Characterization and Computational Insights into the Potential Biological Activity of 4-Hydroxyphenyl 8-Chlorooctanoate Purified from Endophytic Fusarium solani
Abstract
1. Introduction
2. Methodology
2.1. Preliminary Bioactivity Screening and Identification of Endophytic Fungi Isolated from S. surattense
2.2. Biosynthesis of Fungal Natural Products Using Czapek Broth Medium
2.3. Extraction of the Crude Metabolite-Enriched Fraction
2.4. Purification of Fungal Metabolites Using Column and Thin Layer Chromatography
2.5. Sample Preparation and Analysis Using GC–MS
2.6. Fourier Transform Infrared (FTIR) Spectroscopic Analysis
2.7. Structure Elucidation of the Compound Using Nuclear Magnetic Resonance (NMR) Spectroscopy
2.8. ADMET Analysis of the Compound-270
2.9. In Silico Analysis of the Compound-270
2.10. Molecular Dynamics Simulation
3. Results
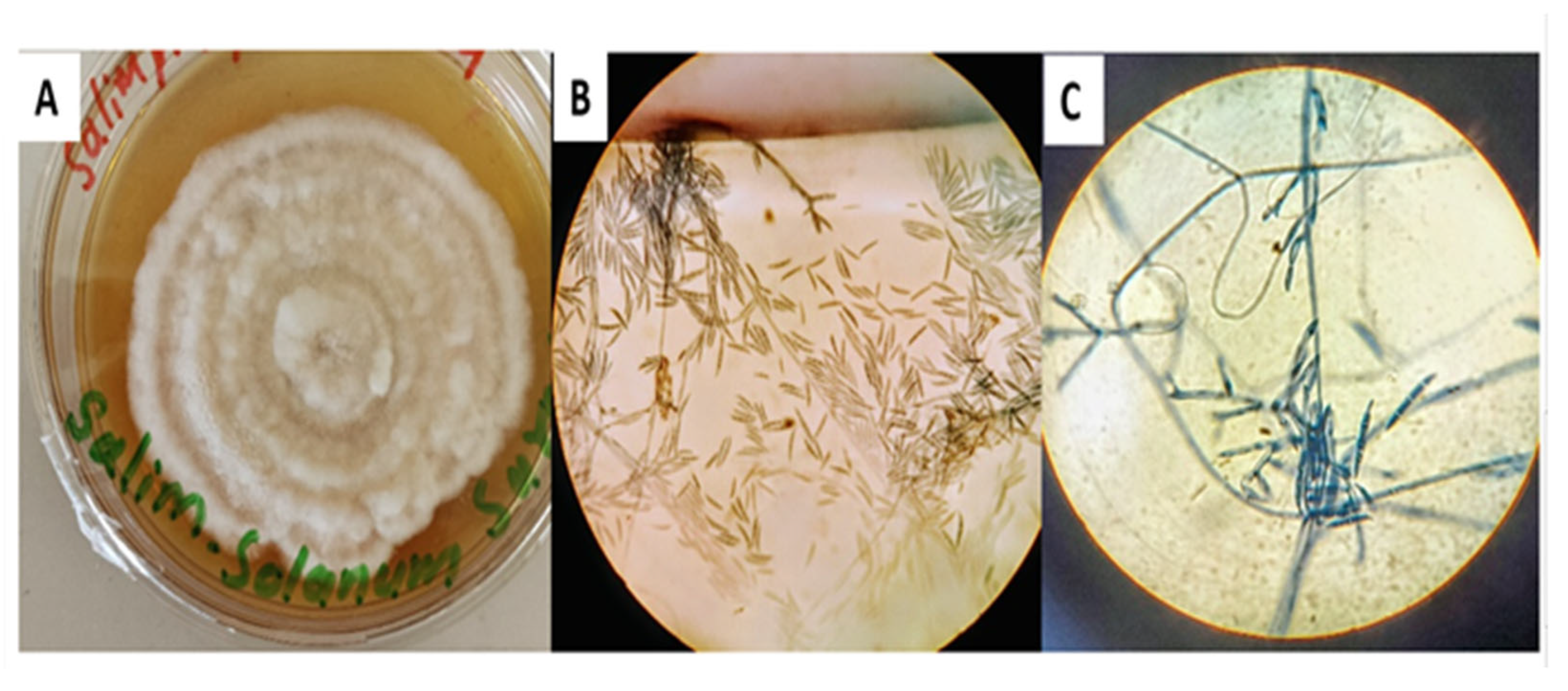
3.1. Culturing and Identification of F. solani Based on Morphological and Microscopic Characteristics
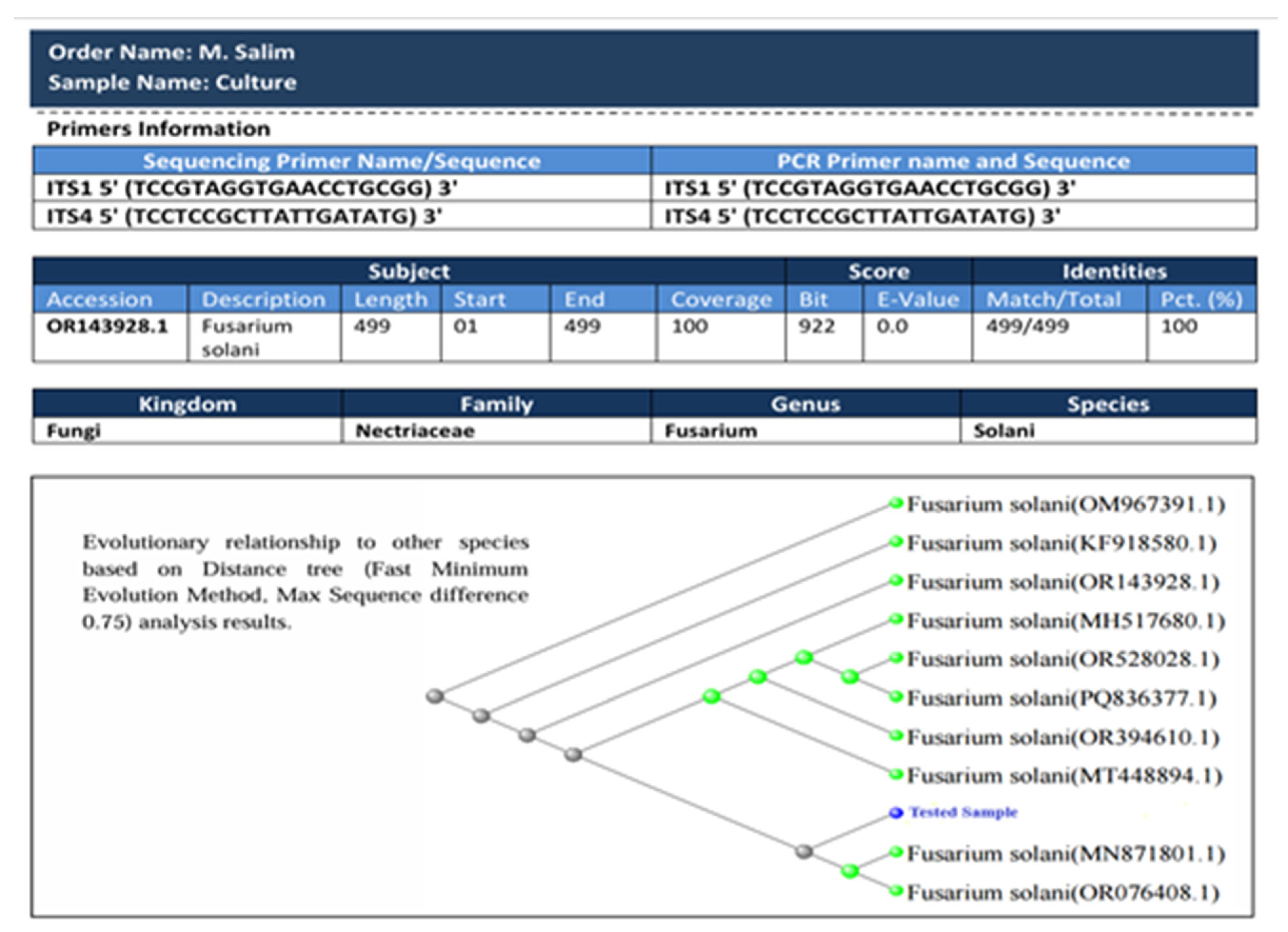
3.2. Molecular Identification
