Monitoring the Transition Period in Dairy Cows through 1H NMR-Based Untargeted Metabolomics
Abstract
:1. Introduction
2. Materials and Methods
2.1. Animals and Sample Collection
2.2. Clinical Biochemistry
2.3. NMR Spectroscopy
2.4. Data Processing
2.5. Metabolite Concentrations
2.6. Statistical Analysis
3. Results and Discussion
3.1. 1H NMR Spectra of Serum Samples
3.2. NMR-Based Serum Metabolomics
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Grummer, R.R. Impact of changes in organic nutrient metabolism on feeding the transition dairy cow. J. Anim. Sci. 1995, 73, 2820–2833. [Google Scholar] [CrossRef]
- Bauman, D.E.; Currie, W.B. Partitioning of nutrients during pregnancy and lactation: A review of mechanisms involving homeostasis and homeorhesis. J. Dairy Sci. 1980, 63, 1514–1529. [Google Scholar] [CrossRef]
- Ingvartsen, K.L. Feeding- and management-related diseases in the transition cow: Physiological adaptations around calving and strategies to reduce feeding-related diseases. Anim. Feed Sci. Technol. 2006, 126, 175–213. [Google Scholar] [CrossRef]
- Butler, W.R. Nutritional interactions with reproductive performance in dairy cattle. Anim. Reprod. Sci. 2000, 60–61, 449–457. [Google Scholar] [CrossRef]
- Drackley, J.K.; Overton, T.R.; Douglas, G.N. Adaptations of glucose and long-chain fatty acid metabolism in liver of dairy cows during the periparturient period. J. Dairy Sci. 2001, 84, E100–E112. [Google Scholar] [CrossRef]
- Meikle, A.; Kulcsar, M.; Chilliard, Y.; Febel, H.; Delavaud, C.; Cavestany, D.; Chilibroste, P. Effects of parity and body condition at parturition on endocrine and reproductive parameters of the cow. Reproduction 2004, 127, 727–737. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ingvartsen, K.L.; Moyes, K. Nutrition, immune function and health of dairy cattle. Animal 2012, 7, 112–122. [Google Scholar] [CrossRef] [Green Version]
- Mulligan, F.J.; Doherty, M.L. Production diseases of the transition cow. Vet. J. 2008, 176, 3–9. [Google Scholar] [CrossRef]
- Ingvartsen, K.L.; Dewhurst, R.J.; Friggens, N.C. On the relationship between lactational performance and health: Is it yield or metabolic imbalance that cause production diseases in dairy cattle? A position paper. Livest. Prod. Sci. 2003, 83, 277–308. [Google Scholar] [CrossRef]
- Oltenacu, P.A.; Broom, D.M. The impact of genetic selection for increased milk yield on the welfare of dairy cows. Anim. Welf. 2010, 19, 39–49. [Google Scholar]
- Ingvartsen, K.L.; Andersen, J.B. Integration of metabolism and intake regulation: A review focusing on periparturient animals. J. Dairy Sci. 2000, 83, 1573–1597. [Google Scholar] [CrossRef]
