1. Introduction
Cognitive ability, whether measured by IQ tests, Piagetian tests, educational/scholastic tests (e.g., PISA, TIMSS, etc.), or other indices of cognitive functioning, differs on average between biogeographic ancestry groups (BGAs). (We use the term “cognitive ability” instead of “general intelligence” or “general cognitive ability” to make clear that we are not committing to a claim about the psychometric nature of the underlying construct. For the purpose of this study, the distinctions between cognitive ability in general and general cognitive ability are not important.) According to Shriver and Kittles [
1], biogeographic ancestry is the personal genetic history, indexed by ancestrally informative autosomal markers, which reflects an individual’s overall ancestry with respect to “population groups” (also referred to as “clusters,” “ancestral groups,” and “ancestral populations”). These reference biogeographic ancestry groups differ owing to the effects of evolutionary factors, such as “isolation by distance,” and barriers that “have all affected human migration and mating patterns in the past.” Biogeographic ancestry groups have commonly been called “races” (see [
2,
3]). Examples of these groups include East Asians, Europeans, sub-Saharan Africans, and Amerindians (see [
4]). Cognitive ability differences appear both between nations (e.g., in predominantly European versus predominantly African national populations), and between self-identified racial and ethnic (SIRE) groups within nations (e.g., Whites versus African Americans in the USA [
5,
6,
7]). The cause(s) of inter- and intra-national cognitive ability gaps is highly uncertain and hotly debated, with some researchers arguing that genetic differences are substantially or even predominantly responsible for at least some of these gaps (e.g., [
8]) and others maintaining that non-genetic environmental factors fully account for the gaps (e.g., [
9,
10]).
Nevertheless, in the most recent survey of intelligence researchers, “genes were rated as the most important cause (17%) [of international differences], followed by educational quality (11.44%), health (10.88%), and educational quantity (10.20%)” [
11]. Further, 90% of the surveyed experts believed that international differences were at least in part genetic in origin. Rindermann, Coyle, and Becker [
12] also reported that 83% of survey respondents believed that the Black-White cognitive gap in the USA is partially due to genetic differences between the relevant populations. While there certainly is no general consensus among researchers, a substantial number of them believe that both international and certain intra-national SIRE differences in cognitive ability have a partial genetic basis [
11,
13].
Although racial and ethnic groups within a country can differ behaviorally for a variety of reasons (e.g., selective self-identification or local selective migration), possible genetic bases of these differences are typically analyzed in an evolutionary framework. When discussing evolved human diversity, one can conceptualize groups in a number of different ways, e.g., as subspecies (taxonomically significant subdivisions of a species), ecotypes (environmentally adapted types), clines (character gradients), and morphs (alternative phenotypes in a population). Following the work of Charles Darwin, communities are frequently delineated by propinquity descent since descent is understood as inductively potent. The preferable term to describe descent-based groups (e.g., variety, genetic population, race, genetic cluster, ancestry group, etc.) is a matter of ongoing semantic dispute. Here we call them “biogeographic ancestry groups” (again, BGAs), as is frequently done in genetic epidemiology. There are phenotypic and genetic differences between these groups, with evolutionary forces acting over relatively short spans of time likely having caused this divergence [
14]. Importantly, these ancestry groups are delimitable using genomic data.
The cause(s) of inter-BGA differences in cognitive ability specifically is the subject of several evolutionary theories, which decompose broadly into two categories: (1) Pleistocene-selection models and (2) Holocene-selection models. The former posit the action of evolutionarily novel factors that differed systematically in the regions to which human groups migrated after leaving Africa around 60 to 100 kya (thousand years ago), and which specifically selected for increased cognitive ability as a mechanism for enhancing survival [
15]. Salient evolutionarily novel challenges likely included the presence of seasonality, specifically cold winters during the main Würm glaciation event 60 kya, when temperatures in Europe and (especially) Northeast Asia were considerably lower on average than they are today. Extreme cold, coupled with the challenge of provisioning for the future such as to anticipate seasonality (e.g., food storage) have been proposed as sources of selection for higher cognitive ability and other somatic traits related to innovation and productivity [
16,
17]. These climatic models predict that North East Asian and European BGA groups will have higher levels of cognitive ability than Pacific Islanders, sub-Saharan Africans, and Amerindians (at least from the tropical and subtropical regions of the Americas).
Research in comparative zoology supports climatic models of this kind. A large body of research has investigated the climatic correlates of cognitive ability—usually measured with brain size as a proxy variable—in non-human animals. Non-migratory birds have been a primary choice of study [
18,
19,
20,
21] given their range of habitat and the ease of studying them. Studies have revealed the expected patterns, namely that birds that live further north and in more seasonally affected areas have larger brains (controlling for body size), more flexible behavior, and more innovative behavior [
18,
20,
22].
