Two- and Three-Dimensional Computer Vision Techniques for More Reliable Body Condition Scoring
Abstract
:1. Introduction
2. Related Research
- Integrating with feeding systems to manage cow nutrition more effectively;
- Alerting the farmer to ill health/lameness promptly;
- Long-term monitoring of each animal attributes which can yield key information for animal husbandry, ethological studies and the development of PLF tools.
2.1. Deep Learning
2.2. Deep Learning
2.3. 3D Deep Learning
3. Materials and Methods
3.1. Camera Installation
3.2. Data Acquisition
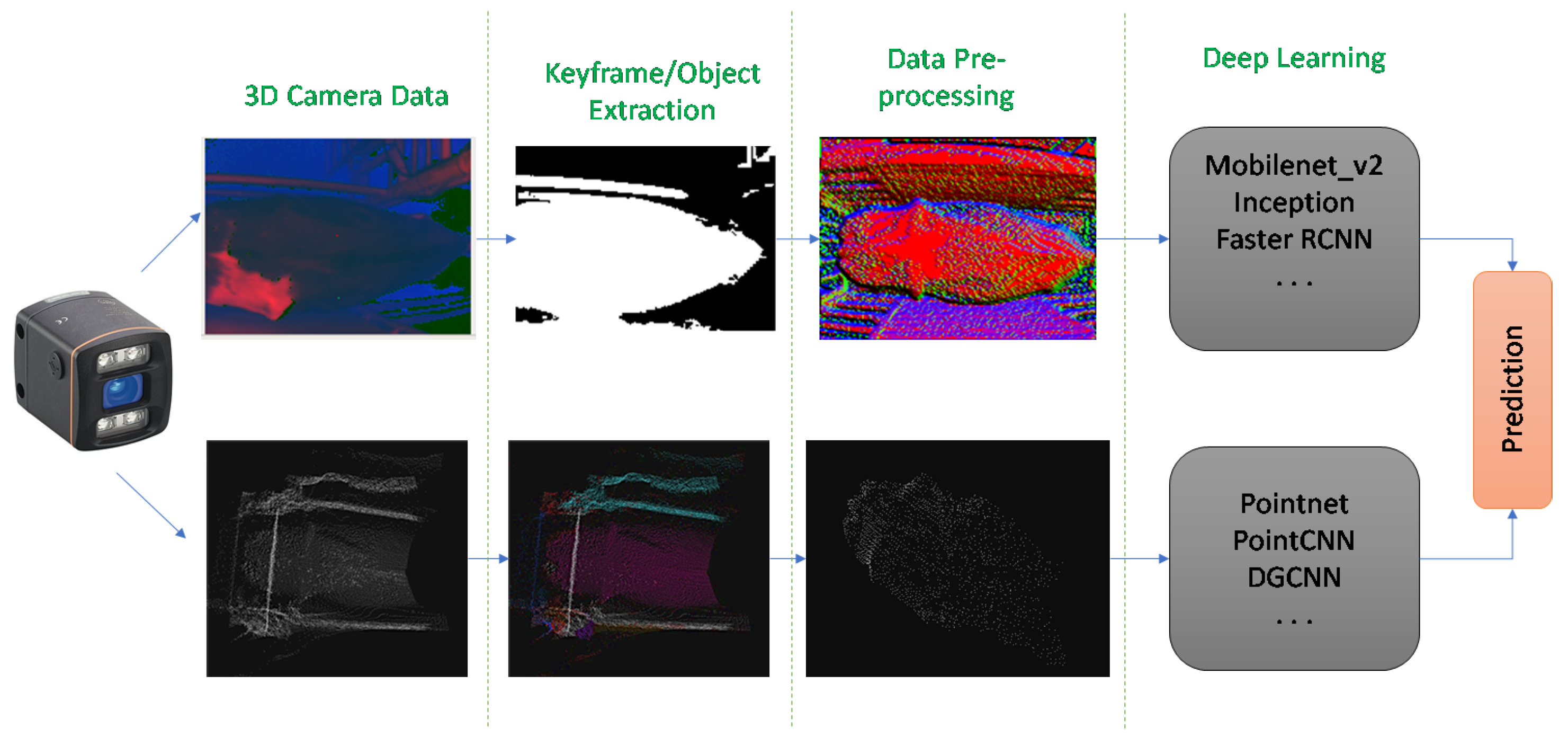
3.3. Image Processing
3.3.1. Keyframe Extraction
Depth-Based Foreground Extraction
Colour Thresholding
Difference Histogram
Depth Measurement at Image Keypoints
Normal Map Calculation
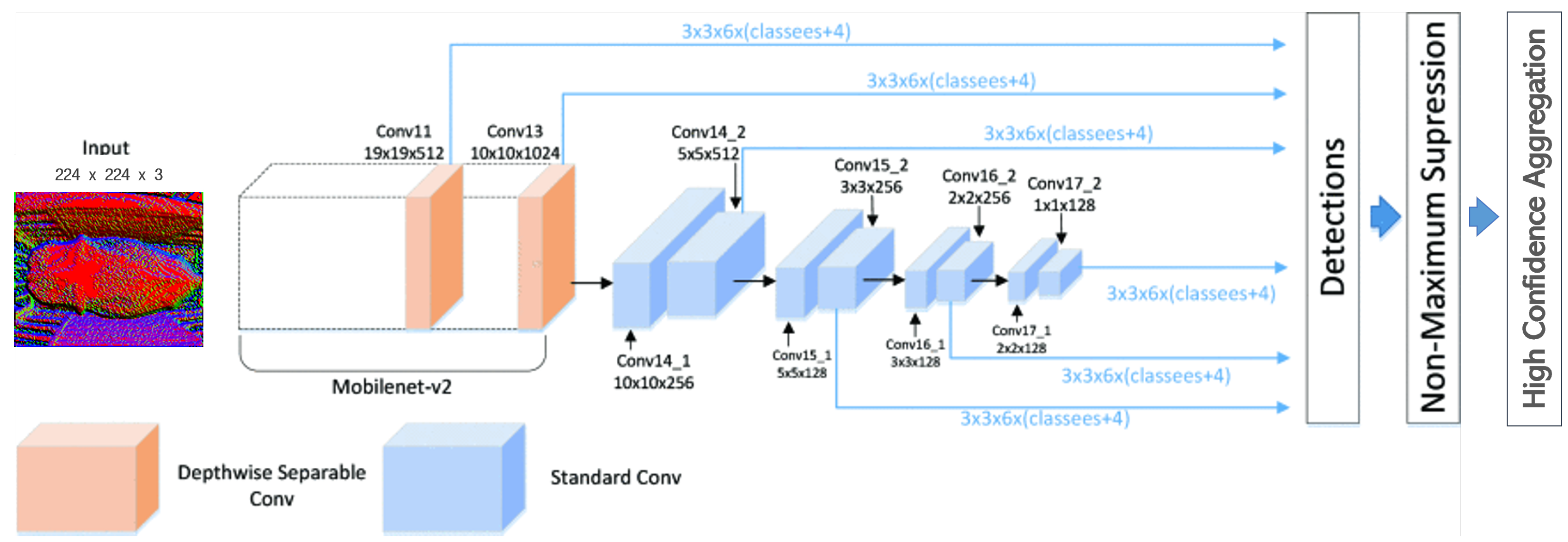
3.3.2. Image Classification
3.4. Point Cloud Processing
3.4.1. Dataset Preparation
Keyframe Extraction
Primitive Shape Matching Segmentation
Region Growing Point Cloud Segmentation
Point Cloud Annotation
3.4.2. Data Pre-Processing/Transformation/Point Cloud Downsampling
3.4.3. Point Cloud Classification
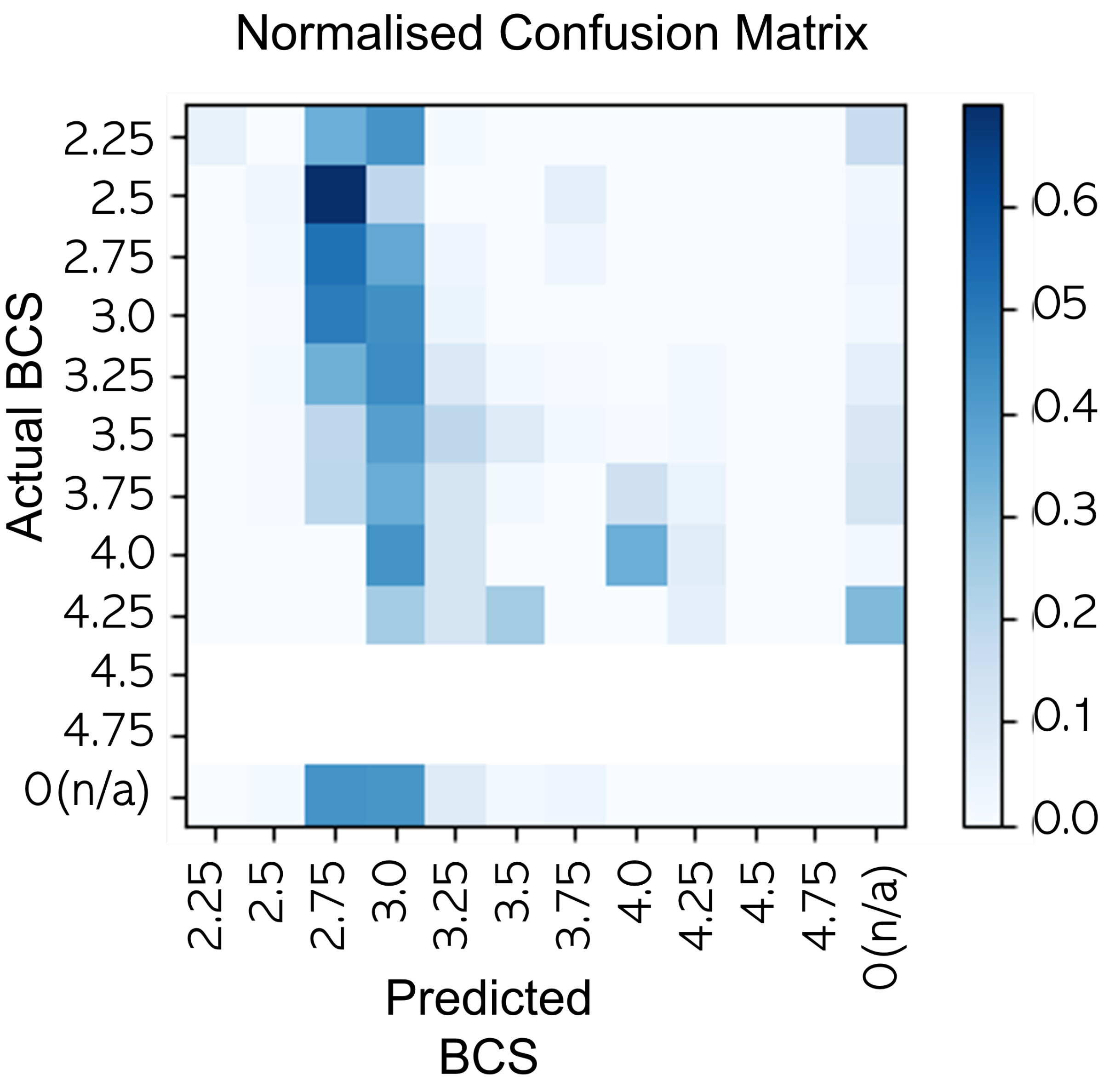
4. Results
5. Repeatability Evaluation for the Scoring Tasks
- The 4 reference BCS sores had a Krippendorff’s of 0.51 (inter-rater reliability)
- Each vet had a Krippendorff’s of 0.39 and 0.79, respectively, (intra-rater reliability)
- The Inferences morning and evening had a Krippendorff’s of 0.70 (machine reliability)
6. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
Abbreviations
| BCS | Body Condition Scoring |
| CNN | Convolutional Neural Network |
| DL | Deep Learning |
| PLF | Precision Livestock Farming |
| RANSAC | Random Sample Consensus |
References