- Burke, C.; Roche, R. Effects of pasture feeding during the periparturient period on postpartum anovulation in grazed dairy cows. J. Dairy Sci. 2007, 90, 4304–4312. [Google Scholar] [CrossRef]
- Enjalbert, F.; Nicot, M.C.; Bayourthe, C.; Moncoulon, R. Ketone bodies in milk and blood of dairy cows: Relationship between concentrations and utilization for detection of subclinical ketosis. J. Dairy Sci. 2001, 84, 583–589. [Google Scholar] [CrossRef]
- Giannesella, M.; Perillo, L.; Fiori, E.; Giudice, E.; Zumbo, A.; Morgante, M.; Piccione, G. Transition period in healthy and diseased dairy cows: Evaluation of metabolic modifications. Large Anim. Rev. 2018, 24, 107–111. [Google Scholar]
- Ceciliani, F.; Lecchi, C.; Urh, C.; Sauerwein, H. Proteomics and metabolomics characterizing the pathophysiology of adaptive reactions to the metabolic challenges during the transition from late pregnancy to early lactation in dairy cows. J. Proteom. 2018, 178, 92–106. [Google Scholar] [CrossRef]
- Zhang, H.; Wu, L.; Xu, C.; Xia, C.; Sun, L.; Shu, S. Plasma metabolomic profiling of dairy cows affected with ketosis using gas chromatography/mass spectrometry. BMC Vet. Res. 2013, 9, 186–199. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Imhasly, S.; Naegeli, H.; Baumann, S.; von Bergen, M.; Luch, A.; Jungnickel, H.; Potratz, S.; Gerspach, C. Metabolomic biomarkers correlating with hepatic lipidosis in dairy cows. BMC Vet. Res. 2014, 10, 122. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Dervishi, E.; Zhang, G.; Dunn, S.M.; Mandal, R.; Wishart, D.S.; Ametaj, B.N. GC-MS metabolomics identifies metabolite alterations that precede subclinical mastitis in the blood of transition dairy cows. J. Proteom. Res. 2017, 16, 433–446. [Google Scholar] [CrossRef]
- Zhang, G.; Dervishi, E.; Dunn, S.M.; Mandal, R.; Liu, P.; Han, B.; Wishart, D.S.; Ametaj, B.N. Metabotyping reveals distinct metabolic alterations in ketotic cows and identifies early predictive serum biomarkers for the risk of disease. Metabolomics 2017, 13, 43–58. [Google Scholar] [CrossRef]
- Zhang, G.; Ametaj, B.N. Ketosis: An old story under a new approach. Dairy 2020, 1, 42–60. [Google Scholar] [CrossRef]
- Sun, L.W.; Zhang, H.Y.; Wu, L.; Shu, S.; Xia, C.; Xu, C.; Zheng, J.S. 1H-Nuclear magnetic resonance-based plasma metabolic profiling of dairy cows with clinical and subclinical ketosis. J. Dairy Sci. 2014, 97, 1552–1562. [Google Scholar] [CrossRef] [PubMed]
- Xu, C.; Sun, L.-W.; Xia, C.; Zhang, H.-Y.; Zheng, J.-S.; Wang, J.-S. 1H-Nuclear magnetic resonance-based plasma metabolic profiling of dairy cows with fatty liver. Asian Australas. J. Anim. Sci. 2016, 29, 219–229. [Google Scholar]