Holocene-selection models stress the observation that the rate of adaptive evolution during the Holocene (beginning roughly 12 kya) was on the order of 100 times that experienced by humans during the preceding Pleistocene epoch, thus the evolutionary factors crucial to the origin of these differences may have arisen relatively recently [
14]. Culture-gene co-evolution theory posits that cognitive ability and cultural complexity arose in tandem via a feedback process. Eurasian populations’ transition into sedentarism during the late Pleistocene and early Holocene would have been associated with a substantial increase in cognitive challenges, such as those related to competition over settled land, management of agriculture, and sustainment of higher standing population densities [
14,
23]. Innovations in modes of production and social organization (such as intensified division of labor) would have led to hierarchical societies involving strong individual-level competition for finite resources, which may have been a major source of social selection favoring higher levels of cognitive ability. Clark [
24,
25] documents the persistence of downward social mobility among the descendants of elites who competed for limited economic and social resources, with the less competitive sinking into lower-status occupational niches. The end result of this process was a persistent “bootstrapping” of the population, as cognitive ability (and other salient “bourgeois” traits) rose across all levels of the social hierarchy, leading to greater degrees of industriousness and innovativeness. This may in turn have boosted the competitiveness of certain European, and perhaps Northeast Asian, BGAs, permitting group-level expansion (or colonialism), which likely enhanced the biocultural “corporate” fitness of these populations—a process that may only have ceased in the West in the mid-19th century. Around this time, the advent of milder climates and concomitantly reduced ecological stress (corresponding to the end of the Maunder Minimum), coupled with both social and technological innovations that allowed those with relatively high cognitive ability to control their fertility, potentially reversed the fitness advantage of those with relatively high cognitive ability compared to those with relatively low cognitive ability [
26]). Given that these Holocene-selection models posit that Eurasian populations had the greatest exposure to environmental challenges favoring the fitness of those with relatively high cognitive ability, they predict that selection for cognitive ability was stronger in Eurasian populations than non-Eurasian ones, such as Pacific Islanders, sub-Saharan Africans, and Amerindians.
A further evolutionary theory of inter-BGA cognitive differences is the disease-stress model. This has assumed forms that posit [
27] and do not posit [
28] genetic differences to explain inter-BGA disparities in cognitive ability. Models of the latter type propose that in the Pleistocene environment of evolutionary adaptedness, humans were repeatedly exposed to periods of high and low parasite stress. They further maintain that this exposure to variable levels of parasite stress favored the evolution of epigenetic adaptations that dynamically regulate tradeoffs of bioenergetic investments into brain/cognitive development or immune system functioning in response to environmental challenges, with greater investments into the latter over the former occurring in high-parasite-stress contexts. Therefore, when humans radiated into novel, more northerly and easterly low-parasite-stress ecologies, they came pre-adapted with the capacity to developmentally trade immune system functioning for higher cognitive ability, which is reflected in very strong inverse ecological correlations between national IQ and parasite-burden indices [
28]. Given the global distribution of parasite burdens, this model predicts, as do the other evolutionary theories, that the populations of Europe and North East Asia have higher cognitive ability relative to those of the Pacific Islands and the tropical and subtropical regions of the Americas, as well as the populations of sub-Saharan Africa. But this version of the disease-stress model is clearly inadequate to account for the total global pattern of inter-BGA cognitive ability variation, since it seemingly cannot explain BGA cognitive ability gaps found within nations, where BGA groups do not differ substantially in their exposure to parasites (since they inhabit the same general environments that the nations encompass). Therefore, our statistical analyses do not endeavor to test predictions of this disease-stress model.
Conversely, genetic disease-stress models posit that globally variable geographical factors covary with disease burdens, and that genetic adaptations to local disease burdens have partially generated inter-BGA differences in social outcomes and related phenotypes, including cognitive ability [
27]. While our data do not permit any direct test of this, or any other, particular evolutionary account of the emergence of inter-BGA cognitive ability variation, global admixture analytic findings indicative of a genetic etiology of such variability would be consistent with all the evolutionary theories reviewed here, apart from the epigenetic disease-stress model.
1.1. Genomic Studies of Selection in Humans
There are several fairly well-documented cases of recent simple selection in humans. Examples include skin tone and high-altitude adaptations. The study of highly polygenic selection via changes in frequencies of many alleles (soft sweeps) in humans has advanced recently due to the availability of large-scale, powerful computer clusters and large genomic datasets. A number of recent studies based on genomic data have found evidence of recent polygenic selection in humans over time [
29,
30] and space [
31,
32,
33,
34]. These findings indicate selection for, e.g., height [
31], body mass [
32], cognitive ability [
33], educational attainment [
34,
35], and schizophrenia [
36].
One significant concern with genomic studies that compare polygenic score (PGS) frequencies between major ancestry groups is the “transethnic validity” of PGS [
37]. Most genome-wide association studies (GWASs) are done solely on people of European ancestry (to avoid population-stratification confounding). These studies unfortunately yield predictive models that are less useful in non-Europeans, particularly Africans [
38,
39]. This problem results from two factors. First, the genetic variants discovered in GWASs are typically not causal (with respect to the trait of interest) but are so-called “tag variants.” These are variants that are close enough on the genome to the causal variants such that their presence in the population is statistically linked (known as linkage disequilibrium; LD). Second, the degree to which two variants on the genome are in LD partly depends on the populations studied and their genetic distance; this is because random genetic patterns can arise over time (i.e., genetic drift in LD).