- Schröder, U.J.; Staufenbiel, R. Invited review: Methods to determine body fat reserves in the dairy cow with special regard to ultrasonographic measurement of backfat thickness. J. Dairy Sci. 2006, 89, 1–14. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Deniz, A.U. The use of new practices for assessment of body condition score. Rev. Mvz CÓRdoba 2016, 21, 5154–5162. [Google Scholar]
- Roche, J.R.; Friggens, N.C.; Kay, J.K.; Fisher, M.W.; Stafford, K.J.; Berry, D.P. Body condition score and its association with dairy cow productivity, health, and welfare. J. Dairy Sci. 2009, 92, 5769–5801. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- O’Mahony, N.; Campbell, S.; Carvalho, A.; Krpalkova, L.; Riordan, D.; Walsh, J. 3D Vision for Precision Dairy Farming. IFAC-PapersOnLine 2019, 52, 312–317. [Google Scholar] [CrossRef]
- Silva, S.R.; Araujo, J.P.; Guedes, C.; Silva, F.; Almeida, M.; Cerqueira, J.L. Precision technologies to address dairy cattle welfare: Focus on lameness, mastitis and body condition. Animals 2021, 11, 2253. [Google Scholar] [CrossRef]
- Bewley, J.; Schutz, M. An Interdisciplinary Review of Body Condition Scoring for Dairy Cattle. Prof. Anim. Sci. 2008, 24, 507–529. [Google Scholar] [CrossRef] [Green Version]
- Halachmi, I.; Klopčič, M.; Polak, P.; Roberts, D.J.; Bewley, J.M. Automatic assessment of dairy cattle body condition score using thermal imaging. Comput. Electron. Agric. 2013, 99, 35–40. [Google Scholar] [CrossRef]
- Weber, A.; Salau, J.; Haas, J.H.; Junge, W.; Bauer, U.; Harms, J.; Suhr, O.; Schönrock, K.; Rothfuß, H.; Bieletzki, S.; et al. Estimation of backfat thickness using extracted traits from an automatic 3D optical system in lactating Holstein-Friesian cows. Livest. Sci. 2014, 165, 129–137. [Google Scholar] [CrossRef]
