + | + | − |
| Farm 1, Winter 4 | + | + | + | − |
| Farm 1, Winter 8 | + | + | + | − |
| Farm 1, Winter 9 | + | + | + | − |
| Farm 1, Winter 19 | + | + | + | − |
| Farm 1, Winter 25 | + | + | + | − |
| Farm 1, Winter 26 | + | + | + | − |
| Farm 1, Winter 27 | + | + | + | − |
| B. licheniformis +ve control | + | + | + | − |
| Farm 2, Summer 1 | + | + | + | − |
| Farm 2, Summer 2 | + | + | + | − |
| Farm 2, Winter 2 | + | + | + | − |
| Farm 2, Winter 9 (B. pumilus) | − | − | − | − |
| B. licheniformis +ve control | + | + | + | − |
| B. pumilus -ve control | − | − | − | − |
| Farm 3, Winter 1 | + | + | + | − |
| Farm 3, Winter 9 | + | + | + | − |
| Farm 3, Winter 10 | + | + | + | − |
| Farm 3, Winter 3 (B. pumilus) | − | − | − | − |
| B. licheniformis +ve control | + | + | + | − |
| B. pumilus -ve control | − | − | − | − |
| Farm 4, Summer 1 | + | + | + | − |
| Farm 4, Summer 5 | + | + | + | − |
| Farm 4, Summer 29 | − | − | − | − |
| B. licheniformis +ve control | + | + | + | − |
| Farm 4, Winter 1 | + | + | + | − |
| Farm 4, Winter 4 | + | + | + | − |
| Farm 4, Winter 5 | + | + | + | − |
| Farm 4, Winter 15 | + | + | + | − |
| Farm 4, Winter 31 | + | + | + | − |
| B. licheniformis +ve control | + | + | + | − |
| Isolates | ||||
|---|---|---|---|---|
| Toxin Genes | B. cereus NCTC 11143 (Positive Control) | Farm 1 Winter 11 (B. mycoides) | Farm 1 Winter 12 (B. mycoides) | Farm 3 Winter 17 (B. cereus) |
| hblA | + | + | + | − |
| hblC | + | + | + | − |
| hblD | + | − | − | + |
| nheA | + | + | + | − |
| nheB | + | + | + | − |
| nheC | + | − | − | − |
| entFM | + | − | − | + |
| cytK1 | + | − | − | − |
| cesB | + | − | − | − |
| Isolate | PCR | ELISA | Toxinotype | |||||
|---|---|---|---|---|---|---|---|---|
| cpa | cpb | etx | iap and ibp | Alpha | Beta | Epsilon | ||
| Farm 3, Winter 1 | + | − | + | − | ++ | − | ++ | D |
| Farm 3, Winter 2 | + | − | + | − | ++ | − | ++ | D |
| Farm 3, Winter 3 | + | − | − | − | ++ | − | A | |
| Farm 3, Winter 4 | + | − | + | − | ++ | − | D# | |
| Farm 3, Winter 5 | + | − | + | − | ++ | − | ++ | D |
| Farm 3, Winter 6 | + | − | − | − | ++ | − | A | |
| Farm 3, Winter 7 | + | − | − | − | ++ | − | A | |
| Farm 3, Winter 8 | + | − | − | − | ++ | − | A | |
| Farm 3, Winter 9 | + | − | − | − | ++ | − | A | |
| Farm 3, Winter 10 | + | − | − | − | ++ | − | A | |
| Farm 3, Winter 11 | + | − | − | − | ++ | − | A | |
| Farm 3, Winter 12 | + | − | − | − | ++ | − | − | A |
| Farm 3, Winter 13 | + | − | − | − | ++ | − | − | A |
| Farm 3, Winter 14 | + | − | − | − | ++ | − | − | A |
| Farm 3, Winter 15 | + | − | − | − | ++ | − | − | A |
| Farm 4, Winter 1 | + | − | − | − | ++ | − | A | |
| Farm 4, Winter 2 | + | − | − | − | ++ | − | A | |
| Farm 4, Winter 3 | + | − | − | − | ++ | − | A | |
| Farm 4, Winter 4 | + | − | − | − | ++ | − | A | |
| Farm 4, Winter 5 | + | − | − | − | ++ | − | A | |
| Farm 4, Winter 6 | + | − | − | − | ++ | − | A | |
| Farm 4, Winter 7 | + | − | − | − | ++ | − | A | |
| Farm 4, Winter 8 | + | − | − | − | ++ | − | A | |
| Farm 4, Winter 9 | + | − | − | − | ++ | − | A | |
| Farm 4, Winter 10 | + | + | + | − | ++ | ++ | ++ | B |
| Farm 4, Winter 11 | + | + | + | − | ++ | ++ | ++ | B |
| Farm 4, Winter 12 | + | + | + | − | ++ | ++ | ++ | B |
| Spoilage Activity | ||||
|---|---|---|---|---|
| Farm, Season | Isolate Number | Proteolytic | Lipolytic | Bacteria |