The effect is that a given variant which tags a causal variant in Europeans may not do so, or not very well, in other populations, thus reducing the validity of European-derived estimates of the true PGS. As a result, the predictive validity of associated variants in one major BGA group frequently does not transfer to others. While a number of methods have been employed to control for drift-related effects [
33,
36], and while robust or partially robust inter-BGA differences in educational attainment and intelligence PGS have been found, until causal variants that are not affected by LD [
38] are identified, PGS-based results will continue to carry a degree of uncertainty.
1.2. Admixture Analysis
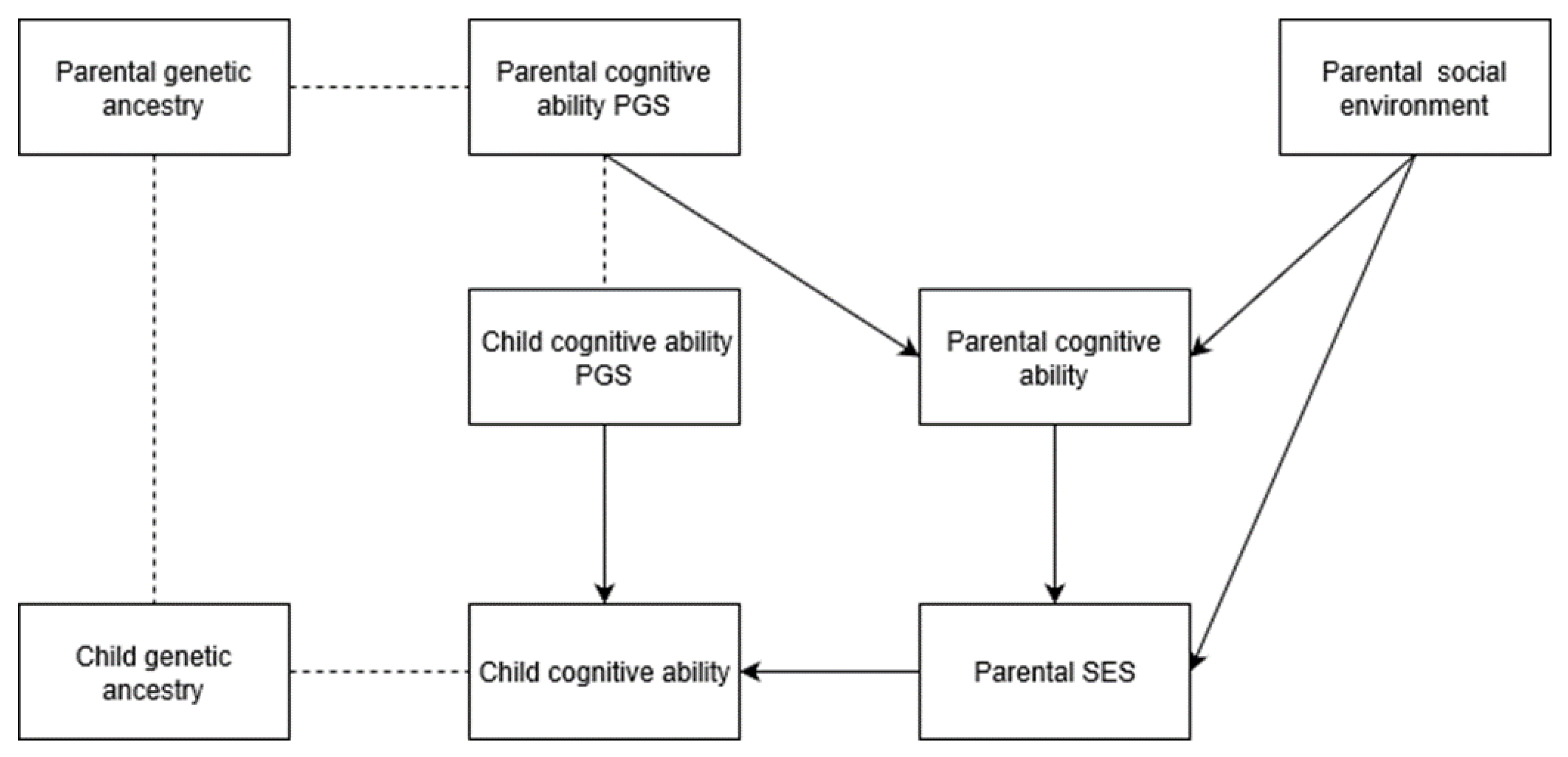
Admixture analysis—analysis of genetic ancestry in previously isolated but recently interbred populations, which relates genetic ancestry to outcomes—is a potent tool used by medical geneticists for the exploration of the source of trait differences and disease disparities in and among admixed populations. The relationships between genomic ancestry and phenotype are treated as indirect evidence of genetic causation, especially when confounds are controlled in regression analysis (e.g., [
40,
41,
42]). Admixture analysis includes global admixture analysis and admixture mapping. When ancestral BGA groups vary in the frequency of genetic variants underlying a trait, in admixed populations the phenotype of interest will be correlated with BGA in genomic regions near the causal genetic variants. This situation allows for the identification of associated loci, a process called admixture mapping [
43,
44]. When the trait has a complex genetic architecture, one where thousands of loci are assumed to contribute to the phenotype, an appropriate first step is global admixture analysis. This process seeks to identify associations between global BGA and phenotype, without attempting to identify local regions of a genome associated with a phenotype. One advantage to global admixture analysis is that it requires much smaller sample sizes compared to admixture mapping.