- Fischer, A.; Luginbühl, T.; Delattre, L.; Delouard, J.; Faverdin, P. Rear shape in 3 dimensions summarized by principal component analysis is a good predictor of body condition score in Holstein dairy cows. J. Dairy Sci. 2015, 98, 4465–4476. [Google Scholar] [CrossRef] [Green Version]
- Spoliansky, R.; Edan, Y.; Parmet, Y.; Halachmi, I. Development of automatic body condition scoring using a low-cost 3-dimensional Kinect camera. J. Dairy Sci. 2016, 99, 7714–7725. [Google Scholar] [CrossRef] [Green Version]
- Lynn, N.C.; Zin, T.T.; Kobayashi, I. Automatic Assessing Body Condition Score from Digital Images by Active Shape Model and Multiple Regression Technique. Proc. Int. Conf. Artif. Life Robot. 2017, 22, 311–314. [Google Scholar] [CrossRef]
- Nir, O.; Parmet, Y.; Werner, D.; Adin, G.; Halachmi, I. 3D Computer-vision system for automatically estimating heifer height and body mass. Biosyst. Eng. 2017, 173, 4–10. [Google Scholar] [CrossRef]
- Hansen, M.F.; Smith, M.L.; Smith, L.N.; Abdul Jabbar, K.; Forbes, D. Automated monitoring of dairy cow body condition, mobility and weight using a single 3D video capture device. Comput. Ind. 2018, 98, 14–22. [Google Scholar] [CrossRef]
- Rodríguez Alvarez, J.; Arroqui, M.; Mangudo, P.; Toloza, J.; Jatip, D.; Rodríguez, J.M.; Teyseyre, A.; Sanz, C.; Zunino, A.; Machado, C.; et al. Body condition estimation on cows from depth images using Convolutional Neural Networks. Comput. Electron. Agric. 2018, 155, 12–22. [Google Scholar] [CrossRef] [Green Version]
- Mullins, I.L.; Truman, C.M.; Campler, M.R.; Bewley, J.M.; Costa, J.H. Validation of a commercial automated body condition scoring system on a commercial dairy farm. Animals 2019, 9, 287. [Google Scholar] [CrossRef] [Green Version]
- An, W.; Jirkof, P.; Hohlbaum, K.; Albornoz, R.I.; Giri, K.; Hannah, M.C.; Wales, W.J. An Improved Approach to Automated Measurement of Body Condition Score in Dairy Cows Using a Three-Dimensional Camera System. Animals 2021, 12, 72. [Google Scholar] [CrossRef]