- Dervishi, E.; Zhang, G.; Hailemariam, D.; Mandal, R.; Wishart, D.S.; Ametaj, B.N. Urine metabolic fingerprinting can be used to predict the risk of metritis and highlight the pathobiology of the disease in dairy cows. Metabolomics 2018, 14, 83. [Google Scholar] [CrossRef]
- Zhang, G.; Dervishi, E.; Zwierzchowski, G.; Mandal, R.; Wishart, D.S.; Ametaj, B.N. Urinary metabolomics around parturition identifies metabolite alterations in dairy cows affected postpartum by lameness: Preliminary study. Diary 2020, 1, 6–19. [Google Scholar] [CrossRef] [Green Version]
- Hailemariam, D.; Mandal, R.; Saleem, F.; Dunn, S.M.; Wishart, D.S.; Ametaj, B.N. Identification of predictive biomarkers of disease state in transition dairy cows. J. Dairy Sci. 2014, 97, 2680–2693. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hailemariam, D.; Mandal, R.; Saleem, F.; Dunn, S.M.; Wishart, D.S.; Ametaj, B.N. Metabolomics approach reveals altered amino acid and sphingolipid profiles associated with pathological state in transition dairy cows. Curr. Metab. 2014, 2, 184–195. [Google Scholar] [CrossRef]
- Kenéz, Á.; Dänicke, S.; Rolle-Kampczyk, U.; von Bergen, M.; Huber, K. A metabolomics approach to characterize phenotypes of metabolic transition from late pregnancy to early lactation in dairy cows. Metabolomics 2016, 12, 165–176. [Google Scholar] [CrossRef]
- Luo, Z.Z.; Shen, L.H.; Jiang, J.; Huang, Y.X.; Bai, L.P.; Yu, S.M.; Yao, X.P.; Ren, Z.H.; Yang, Y.X.; Cao, S.Z. Plasma metabolite changes in dairy cows during parturition identified using untargeted metabolomics. J. Dairy Sci. 2019, 102, 1–12. [Google Scholar] [CrossRef] [Green Version]
- Luke, T.D.W.; Pryce, J.E.; Elkins, A.C.; Wales, W.J.; Rochfort, S.J. Use of large and diverse datasets for 1H NMR serum metabolic profiling of early lactation dairy cows. Metabolites 2019, 10, 180. [Google Scholar] [CrossRef] [PubMed]
- Fariña, S.R.; Chillibroste, P. Opportunities and challenges for the growth of milk production from pasture: The case of farm systems in Uruguay. Agric. Syst. 2019, 176, 102631. [Google Scholar] [CrossRef]
- Kolver, E.S.; Muller, L.D. Performance and nutrient intake of high producing Holstein cows consuming pasture or a total mixed ration. J. Dairy Sci. 1998, 81, 1403–1411. [Google Scholar] [CrossRef]
- Duffield, T.F.; Leslie, K.E.; Sandals, D.; Lissemore, K.; McBride, B.W.; Lumsden, J.H.; Dick, P.; Bagg, R. Effect of prepartum administration of a monensin controlled release capsule on cow health and reproduction. J. Dairy Sci. 1999, 82, 2377–2384. [Google Scholar] [CrossRef]