Templeton [
45] has intricately detailed the logic of global admixture analysis as applied to evolutionary-genetic models. Generally, global admixture analysis within ethnic groups can be viewed as a Mendelian “common garden” experiment, since members of the same ethnic groups experience similar cultures and environments. Factors affecting SIRE groups in general (e.g., stereotype threat, race-based discrimination, segregation, cultural norms, dialect, language, etc.) are either controlled for or attenuated. This basic logic has been adopted by genetic epidemiologists e.g., [
40,
41,
42,
46] who frequently examine the association between global genetic ancestry and traits either within ethnic groups or after controlling for SIRE. Admixture analysis has been applied to study the etiology of inter-BGA differences in, among other traits, height [
47], sleep behaviors [
48], and brain morphology [
49]. The quantitative predictions and theoretical basis of global admixture analysis have often not been explicated. Doing so requires employment of a series of simulations that incrementally increase in model complexity and realism. We do this in Appendix 1 of
Supplementary File 1, which contains the simulations and explanations.
Before the advent of genomic ancestry testing, attempts were made to test evolutionary predictions with respect to inter-BGA cognitive ability differences using other methods (for reviews, see [
50]). Ancestry was estimated using traits such as skin color, blood groups, and reported genealogy. In other words, these studies relied on various proxies or poor measures of genomic ancestry, and further the samples were usually small; moreover, no one has systematically meta-analyzed the data to settle the interpretive dispute. More recent studies have utilized nation-level data on the frequencies of Y-chromosomal haplogroups [
23,
51], estimates of between-country genetic distance [
52], and estimates of regional and national-level ancestry percentages [
5] as predictors of national variation in cognitive ability. These recent studies have been restricted to the national level of analysis, which necessarily reduces sample resolution. As a result of these problems and others, there has been disagreement about whether past studies employing these techniques show, in admixed populations, the expected relations between BGA and cognitive ability [
8,
53,
54].
Thus far, no published study has examined genomic ancestry, assessed using genomic era methods, in a large sample and related the findings to cognitive ability, while taking into account the confounding effect of racial/ethnic identity. One study [
55], using the same sample presently examined, reported ANOVA results for genetic ancestry and test scores. The authors, however, did not report coefficients for specific ancestries, did not use general factor scores, and did not examine the effect of ancestry within SIRE groups. The last of these is a critical aspect of the so-called “common garden” experiment.
A large meta-analysis of pan-American epidemiological studies found that European genetic ancestry was robustly associated with better socioeconomic outcomes relative to African and Amerindian ancestry in admixed populations (European: r = 0.18, k = 28, N = 35,476.5; [
56]). Amerindian and African ancestry were related to poorer socioeconomic outcomes: r = −0.14, k = 31, N = 28,937.5 and r = −0.11, k = 28, N = 32,710.5, respectively. Consistent with evolutionary models, these associations were found within admixed ethnic groups, and when the effect of SIRE was statistically controlled for. Given these associations between genetic ancestry and SES, and the moderate-to-strong relationship between cognitive ability and SES [
57], it is likely that BGA is also associated with higher cognitive ability at least partially independent of SIRE. A related finding is that BGA is a strong predictor of regional cognitive and general socioeconomic outcomes across the Americas [
5,
6,
58] both at the country- and first-order division (e.g., province, state, district) level. This provides ecological support for the expectation that individual BGA will be robustly associated with cognitive ability.
The purpose of the current study is to test the prediction that BGA is associated with cognitive ability after statistically controlling SIRE (which indexes a number of factors commonly invoked to explain cognitive gaps between BGAs, such as racial discrimination), and to quantify the magnitudes of the individual-level associations. More generally, we sought to apply global admixture analysis, via genomic data, on American samples. We aimed to determine if associations between genetic ancestry and IQ are more consistent with commonly proposed evolutionary-genetic or social environment-based explanations for mean cognitive ability differences observed between European and East Asian descent groups relative to African, Amerindian, and Oceanian ones. Based on convergent results from pre-genomic-era studies, national-level analyses of ancestral markers, genomic PGS research, individual-level SES-admixture results, and regional-level SES/cognitive ability-admixture results, we expect that results similar to those found for SES-admixture will be found for cognitive ability.
