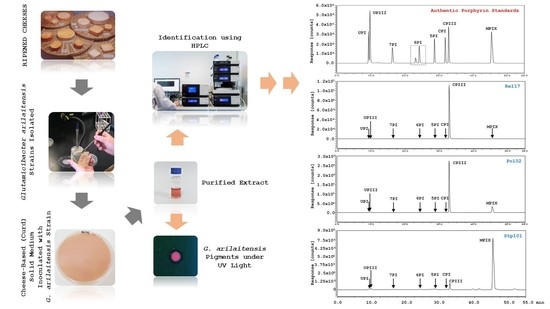
Identification of Red Pigments Produced by Cheese-Ripening Bacterial Strains of Glutamicibacter arilaitensis Using HPLC
Abstract
1. Introduction
2. Materials and Methods
2.1. Strains and Cultures
2.1.1. Bacterial and Yeast Strains
2.1.2. Cultures
2.2. Sample Preparation
2.2.1. Pigment Production
2.2.2. Pigment Extraction and Purification
2.3. High-Performance Liquid Chromatography Analysis
2.3.1. Apparatus
2.3.2. Chromatographic Condition
3. Results
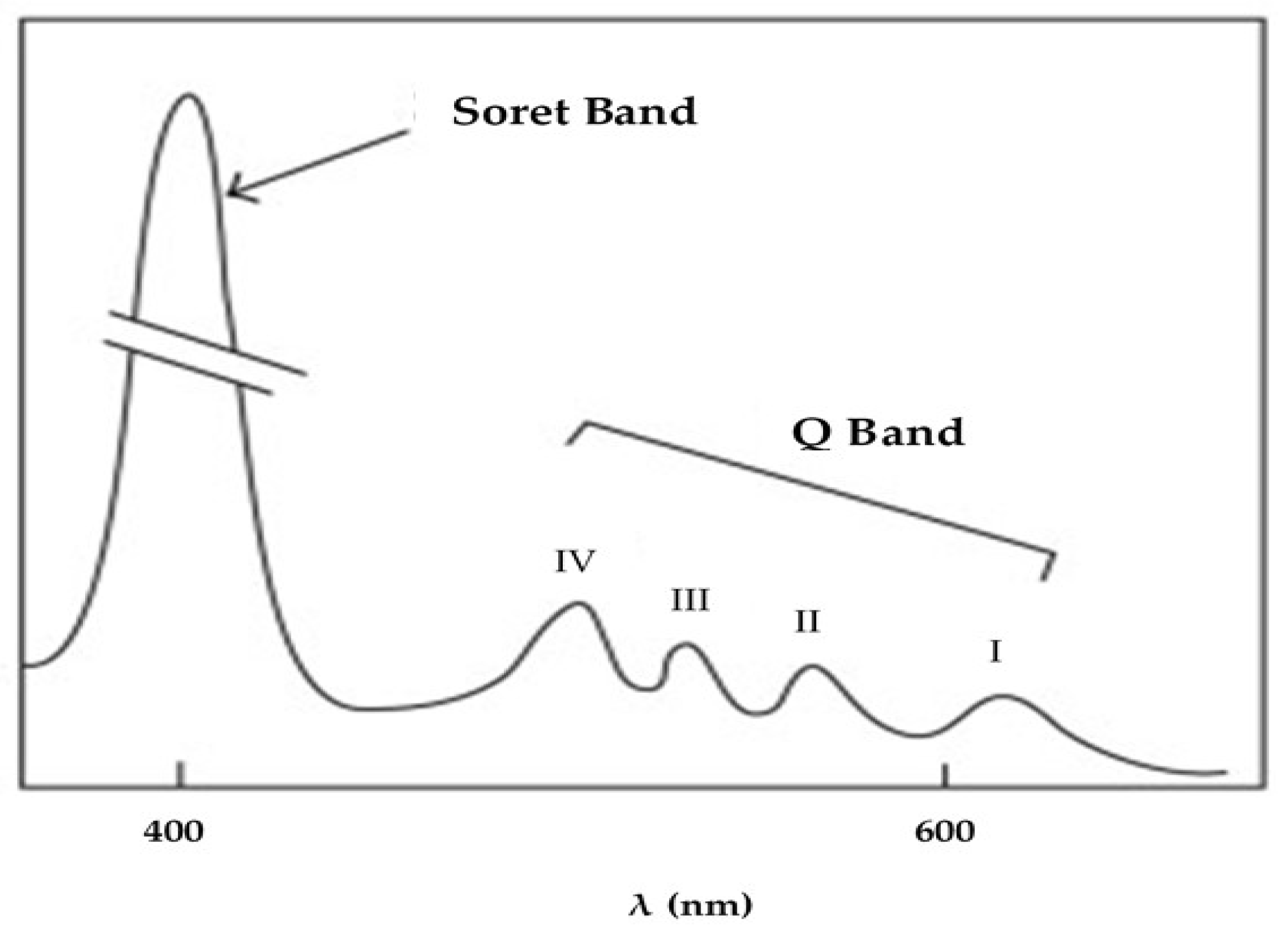
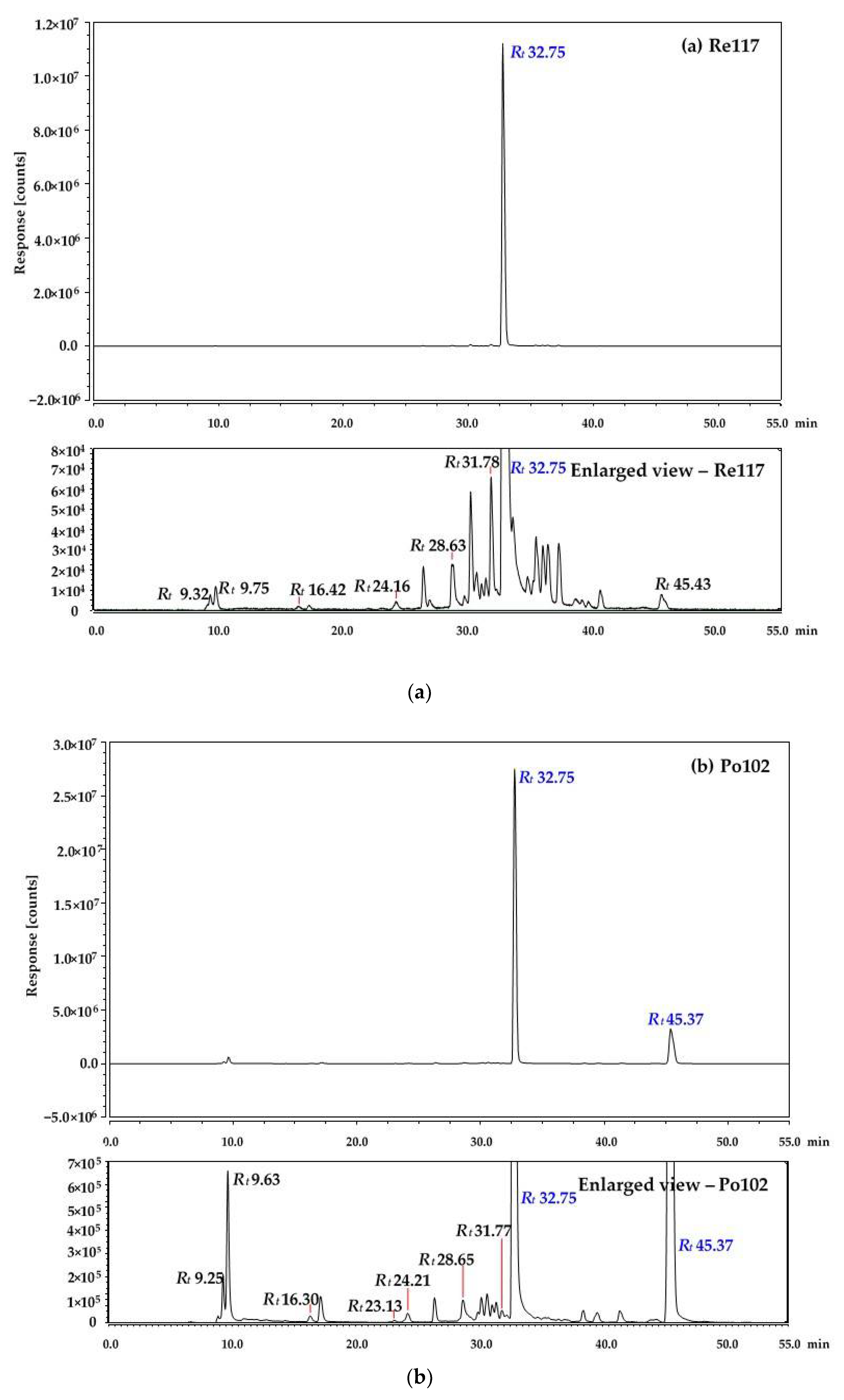
3.1. HPLC Separation of Porphyrins
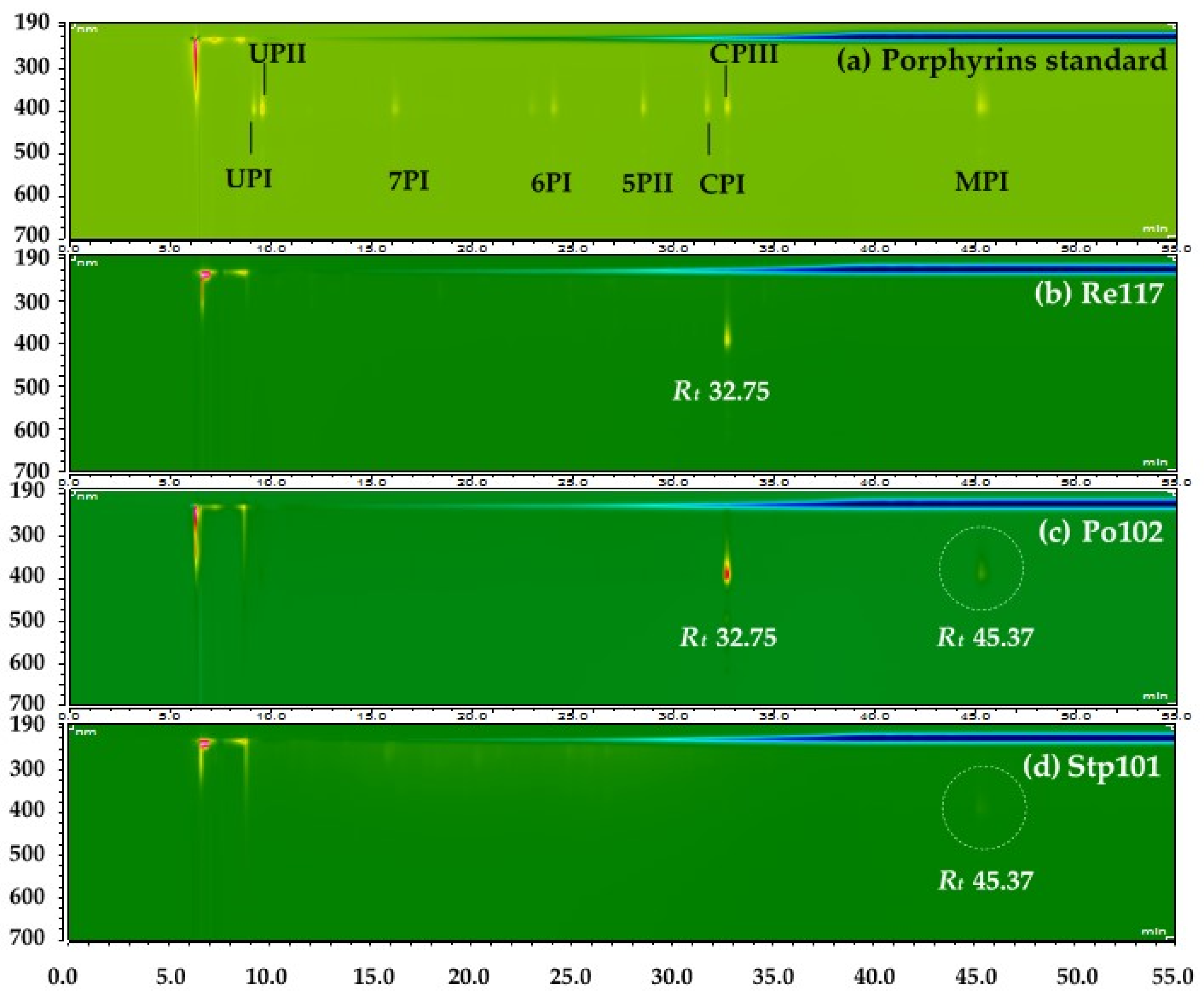
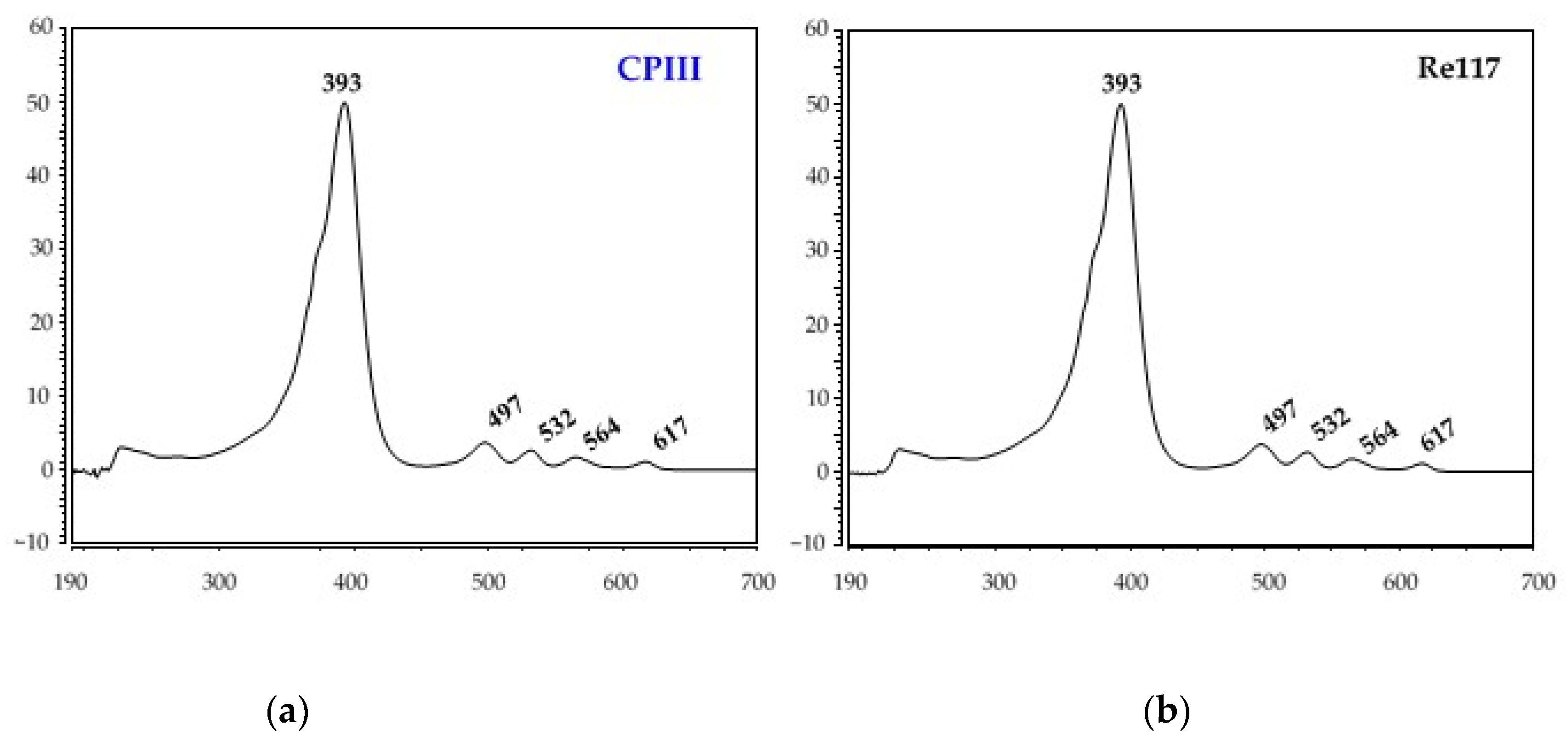
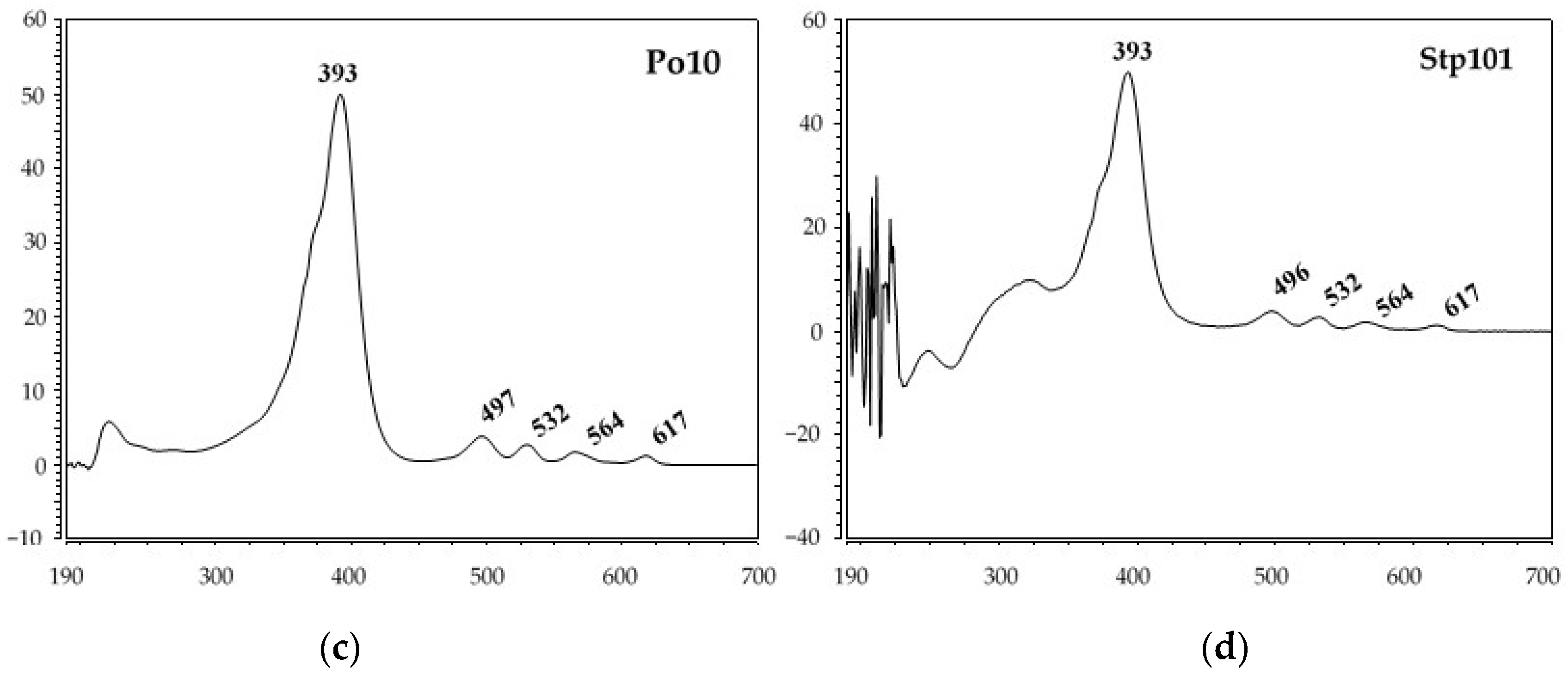
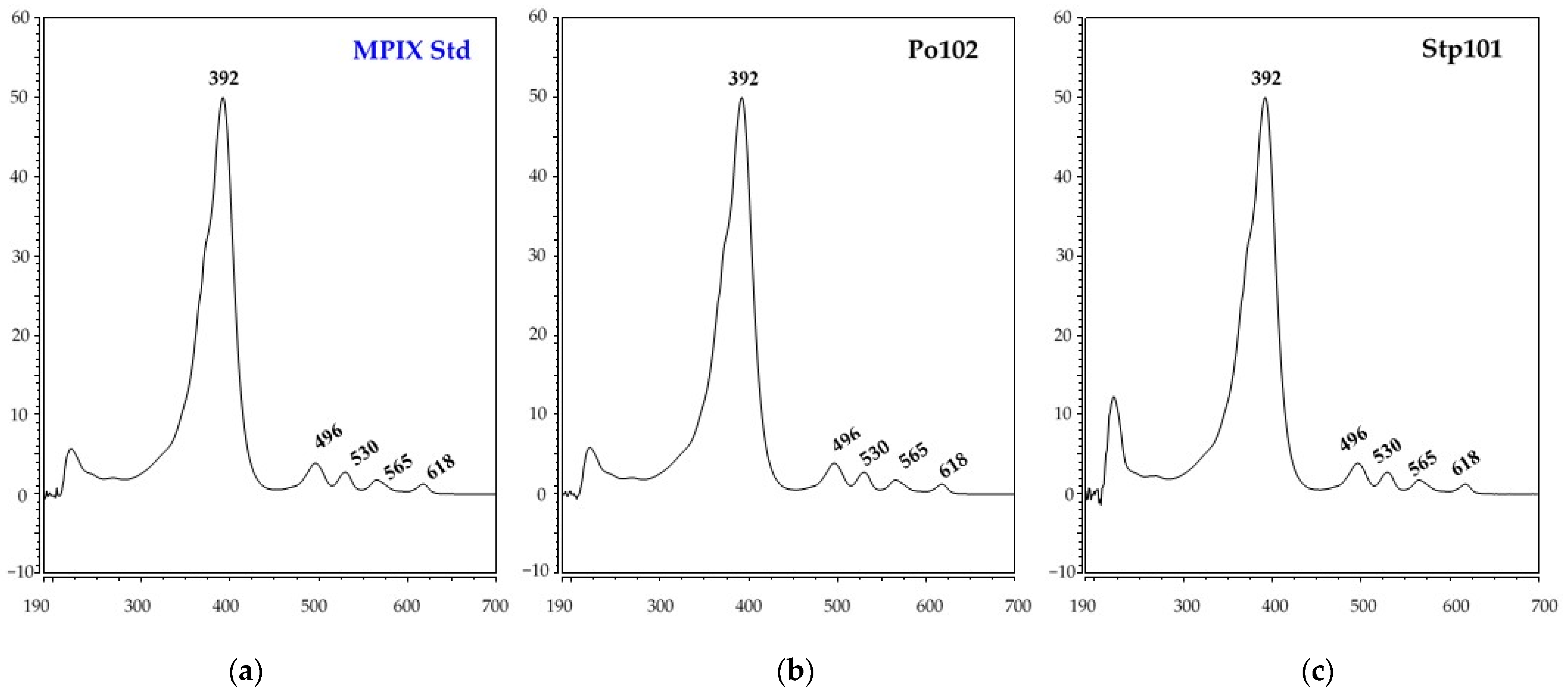
3.2. Identification of Red Pigments Produced by G. arilaitensis Strains