- LeBlanc, S. Postpartum uterine disease and dairy herd reproductive performance: A review. Vet. J. 2008, 176, 102–114. [Google Scholar] [CrossRef]
- Galvão, K.N.; Greco, L.F.; Vilela, J.M.; Sá Filho, M.F.; Santos, J.E. Effect of intrauterine infusion of ceftiofur on uterine health and fertility in dairy cows. J. Dairy Sci. 2009, 92, 1532–1542. [Google Scholar] [CrossRef] [PubMed]
- Ruegg, P.L. New perspectives in udder health management. Vet. Clin. Food Anim. 2012, 28, 149–163. [Google Scholar] [CrossRef]
- Duffield, T.F.; Lissemore, K.D.; McBride, B.W.; Leslie, K.E. Impact of hyperketonemia in early lactation dairy cows on health and production. J. Dairy Sci. 2009, 92, 571–580. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Nicholson, J.K.; Foxall, P.J.D.; Spraul, M.; Farrant, R.D.; Lindon, J.C. 750 MHz 1H and 13C NMR spectroscopy of human blood plasma. Anal. Chem. 1995, 67, 793–811. [Google Scholar] [CrossRef]
- Viant, M.R. Improved methods for the acquisition and interpretation of NMR metabolomic data. Biochem. Biophys. Res. Commun. 2003, 310, 943–948. [Google Scholar] [CrossRef] [PubMed]
- Purohit, P.V.; Rocke, D.M.; Viant, M.R.; Woodruff, D.L. Discrimination models using variance-stabilizing transformation of metabolomic NMR data. Omics 2004, 8, 118–130. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Van den Berg, R.A.; Hoefsloot, H.C.J.; Westerhuis, J.A.; Smilde, A.K.; van der Werf, M.J. Centering, scaling, and transformations: Improving the biological information content of metabolomics data. BMC Genom. 2006, 7, 142. [Google Scholar] [CrossRef] [Green Version]
- Wold, S.; Esbensen, K.; Geladi, P. Principal component analysis. Chemom. Intell. Lab. Syst. 1987, 2, 37–52. [Google Scholar] [CrossRef]
- Trygg, J.; Wold, S. Orthogonal projections to latent structures (O-PLS). J. Chemom. 2002, 16, 119–128. [Google Scholar] [CrossRef]
- Trygg, J.; Holmes, E.; Lundstedt, T. Chemometrics in metabonomics. J. Proteom. Res. 2007, 6, 469–479. [Google Scholar] [CrossRef] [PubMed]
- Simundic, A.-M. Diagnostic accuracy–Part 1. Basic concepts: Sensitivity and specificity, ROC analysis, STARD statement. Point Care 2012, 11, 6–8. [Google Scholar] [CrossRef]
- Ekelund, S. ROC curves–What are they and how are they used? Point Care 2012, 11, 16–21. [Google Scholar] [CrossRef]