5. Discussion and Conclusions
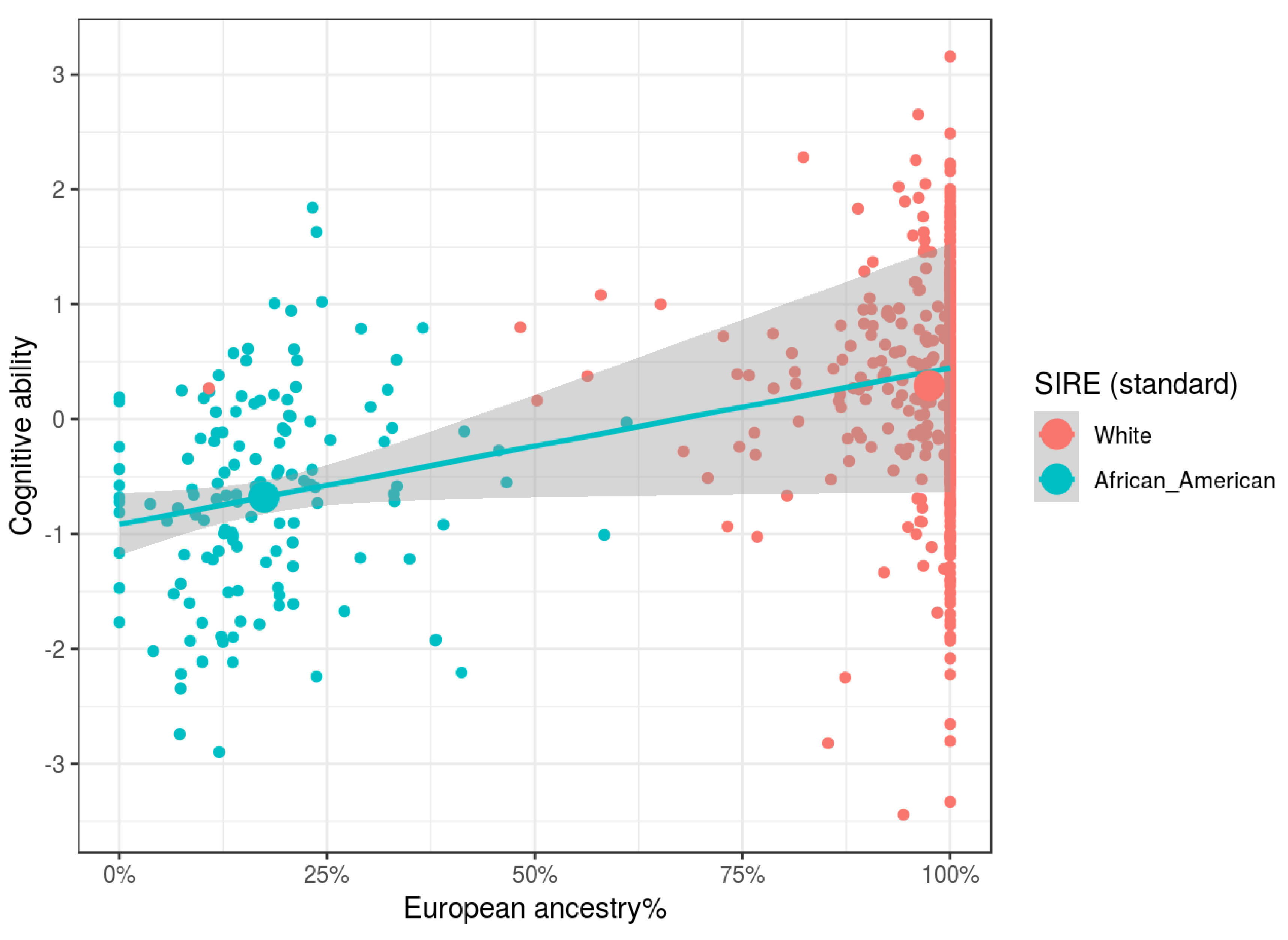
Our results show that independent of SIRE, African, Oceanian, and Amerindian ancestries, relative to Eurasian ones, were associated with lower cognitive ability and parental SES. Relative to European ancestry, there were no clear associations for East Asian ancestry. East Asians in the US are a heterogeneous group, comprised of both South East Asians (e.g., Filipinos and Vietnamese) and North East Asians (e.g., Chinese, Japanese, and Koreans). Since at the national level these groups have different cognitive ability levels [
7], it is difficult to interpret these particular results in light of any global theory of cognitive differences.
Genetic ancestry was found to be related to both cognitive ability and parental socioeconomic status independent of SIRE, as predicted by evolutionary theory. These findings strongly disconfirm claims made by various researchers that there are no statistical relationships between genomic ancestry and cognitive ability when controlling for socially identified racial groups [
9,
53,
54,
75]. Although these findings are congruent with predictions from evolutionary-genetic models, it should be kept in mind that genomic ancestry may also be associated with a number of non-genetic variables that run in families for environmental reasons, some of which may be causal. As such, the apparent validity of genetic ancestry could be due to confounding with these non-genetic variables, or could reflect ancestry-induced social processes. Still, it is worth noting that our results are especially striking in light of the stronger effect of environmental influences on younger compared to older persons’ cognitive ability [
76]. Given this fact, it would be reasonable to expect that at least some environmental factors related to SIRE have their largest potential effects on cognitive ability among young individuals. Yet, in our sample of children, genetic ancestry explained a very great deal of inter-BGA cognitive ability variation, net of SIRE, potentially indicating that environmental effects in general have a limited role in intra-national cognitive ability differences between BGAs.