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
Appendix A
References
- Mounier, J.; Goerges, S.; Gelsomino, R.; Vancanneyt, M.; Vandemeulebroecke, K.; Hoste, B.; Brennen, N.M.; Scherer, S.; Swings, J.; Fitzgerald, G.F.; et al. Sources of the adventitious microflora of a smear-ripened cheese. J. Appl. Microbiol. 2006, 101, 668–681. [Google Scholar] [CrossRef] [PubMed]
- Ritschard, J.S.; Amato, L.; Kumar, Y.; Müller, B.; Meile, L.; Schuppler, M. The role of the surface smear microbiome in the development of defective smear on surface-ripened red-smear cheese. AIMS Microbiol. 2018, 4, 622–641. [Google Scholar] [CrossRef]
- Speight, K.C.; Schiano, A.N.; Harwood, W.S.; Drake, M.A. Consumer insights on prepackaged cheddar cheese shreds using focus groups, conjoint analysis, and qualitative multivariate analysis. J. Dairy Sci. 2019, 102, 6971–6986. [Google Scholar] [CrossRef] [PubMed]
- Daly, D.F.M.; McSweeney, P.L.H.; Sheehan, J.J. Pink discoloration defect in commercial cheese: A review. Dairy Sci. Technol. 2012, 92, 439–453. [Google Scholar] [CrossRef]
- Bottazzi, V.; Cappa, F.; Scolari, G.; Parisi, M.G. Occurring of pink discoloration in Grana cheese made with one-strain starter culture. Sci. Tec. Latt. Casearia 2000, 51, 67–74. [Google Scholar]
- Marley, F.G.; Michel, V. Pinkish colouration in Cheddar cheese—Description and factors contributing to its formation. J. Dairy Res. 2001, 68, 327–332. [Google Scholar] [CrossRef] [PubMed]
- Andersen, C.M.; Wold, J.P.; Mortensen, G. Light-induced changes in semi-hard cheese determined by fluorescence spectroscopy and chemometrics. Int. Dairy J. 2006, 16, 1483–1489. [Google Scholar] [CrossRef]