- Martins, B.; Mendes, A.; Silva, L.; Moreira, T.; Costa, J.; Rotta, P.; Chizzotti, M.; Marcondes, M. Estimating body weight, body condition score, and type traits in dairy cows using three dimensional cameras and manual body measurements. Livest. Sci. 2020, 236, 104054. [Google Scholar] [CrossRef]
- Salau, J.; Haas, J.H.; Junge, W.; Bauer, U.; Harms, J.; Bieletzki, S. Feasibility of automated body trait determination using the SR4K time-of-flight camera in cow barns. SpringerPlus 2014, 3, 225. [Google Scholar] [CrossRef] [Green Version]
- Salau, J.; Haas, J.H.; Junge, W.; Thaller, G. Extrinsic calibration of a multi-Kinect camera scanning passage for measuring functional traits in dairy cows. Biosyst. Eng. 2016, 151, 409–424. [Google Scholar] [CrossRef]
- Salau, J.; Haas, J.H.; Junge, W.; Thaller, G. A multi-Kinect cow scanning system: Calculating linear traits from manually marked recordings of Holstein-Friesian dairy cows. Biosyst. Eng. 2017, 157, 92–98. [Google Scholar] [CrossRef]
- Alvarez, J.R.; Arroqui, M.; Mangudo, P.; Toloza, J.; Jatip, D.; Rodriguez, J.M.; Teyseyre, A.; Sanz, C.; Zunino, A.; Machado, C.; et al. Estimating body condition score in dairy cows from depth images using convolutional neural networks, transfer learning and model ensembling techniques. Agronomy 2019, 9, 90. [Google Scholar] [CrossRef]
- Abdul Jabbar, K.; Hansen, M.F.; Smith, M.L.; Smith, L.N. Early and non-intrusive lameness detection in dairy cows using 3-dimensional video. Biosyst. Eng. 2017, 153, 63–69. [Google Scholar] [CrossRef]
- Rind Thomasen, J.; Lassen, J.; Gunnar Brink Nielsen, G.; Borggard, C.; René, P.; Stentebjerg, B.; Hansen, R.H.; Hansen, N.W.; Borchersen, S. Individual cow identification in a commercial herd using 3D camera technology. In Proceedings of the World Congress on Genetics Applied to Livestock Production, Rotterdam, The Netherlands, 22 June 2018; Volume 11, p. 613. [Google Scholar]