- Ni, Y.; Su, M.; Lin, J.; Wang, X.; Qiu, Y.; Zhao, A.; Chen, T.; Jia, W. Metabolic profiling reveals disorder of amino acid metabolism in four brain regions from a rat model of chronic unpredictable mild stress. FEBS Lett. 2008, 582, 2627–2636. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Sun, H.Z.; Wang, D.M.; Wang, B.; Wang, J.K.; Liu, H.Y.; Guan, L.L.; Liu, J.X. Metabolomics of four biofluids from dairy cows: Potential biomarkers for milk production and quality. J. Proteom. Res. 2015, 14, 1287–1298. [Google Scholar] [CrossRef]
- Thornalley, P.J. Modification of the glyoxalase system in human red blood cells by glucose in vitro. Biochem. J. 1988, 254, 751–755. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Omole, O.O.; Nappert, G.; Naylor, J.M.; Zello, G.A. Both L- and D-lactate contribute to metabolic acidosis in diarrheic calves. J. Nutr. 2001, 131, 2128–2131. [Google Scholar] [CrossRef]
- Sako, T.; Urabe, S.; Kusaba, A.; Kimura, N.; Yoshimura, I.; Tazaki, H.; Imai, S.; Ono, K.; Arai, T. Comparison of plasma metabolite concentrations and lactate dehydrogenase activity in dogs, cats, horses, cattle and sheep. Vet. Res. Commun. 2007, 31, 413–417. [Google Scholar] [CrossRef]
- Krauze, K.M.; Oetzel, G.R. Understanding and preventing subacute ruminal acidosis in dairy herds: A review. Anim. Feed Sci. Technol. 2006, 126, 215–236. [Google Scholar] [CrossRef]
- Asanuma, N.; Hino, T. Regulation of fermentation in a ruminal bacterium, Streptococcus bovis, with special reference to rumen acidosis. Anim. Sci. J. 2002, 73, 313–325. [Google Scholar] [CrossRef]
- Overton, T.R.; Waldron, M.R. Nutritional management of transition dairy cows: Strategies to optimize metabolic health. J. Diary Sci. 2004, 87, E105–E119. [Google Scholar] [CrossRef] [Green Version]
- Aschenbach, J.R.; Kristensen, N.B.; Donkin, S.S.; Hammon, H.M.; Penner, B.G. Gluconeogenesis in dairy cows: The secret of making sweet milk from sour dough. IUBMB Life 2010, 62, 869–877. [Google Scholar] [CrossRef] [PubMed]
- Gonzáles, F.D.; Muiño, R.; Pereira, V.; Campos, R.; Bendito, J.L. Relationship among blood indicators of lipomobilization and hepatic function during early lactation in high-yielding dairy cows. J. Vet. Sci. 2011, 12, 251–255. [Google Scholar] [CrossRef] [Green Version]
- Vercoe, J.E. Urinary allantoine excretion and digestible dry-matter intake in cattle and buffalo. J. Agric. Sci. 1976, 86, 613–615. [Google Scholar] [CrossRef]
- Koch, F.; Lamp, O.; Eslamizad, M.; Weitzel, J.; Kuhla, B. Metabolic response to heat stress in late-pregnant and early lactation dairy cows: Implications to liver-muscle crosstalk. PLoS ONE 2016, 11, e0160912. [Google Scholar] [CrossRef] [PubMed]
- Bell, A.W. Regulation of organic nutrient metabolism during transition from late pregnancy to early lactation. J. Anim. Sci. 1995, 73, 2804–2819. [Google Scholar] [CrossRef]