- Guiliano, G.; Rosati, C.; Bramly, P.M. To dye or not to dye: Biochemistry of annatto unveiled. Trends Biotechnol. 2003, 21, 513–516. [Google Scholar] [CrossRef]
- Sharma, P.; Segat, A.; Kelly, A.; Sheehan, J.J. Colorants in cheese manufacture: Production, chemistry, interaction, and regulation. Compr. Rev. Food Sci. Food Saf. 2020, 19, 1220–1242. [Google Scholar] [CrossRef] [PubMed]
- Quigley, L.; O’Sullivan, D.J.; Daly, D.; O’Sullivan, O.; Burdikova, Z.; Vana, R.; Beresford, T.P.; Ross, R.P.; Fitzgerald, G.F.; McSweeney, P.L.H.; et al. Thermus and the pink discoloration defect in cheese. Appl. Environ. Sci. 2016, 1, e00023-16. [Google Scholar] [CrossRef]
- Bockelmann, W. Smear-ripened cheeses. In Encyclopedia of Dairy Sciences, 2nd ed.; Fox, P.F., McSweeney, P.L.H., Eds.; Elsevier Inc.: Amsterdam, The Netherlands, 2011; pp. 753–766. [Google Scholar]
- Flegler, A.; Runzheimer, K.; Kombeitz, V.; Mänz, A.T.; von Heilborn, D.H.; Etzbach, L.; Schieber, A.; Hőlzl, G.; Hüttler, B.; Woehle, C.; et al. Arthrobacter bussei sp. nov., a pink-colored organism isolated from cheese made of cow’s milk. Int. J. Syst. Evol. Microbiol. 2020, 70, 3027–3036. [Google Scholar] [CrossRef]
- Arai, A.; Igoshi, A.; Inoue, A.; Noda, K.; Tsutsuura, S.; Murata, M. Relationship between lactose utilization of lactic acid bacteria and browning of cheese during storage. Biosci. Biotechnol. Biochem. 2020, 84, 1886–1893. [Google Scholar] [CrossRef]
- Jonnala, B.R.Y. Pink Discoloration in Cheese. Ph.D. Thesis, University College, London, UK, 14 January 2020. Available online: http://hdl.handle.net/10468/9501 (accessed on 22 July 2021).