- Arslan, A.C.; Akar, M.; Alagoz, F. 3D cow identification in cattle farms. In Proceedings of the 2014 22nd Signal Processing and Communications Applications Conference (SIU), Trabzon, Turkey, 23–25 April 2014; pp. 1347–1350. [Google Scholar] [CrossRef]
- O’Mahony, N.; Campbell, S.; Carvalho, A.; Harapanahalli, S.; Velasco-Hernández, G.A.; Riordan, D.; Walsh, J. Adaptive Multimodal Localisation Techniques for Mobile Robots in Unstructured Environments A Review. In Proceedings of the IEEE 5th World Forum on Internet of Things (WF-IoT), Limerick, Ireland, 15–18 April 2019. [Google Scholar]
- Corporation, I. Intel ® RealSense ™ Camera: Depth Testing Methodology; Technical Report; Intel Corporation: Santa Clara, CA, USA, 2018. [Google Scholar]
- IFM Electronic Gmbh. O3D313—3D Camera—ifm; IFM Electronic Gmbh: Essen, Germany, 2018. [Google Scholar]
- Zhang, M.; Zhang, L.; Cheng, H.D. A neutrosophic approach to image segmentation based on watershed method. Signal Process. 2010, 90, 1510–1517. [Google Scholar] [CrossRef]
- Holzer, S.; Rusu, R.B.; Dixon, M.; Gedikli, S.; Navab, N. Adaptive neighborhood selection for real-time surface normal estimation from organized point cloud data using integral images. In Proceedings of the IEEE International Conference on Intelligent Robots and Systems, Vilamoura-Algarve, Portugal, 7–12 October 2012; pp. 2684–2689. [Google Scholar] [CrossRef]
- Keras. Backend—Keras Documentation; Keras.io: San Francisco, CA, USA, 2018. [Google Scholar]
- PyTorch. PyTorch, The PyTorch Foundation; Warsaw: Mazowieckie, Poland, 2019. [Google Scholar]
- Matlab, Unsupervised Learning—MATLAB & Simulink; Matlab: Mathworks, MA, USA, 2016.