| Farm 1, Summer | 1 | B. licheniformis | ||
| 2 | B. pumilus | |||
| 4 | P. marinisediminis | |||
| 5 | B. licheniformis | |||
| 7 | B. licheniformis | |||
| 8 | B. licheniformis | |||
| 9 | B. licheniformis | |||
| 11 | B. licheniformis | |||
| 12 | B. licheniformis | |||
| 13 | B. licheniformis | |||
| Farm 1, Winter | 1 | B. licheniformis | ||
| 2 | P. xylanexedens | |||
| 3 | B. licheniformis | |||
| 4 | B. licheniformis | |||
| 6 | B. clausii | |||
| 8 | B. licheniformis | |||
| 9 | B. licheniformis | |||
| 10 | P. peoriae | |||
| 12 | B. mycoides | |||
| 14 | B. altitudinis | |||
| 17 | B. circulans | |||
| 19 | B. licheniformis | |||
| 24 | S. silvestris | |||
| 25 | B. licheniformis | |||
| 26 | B. licheniformis | |||
| 27 | B. licheniformis | |||
| 28 | B. circulans | |||
| Farm 2, Summer | 1 | B. licheniformis | ||
| 2 | B. licheniformis | |||
| Farm 2, Winter | 1 | B. pumilus | ||
| 2 | B. licheniformis | |||
| 8 | B. kochii | |||
| 9 | B. pumilus | |||
| 10 | S. silvestris | |||
| 11 | B. rigui | |||
| 12 | NT | NT | S. luteola | |
| 13 | P. polymyxa | |||
| Farm 3, Winter | 1 | B. licheniformis | ||
| 2 | B. clausii | |||
| 3 | B. pumilus | |||
| 6 | B. pumilus | |||
| 8 | Ornithinibacillus | |||
| 9 | B. licheniformis | |||
| 10 | B. licheniformis | |||
| 11 | B. licheniformis | |||
| 12 | B. licheniformis | |||
| 13 | B. pumilus | |||
| 17 | B. cereus | |||
| 22 | B. clausii | |||
| 23 | P. amylolyticus | |||
| Farm 4, Summer | 1 | B. licheniformis | ||
| 5 | B. licheniformis | |||
| 6 | B. subtilis | |||
| 17 | B. clausii | |||
| 22 | P. timonensis | |||
| 25 | L. fusiformis | |||
| 27 | B. kochii | |||
| 29 | B. licheniformis | |||
| Farm 4, Winter | 1 | B. licheniformis | ||
| 3 | B. altitudinis | |||
| 4 | B. licheniformis | |||
| 5 | B. licheniformis | |||
| 9 | B. simplex | |||
| 15 | B. licheniformis | |||
| 22 | B. kochii | |||
| 24 | L. fusiformis | |||
| 25 | P. cookii | |||
| 26 | B. clausii | |||
| 29 | B. simplex | |||
| 30 | B. pocheonensis | |||
| 31 | B. licheniformis | |||
| 32 | P. lactis | |||
 intermediate level of spoilage activity (low to medium size clearing zone);
intermediate level of spoilage activity (low to medium size clearing zone);  no spoilage activity detected;
no spoilage activity detected;  and NT not tested.
and NT not tested.Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Gupta, T.B.; Brightwell, G. Identification and Characterisation of Spore-Forming Bacteria in Bovine Raw Milk Collected from Four Dairy Farms in New Zealand. Dairy 2023, 4, 650-671. https://doi.org/10.3390/dairy4040045
Gupta TB, Brightwell G. Identification and Characterisation of Spore-Forming Bacteria in Bovine Raw Milk Collected from Four Dairy Farms in New Zealand. Dairy. 2023; 4(4):650-671. https://doi.org/10.3390/dairy4040045
Chicago/Turabian StyleGupta, Tanushree B., and Gale Brightwell. 2023. "Identification and Characterisation of Spore-Forming Bacteria in Bovine Raw Milk Collected from Four Dairy Farms in New Zealand" Dairy 4, no. 4: 650-671. https://doi.org/10.3390/dairy4040045
APA StyleGupta, T. B., & Brightwell, G. (2023). Identification and Characterisation of Spore-Forming Bacteria in Bovine Raw Milk Collected from Four Dairy Farms in New Zealand. Dairy, 4(4), 650-671. https://doi.org/10.3390/dairy4040045