- Larpin-Laborde, S.; Imran, M.; Bonaïti, C.; Bora, N.; Gelsomino, R.; Goerges, S.; Irlinger, F.; Goodfellow, M.; Ward, A.C.; Vancanneyt, M.; et al. Surface microbial consortia from Livarot, a French smear-ripened cheese. Can. J. Microbiol. 2011, 57, 651–660. [Google Scholar] [CrossRef]
- Irlinger, F.; Bimet, G.; Delettre, J.; Lefèvre, M.; Grimont, P.A.D. Arhtrobacter bergerei sp. nov. and Arthrobacter arilaitensis sp. nov., novel corynform species isolated from the surface of cheeses. Int. J. Syst. Evol. Microbiol. 2005, 55, 457–462. [Google Scholar] [CrossRef] [PubMed]
- Sutthiwong, S.; Dufossé, L. Production of carotenoids by Arthrobacter arilaitensis strains isolated from smear-ripened cheeses. FEMS Microbiol. Lett. 2014, 174–181. [Google Scholar] [CrossRef] [PubMed]
- Guiffrida, D.; Sutthiwong, N.; Dugo, P.; Donato, P.; Cacciola, F.; Girad-Valenciennes, E.; Le Mao, Y.; Monnet, C.; Fouillaud, M.; Caro, Y.; et al. Characterisation of the C50 carotenoids produced by strains of the cheese-ripening bacterium Arthrobacter arilaitensis. Int. Dairy J. 2016, 55, 10–16. [Google Scholar] [CrossRef]
- Sutthiwong, N.; Fouillaud, M.; Dufossé, L. The influence of pH, NaCl and the deacidifying yeasts Debaryomyces hansenii and Kluyveromyces marxianus on the production of pigments by the cheese-ripening bacteria Arthobacter arilaitensis. Foods 2018, 7, 190. [Google Scholar] [CrossRef] [PubMed]
- Cleary, J.L.; Kolachina, S.; Wolfe, B.E.; Sanchez, L.M. Coproporphyrins III produced by the bacterium Glutamicibacter arilaitensis binds zinc and is upregulated by fungi in cheese rinds. Appl. Environ. Sci. 2018, 3, e0036-18. [Google Scholar] [CrossRef]
- Dong, M.W. The essence of modern HPLC: Advantages, limitations, fundamentals, and opportunities. LCGC North Am. 2013, 31, 472–479. [Google Scholar]
- Núñez, O.; Lucci, P. Application of liquid chromatography in food analysis. Foods 2020, 9, 1277. [Google Scholar] [CrossRef]
- Galaup, P.; Sutthiwong, N.; Leclercq-Perlat, M.-N.; Valla, A.; Caro, Y.; Fouillaud, M.; Guérard, F.; Dufossé, L. First isolation of Brevibacterium sp. pigments in the rind of an industrial red-smear-ripened soft cheese. Int. J. Dairy Technol. 2015, 68, 144–147. [Google Scholar] [CrossRef]
- Kumura, H.; Ohtsuyama, T.; Taitoh, M.; Koyanagi, H.; Kobayashi, K.; Wakamatsu, J.-I.; Ishizuka, S. Application of red pigment producing edible fungi for development of a novel type of functional cheese. J. Food Process Preserv. 2018, e13707. [Google Scholar] [CrossRef]
- Larroude, M.; Onésime, D.; Rué, O.; Nicaud, J.-M.; Rossignol, T. A Yarrowia lipolytica engineered for pyomelanin production. Microorganisms 2021, 9, 838. [Google Scholar] [CrossRef]
- Benton, C.M.; Lim, C.K. Liquid chromatography: Porphyrins. In Encyclopedia of Analytical Science, 3rd ed.; Worsfold, P.J., Poole, C.F., Townshend, A., Miró, M., Eds.; Elsevier Inc.: Amsterdam, The Netherlands, 2013; pp. 209–230. [Google Scholar]
- Vogelsang, D.F.; Maleczka, R.E., Jr.; Lee, A. HPLC characterization of cis and trans mixtures of double-decker shaped silsesquioxanes. Silicon 2018, 11, 5–13. [Google Scholar] [CrossRef]
- Irlinger, F.; Layec, S.; Hélinck, S.; Dugat-Bony, E. Cheese rind microbial communities: Diversity, composition and origin. FEMS Microbiol. Lett. 2015, 362, 1–11. [Google Scholar] [CrossRef]
- Giovannetti, R. The use of spectrophotometry UV-Vis for the study of porphyrins. In Macro to Nano Spectroscopy, 1st ed.; Uddin, J., Ed.; IntechOpen Ltd.: London, UK, 2012. [Google Scholar] [CrossRef]