- Google. Google AI Blog: MobileNets: Open-Source Models for Efficient On-Device Vision; Technical Report; Google: San Francisco, CA, USA, 2017. [Google Scholar]
- Der Chien, W. An Evaluation of TensorFlow as a Programming Framework for HPC Applications. Masters Thesis, KTH Royal Institute of Technology, Stockholm, Sweden, 2018. [Google Scholar]
- Chiu, Y.C.; Tsai, C.Y.; Ruan, M.D.; Shen, G.Y.; Lee, T.T. Mobilenet-SSDv2: An Improved Object Detection Model for Embedded Systems. In Proceedings of the 2020 International Conference on System Science and Engineering (ICSSE), Kagawa, Japan, 3 September 2020. [Google Scholar] [CrossRef]
- Rusu, R.B.; Cousins, S. 3D is Here: Point Cloud Library (PCL). In Proceedings of the Proceedings—IEEE International Conference on Robotics and Automation, Shanghai, China, 9–13 May 2011; pp. 1–4. [Google Scholar] [CrossRef] [Green Version]
- Goldenshluger, A.; Zeevi, A. Hough Transform. Estim. Ann. Stat. 2004, 32, 1908–1932. [Google Scholar] [CrossRef] [Green Version]
- Li, L.; Yang, F.; Zhu, H.; Li, D.; Li, Y.; Tang, L. An improved RANSAC for 3D point cloud plane segmentation basedon normal distribution transformation cells. Remote. Sens. 2017, 9, 433. [Google Scholar] [CrossRef] [Green Version]
- Jin, Y.H.; Lee, W.H. Fast cylinder shape matching using random sample consensus in large scale point cloud. Appl. Sci. 2019, 9, 974. [Google Scholar] [CrossRef] [Green Version]
- Vo, A.V.; Truong-Hong, L.; Laefer, D.F.; Bertolotto, M. Octree-based region growing for point cloud segmentation. ISPRS J. Photogramm. Remote. Sens. 2015, 104, 88–100. [Google Scholar] [CrossRef]
- O’Mahony, N.; Campbell, S.; Carvalho, A.; Krpalkova, L.; Riordan, D.; Walsh, J. Point cloud annotation methods for 3D deep learning. In Proceedings of the International Conference on Sensing Technology, ICST, Sydney, Australia, 2–4 December 2019; pp. 274–279. [Google Scholar] [CrossRef]
- Jain, S.; Munukutla, S.; Held, D. Few-Shot Point Cloud Region Annotation with Human in the Loop. In Proceedings of the ICML Workshop on Human in the Loop Learning (HILL 2019), Long Beach, CA, USA, 14 June 2019. [Google Scholar]
- Jiang, B.; Wu, Q.; Yin, X.; Wu, D.; Song, H.; He, D. FLYOLOv3 deep learning for key parts of dairy cow body detection. Comput. Electron. Agric. 2019, 166, 104982. [Google Scholar] [CrossRef]
- Qi, C.R.; Liu, W.; Wu, C.; Su, H.; Guibas, L.J. Frustum PointNets for 3D Object Detection from RGB-D Data. In Proceedings of the IEEE Computer Society Conference on Computer Vision and Pattern Recognition, Salt Lake City, UT, USA, 22 June 2018; pp. 918–927. [Google Scholar] [CrossRef] [Green Version]
- Wang, Y.; Sun, Y.; Liu, Z.; Sarma, S.E.; Bronstein, M.M.; Solomon, J.M. Dynamic Graph Cnn for Learning on Point Clouds. ACM Trans. Graph. 2019, 38, 5. [Google Scholar] [CrossRef] [Green Version]