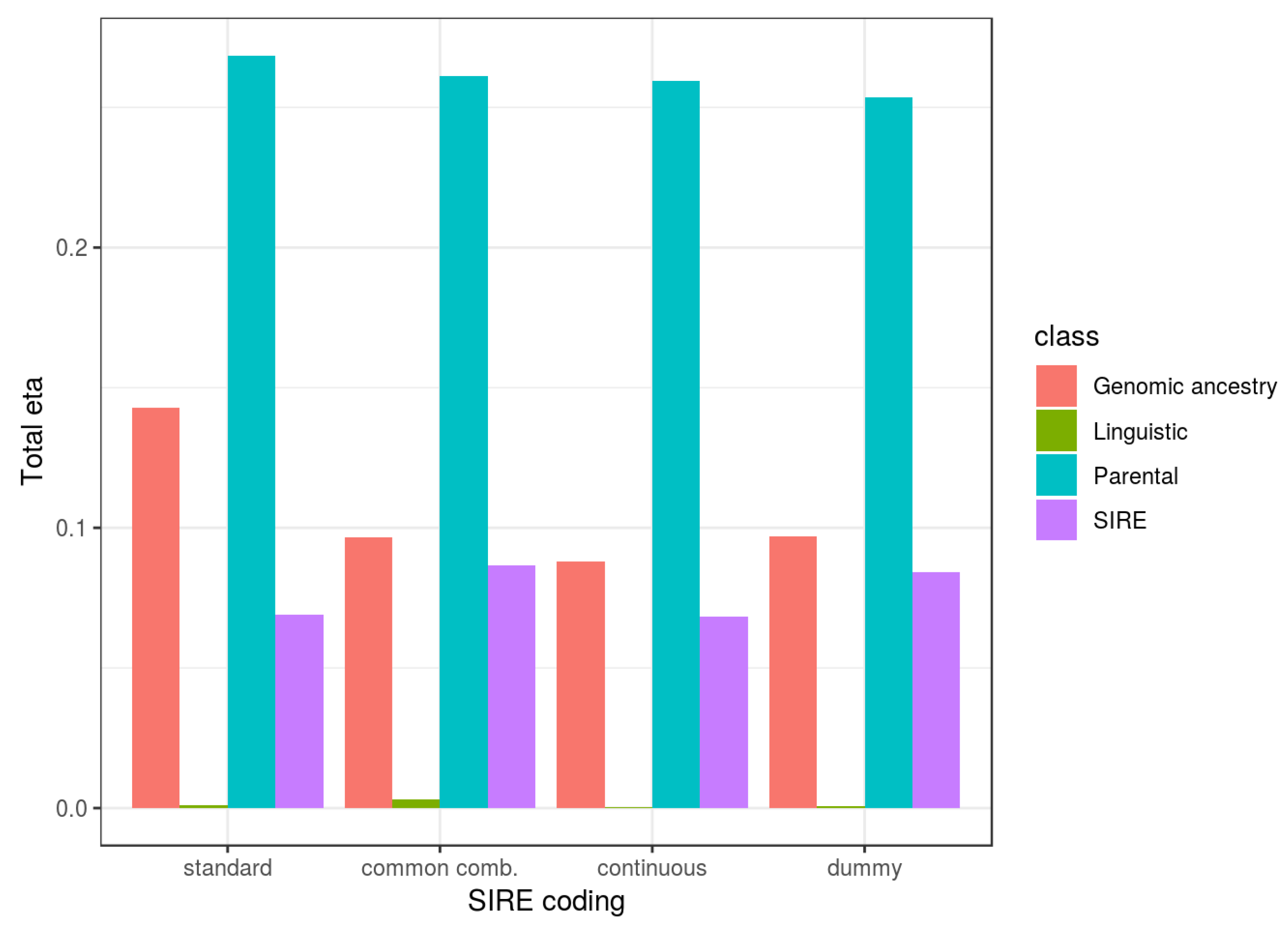
One set of the analyses included parental SES as a predictor of children’s cognitive ability, and it was found to be a useful predictor. Inclusion of this in the models reduced the validity of ancestry predictors by 36–48% (cf.
Section 4.2). As discussed above, a reduction in the effect size is expected on both genetic and non-genetic models. This is because in genetic models, controlling for parental SES indirectly controls for parental cognitive ability, parental genetic influences on cognitive ability, and ultimately children’s genetic influences on cognitive ability, and thus the association between genomic ancestry and the relevant genetic factors is weakened.
Overall, genetic ancestry was a better predictor of outcomes than was SIRE membership. As the US population becomes more ancestrally heterogeneous (owing to admixture), and SIRE and genetic ancestry become less related, genetic ancestry may turn out to be an even better tool for studying race-related social differences. Research suggests this may be the case for very-admixed countries such as Brazil (e.g., [
77]).
5.1. Non-Evolutionary Explanations
While the results we found are consistent with an evolutionary model, there are some potential alternative explanations, namely: phenotypic discrimination, confounding due to immigration status, confounding due to geographic location, and intergenerational environmental transmission.