- Hayirli, A.; Grummer, R.R.; Nordheim, E.V.; Crump, P.M. Animal and dietary factors affecting feed intake during the prefresh transition period in Holsteins. J. Dairy Sci. 2002, 85, 3430–3443. [Google Scholar] [CrossRef]
- Grant, R.J.; Albright, J.L. Effect of animal grouping on feeding behavior and intake of dairy cattle. J. Dairy Sci. 2001, 85, E156–E163. [Google Scholar] [CrossRef]
- Meikle, A.; de Brun, V.; Carriquiry, M.; Soca, P.; Sosa, C.; Adrien, M.L.; Chilibroste, P.; Abecia, J.A. Influences of nutrition and metabolism on reproduction of the female ruminant. Anim. Reprod. 2018, 15, 899–911. [Google Scholar] [CrossRef]
- Kim, H.S.; Kim, E.D.T.; Eom, J.S.; Choi, Y.Y.; Lee, S.J.; Lee, S.S.; Chung, C.D.; Lee, S.S. Exploration of metabolite profiles in the biofluids of dairy cows by proton nuclear magnetic resonance analysis. PLoS ONE 2021, 16, e0246290. [Google Scholar]
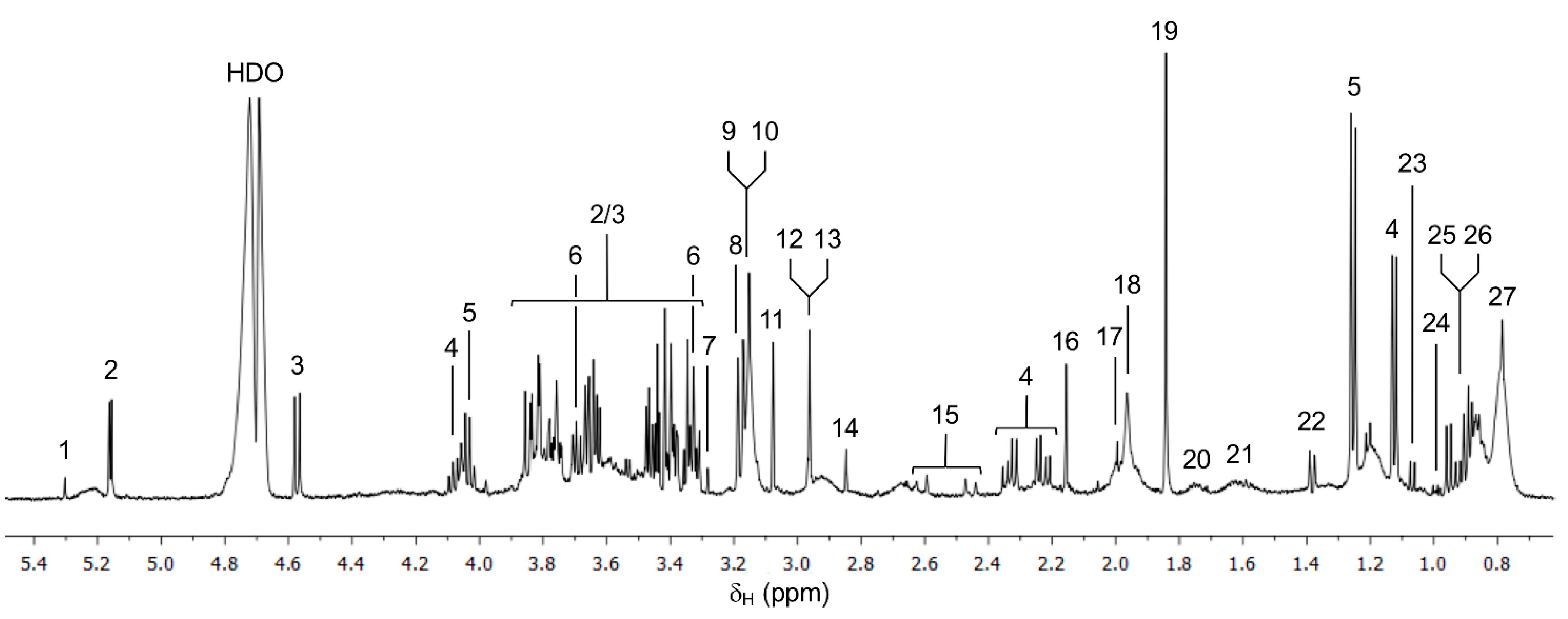
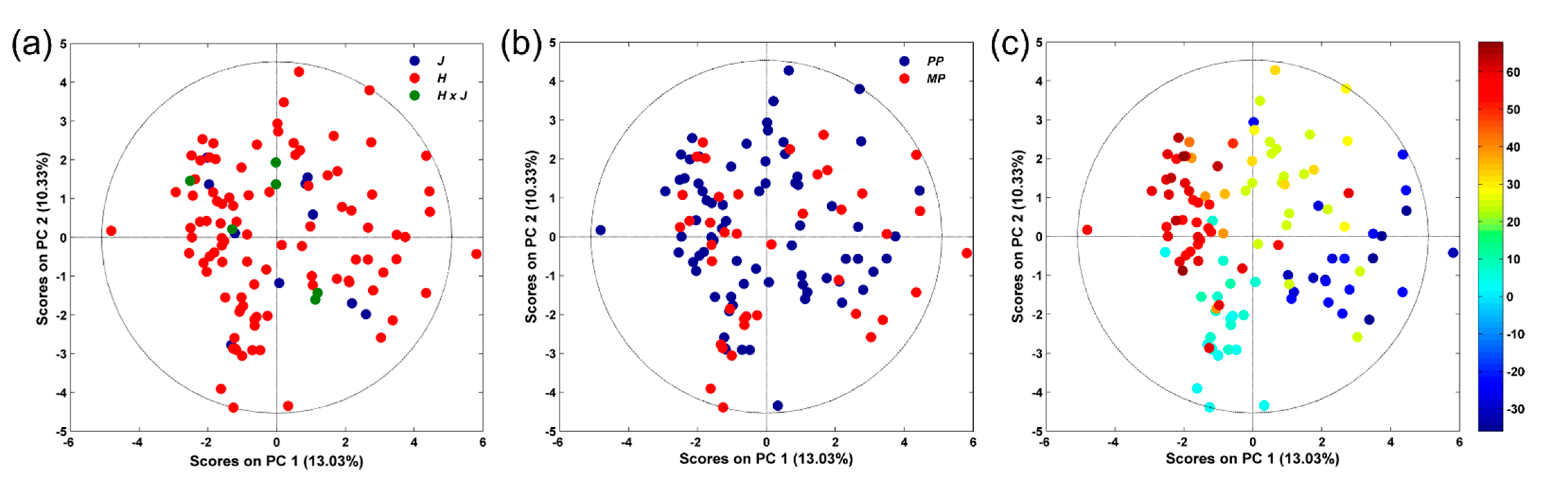


| Animal Data | Stage | |||
|---|---|---|---|---|
| C − 4 | C + 1 | C + 4 | C + 8 | |
| Days in stage | −36 to −22 | 3 to 11 | 23 to 34 | 39 to 68 |
| N (H, J, H × J) a | 23 (19, 2, 2) | 20 (18, 0, 2) | 24 (20, 2, 2) | 20 (17, 1, 2) |
| Parity (PP, MP) b | 15, 8 | 12, 8 | 15, 9 | 13, 7 |
| Disease (SCK, MCM) c | 0, 9 | 2, 6 | 6, 10 | 2, 11 |
| Milk production (L/day) | - | 23 ± 7 | 28 ± 6 | 25 ± 7 |
| Biochemistry | ||||
| Albumin (g/L) | 31.00 ± 2.19 | 29.64 ± 2.91 | 29.44 ± 5.67 | 31.42 ± 2.78 |
| Urea (mM) | 2.40 ± 1.31 | 2.82 ± 1.10 | 2.65 ± 1.11 | 3.43 ± 1.46 |
| Cholesterol (mM) | 2.71 ± 0.43 | 2.29 ± 0.50 | 3.81 ± 0.75 | 5.09 ± 0.93 |
| Total protein (g/L) | 79.61 ± 7.00 | 74.23 ± 7.19 | 78.13 ± 7.72 | 80.52 ± 6.64 |
| Globuline (g/L) | 48.90 ± 6.50 | 44.59 ± 5.17 | 48.69 ± 5.12 | 49.10 ± 4.60 |
| BHB (mM) | 0.38 ± 0.21 | 0.56 ± 0.32 | 0.78 ± 0.63 | 0.65 ± 0.35 |
| NEFA (mM) | 0.29 ± 0.18 | 0.63 ± 0.22 | 0.68 ± 0.36 | 0.47 ± 0.20 |
| Metabolite | Concentration (mM) | Fold Change | ||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| C − 4 | C + 1 | C + 4 | C + 8 | C + 1/C − 4 | p | C + 4/C − 4 | p | C + 8/C − 4 | p | |
| Acetate | 0.83 ± 0.80 | 1.36 ± 1.22 | 1.99 ± 2.17 | 1.86 ± 1.13 | 1.63 | 0.0494 | 2.40 | 0.0091 | 2.25 | 0.0002 |
| Acetone | 0.11 ± 0.15 | 0.12 ± 0.08 | 0.17 ± 0.16 | 0.12 ± 0.09 | 1.14 | 0.3519 | 1.62 | 0.0724 | 1.10 | 0.3734 |
| Alanine | 0.18 ± 0.17 | 0.26 ± 0.22 | 0.43 ± 0.46 | 0.51 ± 0.39 | 1.50 | 0.0788 | 2.48 | 0.0071 | 2.88 | 0.0002 |
| Allantoin | 0.07 ± 0.07 | 0.16 ± 0.16 | 0.24 ± 0.27 | 0.20 ± 0.12 | 2.16 | 0.0167 | 3.41 | 0.0021 | 2.81 | <0.0001 |