- Liu, Y.; Fan, B.; Xiang, S.; Pan, C. Relation-shape convolutional neural network for point cloud analysis. In Proceedings of the IEEE Computer Society Conference on Computer Vision and Pattern Recognition, Long Beach, CA, USA, 20 June 2019; pp. 8887–8896. [Google Scholar] [CrossRef] [Green Version]
- Li, Y.; Bu, R.; Sun, M.; Wu, W.; Di, X.; Chen, B. PointCNN: Convolution on X-transformed points. Adv. Neural Inf. Process. Syst. 2018, 31, 820–830. [Google Scholar]
- Kristensen, E.; Dueholm, L.; Vink, D.; Andersen, J.E.; Jakobsen, E.B.; Illum-Nielsen, S.; Petersen, F.A.; Enevoldsen, C. Within- and across-person uniformity of body condition scoringin Danish Holstein cattle. J. Dairy Sci. 2006, 89, 3721–3728. [Google Scholar] [CrossRef]
- Gwet, K.L. On The Krippendorff’s Alpha Coefficient; Technical Report; 2011; Manuscript submitted for publication. [Google Scholar]
















| Features | Reference | Images in Dataset | Automated/ 3D/2D | Performance |
|---|---|---|---|---|
| Hook angle, posterior hook angle, depression | [6] | 834 | No/2D | 92.79 (note manual input is required) |
| Goodness of fit of a parabolic shape of the segmented image | [7] | 172 | Yes/2D | R = 0.94 |
| Measurement between specific points on a cows back | [8] | - | No/3D | Area estimate only |
| Principal Component Analysis | [9] | 25 | No/3D | R = 0.96 |
| 14 individual features per cow, derived from cows’ topography | [10] | 2650 | Yes/3D | 74% accurate within 0.25 |
| Area around the tailhead and left and right hooks | [11] | 130 | Yes/2D | Area estimate only |
| Body mass, hip height and withers height | [12] | 107 | Yes/3D | R2 = 0.946 (body mass estimation) |
| 3D surface of cows back and fitted sphere | [13] | 95 | Yes/3D | Area estimate only |
| Features determined by CNN on pre-processed depth images | [14] | 503 | Yes/3D | 0.78 accurate within 0.25 |
| Proprietary BCS system | [15] | 344 | Yes | 0.76 correlation |
| Refinement of [15] with smoothing filter | [16] | 32 | Yes | 0.86 Pearson correlation |
| Manual body measurements | [17] | 55 | No | R2 of 0.63 and RMSE of 0.16 |
| Input Data | Location | Model | Inference Time (ms) | Classification Accuracy |
|---|---|---|---|---|
| IP Camera RGB Image | Drafting crate | Mobilenet V2 | 62 | 0.25 |
| Depth Image | Drafting crate | Mobilenet V2 | 30 | 0.26 |
| Depth Image | Drafting crate | Inception | 50 | 0.24 |
| Composite Image | Drafting crate | Mobilenet V2 | 30 | 0.29 |
| Normal Map | Drafting crate | Mobilenet V2 | 30 | 0.39 |
| Normal Map | Drafting crate | Mobilenet V1 FPN | 30 | 0.38 |
| Input Data | Location | Preprocessing | Model | Classification Accuracy |
|---|---|---|---|---|
| Convolution-based | Rotary | Region-growing segmentation + outlier removal + Normal-Space Subsampling | Relation-Shape CNN | 0.185 |
| Graph-based | Rotary | Region-growing segmentation + outlier removal + Normal-Space Subsampling | DGCNN | 0.205 |
| Convolution-based | Rotary | Region-growing segmentation + outlier removal + Normal-Space Subsampling | PointCNN | 0.394 |