- Valicsek, Z.; Horváth, O. Application of the electronic spectra of porphyrins for analytical purposes: The effects of metal ions and structural distortions. Microchem. J. 2013, 107, 47–62. [Google Scholar] [CrossRef]
- Mamardashvili, G.M.; Lazovskiy, D.A.; Khodov, I.A.; Efimov, A.E.; Mamardashvil, N.Z. New polyporphyrin arrays with controlled fluorescence obtained by diaxial Sn(IV)-porphyrin phenolates chelation with Cu2+ cation. Polymers 2021, 13, 829. [Google Scholar] [CrossRef] [PubMed]
- Mason, M.G.; Ball, A.S.; Reeder, B.J.; Silkstone, G.; Nicholls, P.; Wilson, M.T. Extracellular heme peroxidases in Actinomycetes: A case of mistaken identity. Appl. Environ. Microbiol. 2001, 67, 4512–4519. [Google Scholar] [CrossRef][Green Version]
- Rostami, M.; Rafiee, L.; Hassanzadeh, F.; Dadrass, A.R.; Ghadam, A.K. Synthesis of some new porphyrins and their metalloderivatives as potential sensitizers in photo-dynamic therapy. Res. Pharm. Sci. 2015, 10, 504–513. [Google Scholar]
- Sutthiwong, N.; Fouillaud, M.; Valla, A.; Caro, Y.; Dufossé, L. Bacteria belonging to the extremely versatile genus Arthrobacter as novel source of natural pigments with extended hue range. Food Res. Int. 2014, 65, 156–162. [Google Scholar] [CrossRef]
- Lesage, S.; Xu, H.; Durham, L. The occurrence and roles of porphyrins in the environment: Possible implications for bioremediation. Hydrol. Sci. J. 1993, 38, 343–354. [Google Scholar] [CrossRef]
- Kojima, I.; Maruhashi, K.; Fujiwara, Y.; Saito, T.; Kajiwara, M.; Mizutani, M. Identification of porphyrins produced from isopropanol by Arthrobacter hyalinus. J. Ferment. Bioeng. 1993, 75, 353–358. [Google Scholar] [CrossRef]
- Scharf, R.; Mamet, R.; Zimmels, Y.; Kimchie, S.; Schoenfeld, N. Evidence for the interference of aluminum with bacterial porphyrin biosynthesis. Biometals 1994, 7, 135–141. [Google Scholar] [CrossRef] [PubMed]
- Kajiwara, M.; Tokiwa, S.; Takatori, K.; Kojima, I. Properties of zinc uroporphyrin III produced from isopropanol by Arthrobacter hyalinus. J. Ferment. Bioeng. 1995, 79, 174–176. [Google Scholar] [CrossRef]
- Yang, H.-S.; Hoober, J.K. Divergent pathways for δ-aminolevulinic acid synthesis in two species of Arthrobacter. FEMS Microbiol. Lett. 1995, 134, 259–263. [Google Scholar] [CrossRef]
- Chaves, J.C.A.; Gregorutti dos Santos, C.; Aparecida de Miranda, E.G.; Arantes Junior, J.T.; Nantes, I.L. Free-base and metal complexes of 5,10,15,20-tetrakis(NMethyl Pyridinium L)porphyrin: Catalytic and therapeutic properties. In Phthalocyanines and Some Current Applications; Yilmaz, Y., Ed.; IntechOpen Ltd.: London, UK, 2017. [Google Scholar] [CrossRef]
- Pérez-Urria, E.; Avalos, A. Approach to a comparative study of the metabolism of porphyrins and chlorophylls. J. Nat. Sci. 2015, 3, 1–16. [Google Scholar] [CrossRef]
- Schobert, M.; Jahn, D. Regulation of heme biosynthesis in non-phototrophic bacteria. J. Mol. Microbiol. Biotechnol. 2002, 4, 287–294. [Google Scholar] [PubMed]
- Zhang, J.; Kang, Z.; Chen, J.; Du, G. Optimization of the heme biosynthesis pathway for the production of 5-aminolevulinic acid in Escherichia coli. Sci. Rep. 2015, 5, 8584. [Google Scholar] [CrossRef]
- Wolfe, B.E.; Button, J.E.; Santarelli, M.; Dutton, R.J. Cheese rind communities provide tractable systems for in situ and in vitro studies of microbial diversity. Cells 2014, 158, 422–433. [Google Scholar] [CrossRef] [PubMed]