| Asparagine | 0.61 ± 0.42 | 1.71 ± 2.20 | 2.36 ± 2.90 | 1.92 ± 1.27 | 2.82 | 0.0116 | 3.88 | 0.0074 | 3.16 | <0.0001 |
| Betaine | 0.18 ± 0.15 | 0.34 ± 0.38 | 0.56 ± 0.63 | 0.43 ± 0.26 | 1.92 | 0.0329 | 3.15 | 0.0035 | 2.44 | 0.0001 |
| BHB | 0.38 ± 0.20 | 0.56 ± 0.32 | 0.78 ± 0.62 | 0.65 ± 0.35 | 1.46 | 0.0175 | 2.04 | 0.0028 | 1.71 | 0.0009 |
| Citrate | 0.10 ± 0.09 | 0.26 ± 0.25 | 0.39 ± 0.50 | 0.22 ± 0.18 | 2.59 | 0.0097 | 4.01 | 0.0085 | 2.29 | 0.0035 |
| Citrulline | 0.13 ± 0.12 | 0.26 ± 0.31 | 0.48 ± 0.65 | 0.28 ± 0.23 | 2.01 | 0.0442 | 3.75 | 0.0069 | 2.18 | 0.0039 |
| Creatine | 0.04 ± 0.04 | 0.10 ± 0.14 | 0.15 ± 0.22 | 0.10 ± 0.09 | 2.51 | 0.0201 | 3.70 | 0.0105 | 2.33 | 0.0029 |
| TMA | 0.06 ± 0.04 | 0.23 ± 0.67 | 0.13 ± 0.19 | 0.10 ± 0.09 | 1.54 | 0.0325 | 2.13 | 0.0484 | 1.68 | 0.0229 |
| α-Glucose | 1.00 ± 0.88 | 2.27 ± 2.54 | 3.50 ± 4.15 | 2.91 ± 2.04 | 2.28 | 0.0145 | 3.51 | 0.0033 | 2.29 | <0.0001 |
| β-Glucose | 0.92 ± 0.81 | 2.19 ± 2.61 | 3.49 ± 4.74 | 2.81 ± 2.08 | 2.38 | 0.0415 | 3.77 | 0.0068 | 3.04 | <0.0001 |
| Lactate | 3.60 ± 3.10 | 1.16 ± 0.63 | 1.67 ± 1.13 | 1.60 ± 0.77 | −2.23 | 0.0038 | −2.16 | 0.0015 | −2.26 | 0.0006 |
| Leucine | 0.12 ± 0.10 | 0.06 ± 0.03 | 0.06 ± 0.03 | 0.03 ± 0.03 | −1.83 | 0.0166 | −1.93 | 0.0036 | −3.80 | < 0.0001 |
| Lysine | 0.08 ± 0.07 | 0.23 ± 0.31 | 0.37 ± 0.48 | 0.26 ± 0.20 | 2.63 | 0.0169 | 4.16 | 0.0037 | 2.97 | 0.0001 |
| Valine | 0.23 ± 0.17 | 0.22 ± 0.07 | 0.24 ± 0.08 | 0.26 ± 0.09 | −1.02 | 0.4583 | 1.04 | 0.3938 | 1.14 | 0.1901 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
López Radcenco, A.; Adrien, M.d.L.; Ruprechter, G.; de Torres, E.; Meikle, A.; Moyna, G. Monitoring the Transition Period in Dairy Cows through 1H NMR-Based Untargeted Metabolomics. Dairy 2021, 2, 356-366. https://doi.org/10.3390/dairy2030028
López Radcenco A, Adrien MdL, Ruprechter G, de Torres E, Meikle A, Moyna G. Monitoring the Transition Period in Dairy Cows through 1H NMR-Based Untargeted Metabolomics. Dairy. 2021; 2(3):356-366. https://doi.org/10.3390/dairy2030028
Chicago/Turabian StyleLópez Radcenco, Andrés, María de Lourdes Adrien, Gretel Ruprechter, Elena de Torres, Ana Meikle, and Guillermo Moyna. 2021. "Monitoring the Transition Period in Dairy Cows through 1H NMR-Based Untargeted Metabolomics" Dairy 2, no. 3: 356-366. https://doi.org/10.3390/dairy2030028
APA StyleLópez Radcenco, A., Adrien, M. d. L., Ruprechter, G., de Torres, E., Meikle, A., & Moyna, G. (2021). Monitoring the Transition Period in Dairy Cows through 1H NMR-Based Untargeted Metabolomics. Dairy, 2(3), 356-366. https://doi.org/10.3390/dairy2030028