Some have posited discrimination based on stereotypical race-phenotype, called colorism [
78]. It has been argued that such discrimination could account for covariances between BGA, cognitive ability, and SES (e.g., [
79]). Unfortunately, this dataset does not have appearance data (e.g., skin color), so we could not test whether the associations found are statistically mediated by phenotypic differences. With regard to this sample specifically, we find it unlikely that colorism could directly lead to the association between ancestry and cognitive ability given the ages of the participants. This is because most colorism models propose market-based discrimination (e.g., [
80]). A theoretical possibility is that such discrimination induces associations between parental SES and BGA and that parental SES differences influence offspring cognitive ability. However, some of our associations were only partially reduced in strength when controlling for parental SES, downgrading the likelihood of this scenario.
More generally, it is not clear that colorism is actually a potent force, at least in the USA. Consider research based on sibling designs, which can distinguish between discriminatory and intergenerational effects. A number of studies in the economics literature have utilized sibling control designs in this fashion [
81,
82,
83,
84,
85,
86]. Unfortunately, they differ somewhat in design (e.g., raw vs. SES-controlled results for between-family regressions), and do not report standardized effect measures, so we were unable to quantitatively meta-analyze them. However, generally speaking, when family characteristics are controlled for, residual associations between racial appearance and social outcomes are small. In the words of one researcher who studied a large dataset from Brazil: “[T]he estimated coefficients are small in magnitude, implying that individual discrimination is not the primary determinant of interracial disparities. Instead, racial differences are largely explained by the family and community that one is born into” [
81]. Mill and Stein [
83] make statements to the same effect based on an analysis of a large dataset from the USA.
Another possibility is confounding due to immigration status. Some SIRE groups in our study (specifically East Asians and Hispanics) contain substantial numbers of new immigrants. For these, possible interactions between generational status and admixture complicate interpretations of associations between BGA and social outcomes. For example, the US Hispanic population is comprised of ongoing waves of migrants primarily from Central America and the Caribbean. Since there is an association between immigrants’ generational status and social outcomes (see [
87]), if there is likewise an association between ancestry and generation status, this could lead to biases in parameter estimates. It is difficult to untangle these effects without detailed information about migrant status and specific population histories. That said, Kirkegaard et al. [
56] showed that associations between SES and ancestry can be found across the Americas. It seems unlikely that Amerindian ancestry would be related to SES among native Mexicans, and that African ancestry would be related to SES among native Puerto Ricans, but that in the USA the associations within Latin-American-origin populations would only be due to migrant status confounding. And it also seems unlikely that the cause of the association between ancestry and cognitive ability would be radically different from the cause of the association between ancestry and SES. Furthermore, migrant status confounding cannot non-negligibly factor into associations with African ancestry found in our sample, indicating that non-genetic environmental explanations that rely on migrant status confounding would not be parsimonious.
Regarding geography, we were unable to directly control for the specific locations of the participants. However, when we ran the analyses adding controls for MRI scanner location, as a proxy for geographic location, there was no substantive effect. Overall, the three explanations just discussed seem unlikely to account for our findings. However, the potential confounds that they invoke warrant investigation in future research.
5.2. Related Research
While few studies have looked at the association between genetic ancestry and cognitive outcomes, a large number of older ones have examined the relationship between genealogical and phenotypic indexes of ancestry and cognitive performance. Summarizing research on the relationship between indexes of Amerindian ancestry and outcomes, Berry [
88] notes:
Nevertheless, many other researchers have explored the relationship between academic achievement and certain indices of assimilation and have reached the same conclusion. Coombs (123:6) reports: “Amazingly consistent relationship between the degree of Indian blood and the pre-school language on the one hand and level of achievement on the other. These two characteristics are the best indices of the degree of acculturation.
Atkinson (22) tested students at Union High School, Roosevelt, Utah, and found whites superior, mixed-bloods second, and full-bloods third—a fact frequently encountered in the literature. As many writers have pointed out … such terms as full-blood and mixed-blood refer to social rather than biological groups.
Berry [
88] and many of the researchers he cites interpret the apparent outcome-by-admixture associations as owing to cultural factors; notably, they also interpret the purported ancestry divisions as being delineated culturally, not genetically. Their view suggests that the factors relevant to performance differences can readily diffuse horizontally across cultural groups. Our results do not exclude this possibility but suggest that the relevant factors are being vertically transmitted along genealogical lines – the most plausible candidate being genes.
Loehlin, Lindzey, and Spuhler [
50] summarized research on the relationship between phenotypic indexes of African ancestry and outcomes:
The majority of the studies with persons of mixed African-European ancestry found that groups of subjects judged to be of more African ancestry were on the average slightly inferior on the tests of intellectual functioning employed.
Because of the probability of complex and differential environmental response to physical differences and the likelihood of assortative mating complicating the genetic picture, in addition to the questionable reliability of the racial measures themselves, it is easy to decide that these findings lend themselves to no firm conclusions. One might go beyond this generalization and suggest that the observed findings provide little solace for extremes of either environmentalism or genetical determinism.
Our research design addresses many of the problems noted by Loehlin et al. [
50]. First, we use a multilocus index of genetic ancestry, so cross-assortative mating for specific phenotypic characters (such as skin color) and cognitive ability is not a possible confound. Next, our index of ancestry is highly reliable, unlike the blood group studies that Loehlin et al. [
50] discuss. Moreover, we show in our supplementary simulation that, owing to range restriction in ancestry, the typically small correlations are consistent with large between-group differences. Since there is disagreement about the interpretation of these pre-genomic studies, a systematic meta-analysis is warranted to confirm that indexes of ancestry are generally correlated with cognitive ability.