- Kastman, E.K.; Kamelamela, N.; Norville, J.W.; Cosetta, C.M.; Dutton, R.J.; Wolfe, B.E. Biotic interactions shape the ecological distributions of staphylococcus species. mBio 2016, 7, e01157-16. [Google Scholar] [CrossRef] [PubMed]
- Frankenberg, N.; Moser, J.; Jahn, D. Bacterial heme biosynthesis and its biotechnological application. Appl. Microbiol. Biotechnol. 2003, 63, 115–127. [Google Scholar] [CrossRef]
- Middleton, K.S.; Gunner, H.B. Porphyrin production by Arthrobacter globiformis. Can. J. Microbiol. 1968, 14, 1095–1103. [Google Scholar] [CrossRef]
- Kortsee, G.J.J. Porphyrin Formation and Its Regulation in Arthrobacter. Ph.D. Thesis, Agricultural University, Wageningen, The Netherlands, 1969. [Google Scholar]
- Bonkovsky, H.L.; Guo, J.-T.; Hou, W.; Li, T.; Narang, T.; Thapar, M. Porphyrin and Heme Metabolism and the Porphyrias. Compr. Physiol. 2013, 3, 365–401. [Google Scholar] [CrossRef] [PubMed]
- Kwon, S.J.; de Boer, A.L.; Petri, R.; Schmidt-Dannert, C. High-level production of porphyrins in metabolically engineered Escherichia coli: Systematic extension of a pathway assembled from overexpressed genes involved in heme biosynthesis. Appl. Environ. Microbiol. 2003, 69, 4875–4883. [Google Scholar] [CrossRef] [PubMed]
- Brazier, J.S. Analysis of the porphyrin content of fluorescent pus by absorption spectrophotometry and high performance liquid chromatography. J. Med. Microbiol. 1990, 33, 29–34. [Google Scholar] [CrossRef]
- Yamane, Y.; Ooshima, H.; Kato, J. Overproduction of riboflavin by an Arthrobacter sp. mutant resistant to 5-fluorouracil. Enzyme Microb. Technol. 1993, 15, 877–880. [Google Scholar] [CrossRef]
- Josefsen, L.B.; Boyle, R.W. Photodynamic therapy and the development of metal-based photosensitisers. Met. Based Drugs 2008. [Google Scholar] [CrossRef] [PubMed]
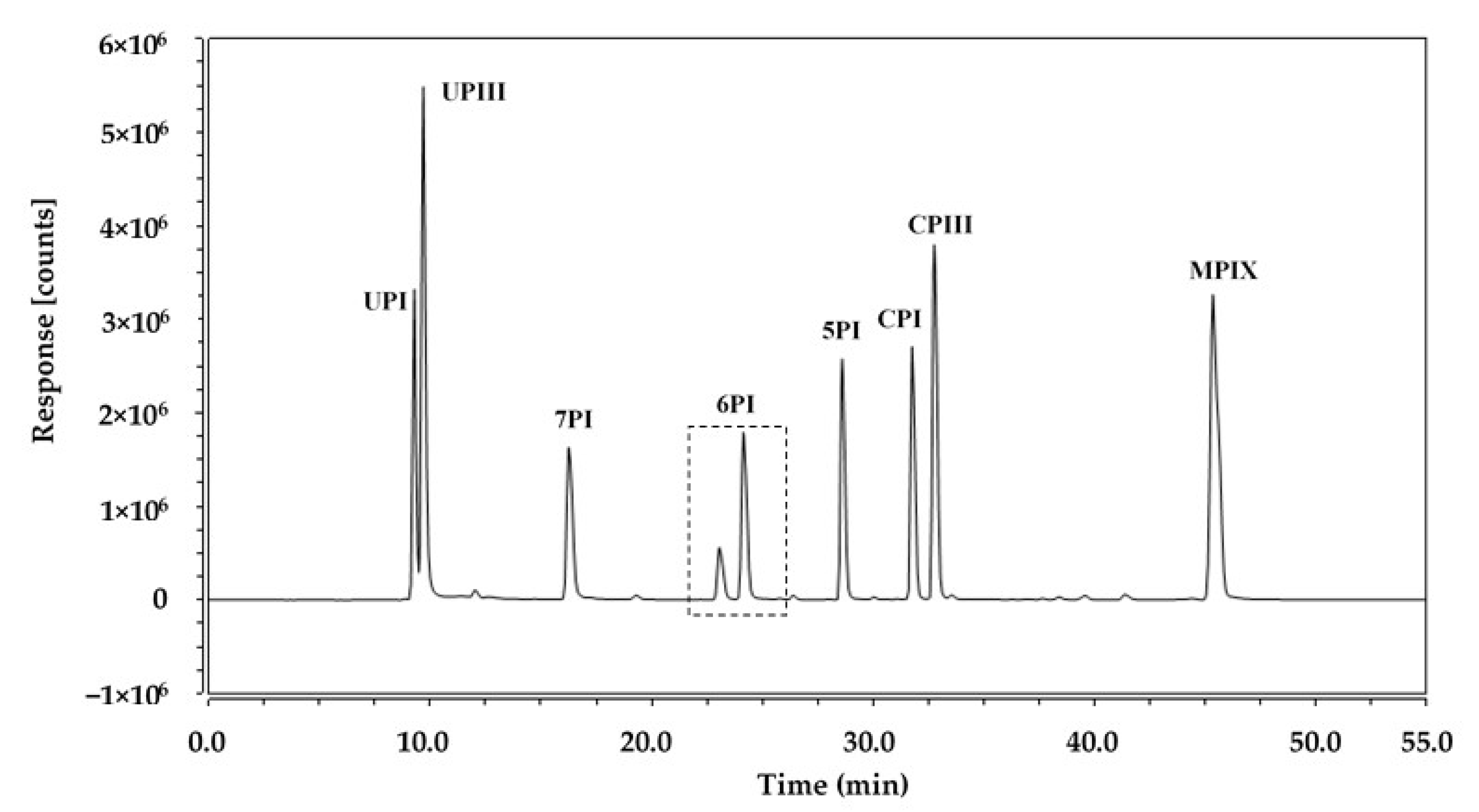


| Porphyrins | Rt (min) | Soret Band λ (nm) | Q Band λ (nm) |
|---|---|---|---|
| UPI | 9.28 | 398 | 501, 537, 563, 615 |
| UPIII | 9.68 | 398 | 501, 537, 563, 615 |
| 7PI | 16.26 | 397 | 500, 536, 563, 615 |
| 6PI | 23.06 | 396 | 500, 535, 564, 616 |
| 24.14 | 396 | 500, 534, 564, 616 | |
| 5PI | 28.60 | 394 | 450, 533, 564, 617 |
| CPI | 31.77 | 393 | 497, 532, 564, 616 |
| CPIII | 32.76 | 393 | 497, 532, 564, 617 |
| MPIX | 43.35 | 392 | 496, 530, 565, 618 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Sutthiwong, N.; Sukdee, P.; Lekhavat, S.; Dufossé, L. Identification of Red Pigments Produced by Cheese-Ripening Bacterial Strains of Glutamicibacter arilaitensis Using HPLC. Dairy 2021, 2, 396-409. https://doi.org/10.3390/dairy2030031
Sutthiwong N, Sukdee P, Lekhavat S, Dufossé L. Identification of Red Pigments Produced by Cheese-Ripening Bacterial Strains of Glutamicibacter arilaitensis Using HPLC. Dairy. 2021; 2(3):396-409. https://doi.org/10.3390/dairy2030031
Chicago/Turabian StyleSutthiwong, Nuthathai, Piyada Sukdee, Supaporn Lekhavat, and Laurent Dufossé. 2021. "Identification of Red Pigments Produced by Cheese-Ripening Bacterial Strains of Glutamicibacter arilaitensis Using HPLC" Dairy 2, no. 3: 396-409. https://doi.org/10.3390/dairy2030031
APA StyleSutthiwong, N., Sukdee, P., Lekhavat, S., & Dufossé, L. (2021). Identification of Red Pigments Produced by Cheese-Ripening Bacterial Strains of Glutamicibacter arilaitensis Using HPLC. Dairy, 2(3), 396-409. https://doi.org/10.3390/dairy2030031