| Point-based | Draft | Region-of-Interest segmentation + minor Normal-Space Subsampling + block merging | Pointnet++ | 0.53 |
| Rater 1a | Rater 1b | Rater 2a | Rater 2b | Algorithm 1a | Algorithm 1b | Algorithm 2a | Algorithm 2b | |
|---|---|---|---|---|---|---|---|---|
| Rater 1a | 1.0000 | |||||||
| Rater 1b | 0.4920 | 1.0000 | ||||||
| 0.6890 | ||||||||
| 1.0000 | ||||||||
| 1.0000 | ||||||||
| 1.0000 | ||||||||
| Rater 2a | 0.2270 | 0.2850 | 1.0000 | |||||
| 0.4750 | 0.5250 | |||||||
| 0.9340 | 0.9180 | |||||||
| 0.9840 | 1.0000 | |||||||
| 1.0000 | 1.0000 | |||||||
| Rater 2b | 0.1570 | 0.1900 | 0.3950 | 1.0000 | ||||
| 0.4750 | 0.5080 | 0.6070 | ||||||
| 0.9180 | 0.9180 | 0.9020 | ||||||
| 1.0000 | 0.9840 | 0.9670 | ||||||
| 1.0000 | 1.0000 | 1.0000 | ||||||
| Algorithm 1a | −0.1683 | −0.2307 | −0.2228 | −0.2228 | 1.0000 | |||
| 0.1633 | 0.1224 | 0.0612 | 0.0612 | |||||
| 0.6735 | 0.6531 | 0.4286 | 0.4286 | |||||
| 0.8776 | 0.8980 | 0.7959 | 0.7959 | |||||
| 0.9796 | 1.0000 | 0.9388 | 0.9388 | |||||
| Algorithm 1b | −0.1412 | −0.2074 | −0.1950 | −0.2076 | 0.1570 | 1.0000 | ||
| 0.1429 | 0.1020 | 0.0408 | 0.0612 | 0.4750 | ||||
| 0.4082 | 0.4490 | 0.2857 | 0.2653 | 0.9180 | ||||
| 0.8571 | 0.8776 | 0.6939 | 0.6531 | 1.0000 | ||||
| 1.0000 | 1.0000 | 0.8980 | 0.9184 | 1.0000 | ||||
| Algorithm 2a | −0.0530 | 0.0267 | −0.0339 | −0.0415 | −0.0003 | 0.0452 | 1.0000 | |
| 0.2857 | 0.3469 | 0.2245 | 0.2653 | 0.3265 | 0.3469 | |||
| 0.7551 | 0.7755 | 0.5510 | 0.5306 | 0.8571 | 0.7755 | |||
| 0.9796 | 1.0000 | 0.9184 | 0.8776 | 1.0000 | 0.9388 | |||
| 1.0000 | 1.0000 | 1.0000 | 1.0000 | 1.0000 | 1.0000 | |||
| Algorithm 2b | 0.0484 | 0.0118 | −0.0762 | 0.0290 | 0.3878 | 0.2245 | 0.3720 | 1.0000 |
| 0.3878 | 0.3673 | 0.2245 | 0.3878 | 0.8776 | 0.6939 | 0.5918 | ||
| 0.7551 | 0.8163 | 0.6327 | 0.8776 | 0.9592 | 0.9388 | 0.9592 | ||
| 1.0000 | 0.9796 | 0.8776 | 0.9592 | 1.0000 | 1.0000 | 1.0000 | ||
| 1.0000 | 1.0000 | 1.0000 | 1.0000 | 1.0000 | 1.0000 | 1.0000 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
O’Mahony, N.; Krpalkova, L.; Sayers, G.; Krump, L.; Walsh, J.; Riordan, D. Two- and Three-Dimensional Computer Vision Techniques for More Reliable Body Condition Scoring. Dairy 2023, 4, 1-25. https://doi.org/10.3390/dairy4010001
O’Mahony N, Krpalkova L, Sayers G, Krump L, Walsh J, Riordan D. Two- and Three-Dimensional Computer Vision Techniques for More Reliable Body Condition Scoring. Dairy. 2023; 4(1):1-25. https://doi.org/10.3390/dairy4010001
Chicago/Turabian StyleO’Mahony, Niall, Lenka Krpalkova, Gearoid Sayers, Lea Krump, Joseph Walsh, and Daniel Riordan. 2023. "Two- and Three-Dimensional Computer Vision Techniques for More Reliable Body Condition Scoring" Dairy 4, no. 1: 1-25. https://doi.org/10.3390/dairy4010001
APA StyleO’Mahony, N., Krpalkova, L., Sayers, G., Krump, L., Walsh, J., & Riordan, D. (2023). Two- and Three-Dimensional Computer Vision Techniques for More Reliable Body Condition Scoring. Dairy, 4(1), 1-25. https://doi.org/10.3390/dairy4010001