A number of studies have addressed the issue of the association between genetic ancestry and cognitive ability in more indirect ways (e.g., [
8]). A popular approach involves testing Spearman’s hypothesis, which posits that the magnitude of measured cognitive ability gaps between BGAs is a function of the
g loadings of the test instruments (studies generally support Spearman’s hypothesis; see [
89,
90,
91]). The extent to which
g moderates the magnitude of inter-BGA cognitive ability gaps may be relevant insofar as the
g loadings of tests potentially correlate perfectly with their heritabilities [
92]. Nonetheless, some argue that the truth of Spearman’s hypothesis does not exclude the possibility that inter-BGA cognitive ability gaps have non-genetic environmental origins (e.g., [
93]). Future admixture analyses could resolve this debate over the implications of the truth of Spearman’s hypothesis. For example, if the present model were run for separate subtests, the resultant vector of model beta values could be correlated with the vector of subtest
g loadings to determine whether more
g-loaded subtests involve more strongly genetic inter-BGA ability gaps.
5.3. Evolutionary-Genetic Explanations versus Familial Environmental Models
In the context of admixture analyses of morphological and health-related traits, it is generally accepted (e.g., in genetic epidemiology) that the finding of a correlation between outcomes and genetic ancestry constitutes support for evolutionary-genetic models of inter-BGA differences, especially if potential environmental confounds are statistically controlled (see Introduction). Logically the same inference should apply to behavioral traits and also to outcomes under partial genetic influence, such as educational attainment. (Others make the same point: Nisbett [
53] suggests that a relation between ancestry and IQ within SIRE groups would constitute “direct evidence” supporting hereditarian models of inter-BGA cognitive differences.) But many researchers are inconsistent in their treatment of these outcomes (see, e.g., [
77]), falling for the so-called sociologist’s fallacy [
94,
95]. (Specifically, the sociologist’s fallacy refers to the hasty inference—made without considering the possible role of genetic factors—that a correlation between a social variable (such as SES) and a phenotype (such as cognitive ability) implies that the social variable is causal with respect to the phenotype.)
Support for evolutionary-genetic models provided by global admixture analysis is indirect because there are possible confounds, as discussed above. It is also possible that non-genetic familial models (as noted) make the same predictions. Evolutionary-genetic models, though, can be directly tested using admixture mapping [
46], which in genetic epidemiology is usually the next step taken after consistent findings of an association between outcomes and global genetic ancestry.
The idea behind the method is fairly simple: though we may not know exactly where the causal variants are on the genome, we know roughly where they are located (i.e., in which genes), and they can plausibly be assumed to be the same loci across populations [
39]. This means that if we take cases and controls (or high/low trait groups) from a given admixed population where one ancestry (A) has a higher polygenic score for a trait (e.g., higher disease risk), the cases will have an overall higher proportion of that A ancestry than the controls. However, this increase in ancestry will not be randomly distributed along the genome but will be concentrated in regions around the causal variants. Thus, the test for evolutionary-genetic models is whether such local ancestry enrichment can be found for variants identified via GWASs for cognitive ability. As cognitive ability is strongly polygenic, this test will likely require a large sample size. This method can also be used to test racial-phenotypic discrimination models, as these models predict that associations between genetic ancestry and both behavioral and social outcomes will be more pronounced on regions of the genome related to visible phenotypes. Such analyses have been conducted in relation to assortative mating [
96].
Following methods in genetic epidemiology, we began with a preliminary global BGA analysis, and have shown that non-White (except East Asian) ancestry in the United States is correlated with lower cognitive ability and parental SES. Before drawing conclusions, however, it is important to replicate these results. A single dataset or study cannot settle a debate [
97], owing to potentially unidentified confounds. However, if our results can be replicated, admixture mapping, which can discriminate between phenotypic discrimination, environmental familial, and evolutionary-genetic models, is the next warranted step.
5.4. Implications for Research on Differences
Insofar as there is interest in ameliorating ethnic and racial differences in important behavioral and social outcomes, it is important to understand their causation. Results in this paper are largely in agreement with a familial model of differences in the Americas. These results suggest that outcome differences are being passed on along family lines, and that they are not due to common ethno-cultural factors. This is consistent with an evolutionary-genetic model, but environmental familial models cannot be ruled out at this stage.
These results clearly indicate that using standard categorical SIRE classifications fails to capture the full extent of BGA-associated disadvantages. As for other general models of American race/ethnic differences, contra [
98], these results suggest the possibility of non-trivial omitted variable bias in discrimination-based models. Proponents of such models should try to show that their associations are independent of genetic ancestry. It is not clear that this is generally the case. (For example, see the 1982 Pelotas-birth cohort results reported in Supplementary File 2 of [
56], which show no consistent association between interviewer-reported color and social outcomes after genetic ancestry is statistically controlled.) Proponents of cultural identification models are also encouraged to take genetic ancestry into account.