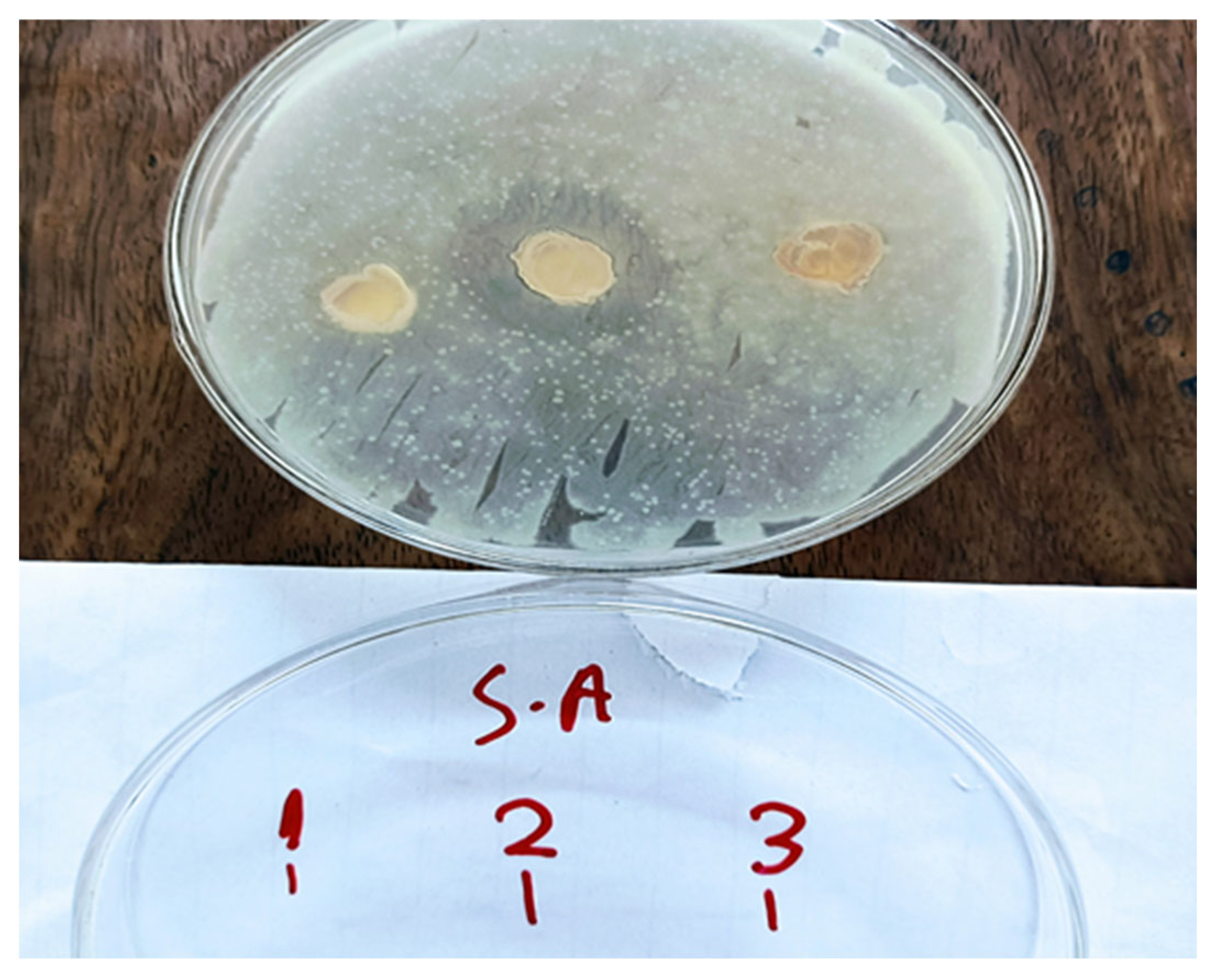
3.3. Preliminary Bioactivity Testing of the Fungal Isolates
3.4. Purification of the Compound Using Chromatographic Techniques
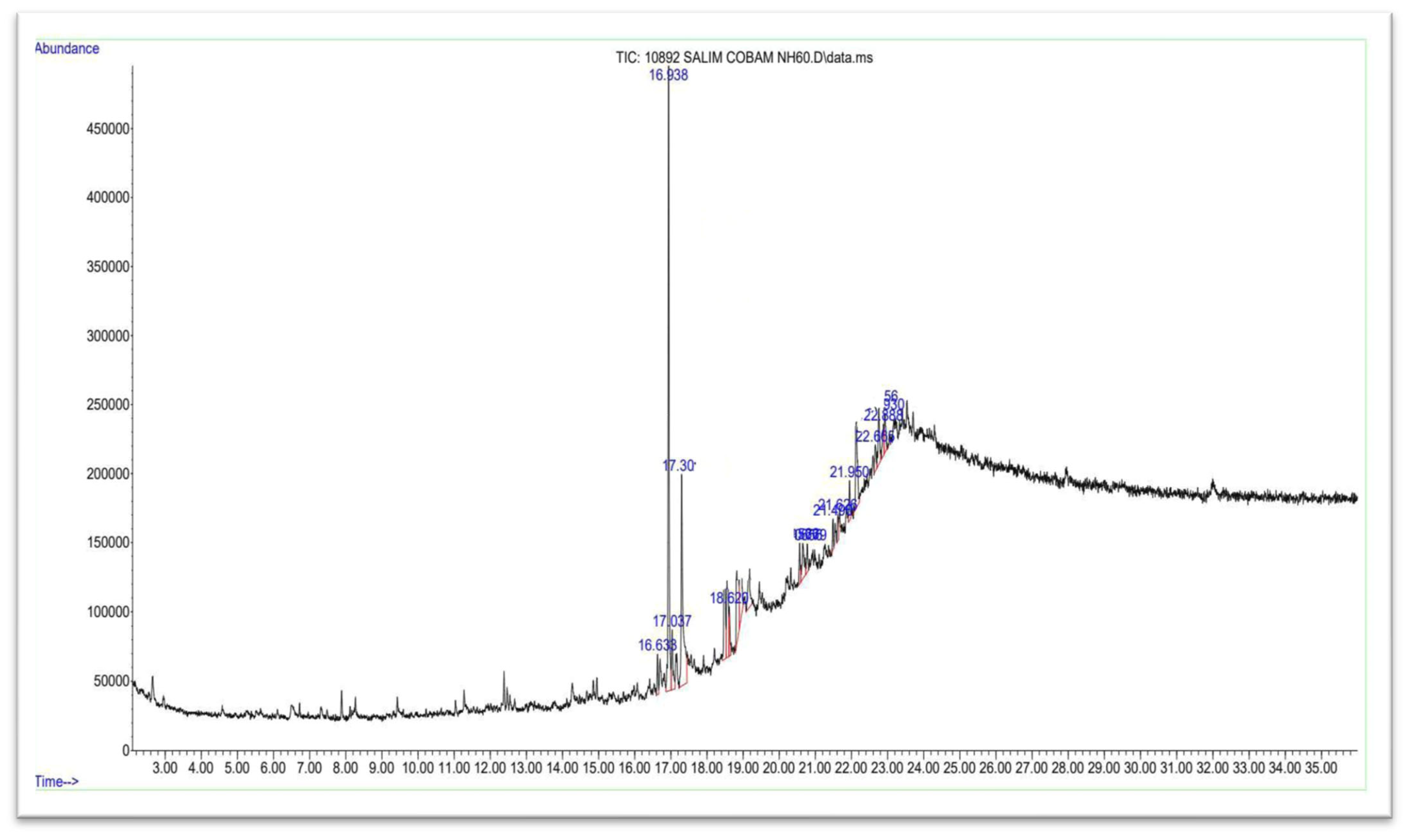
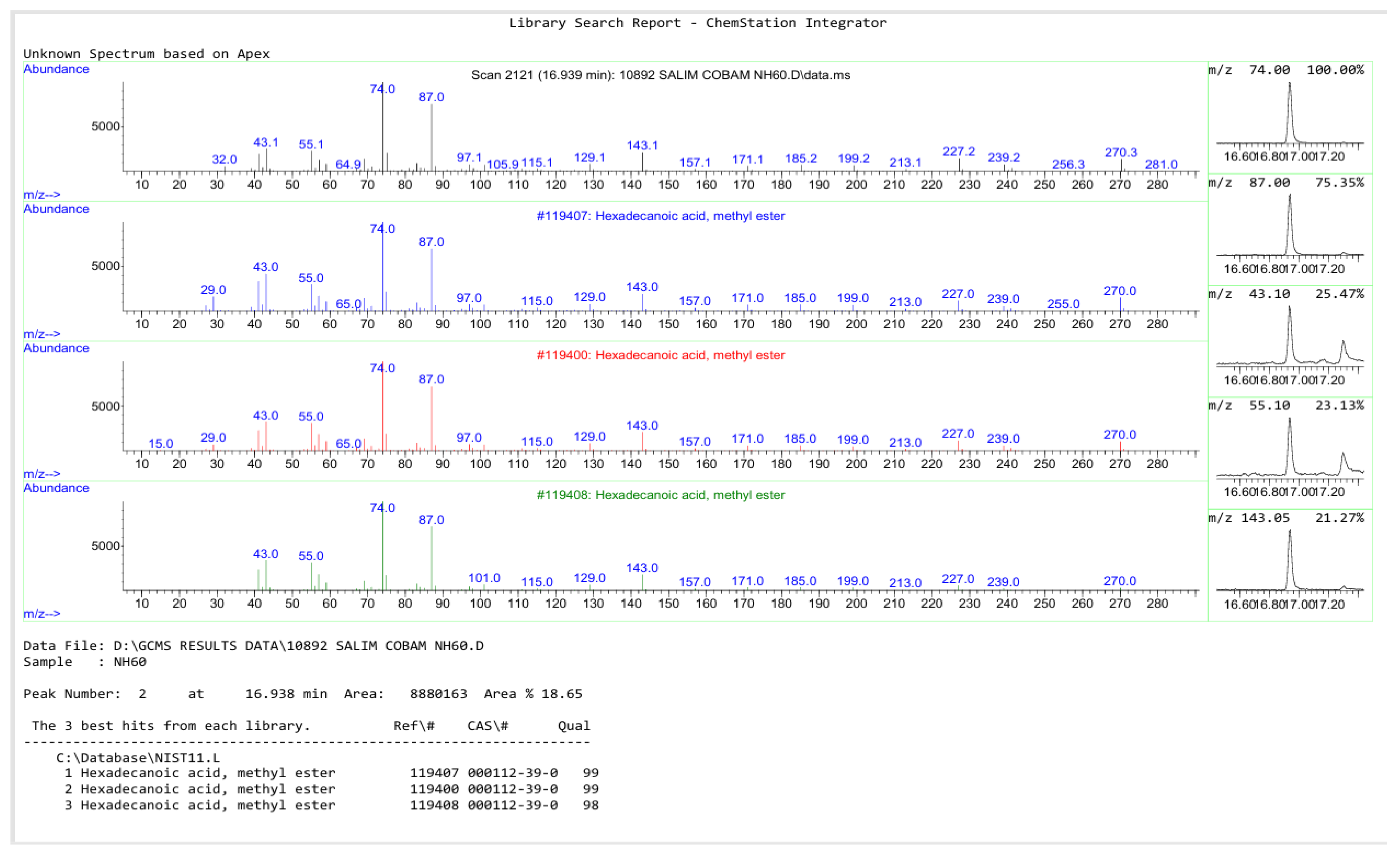
3.5. GC–MS Analysis of the Pure Compound
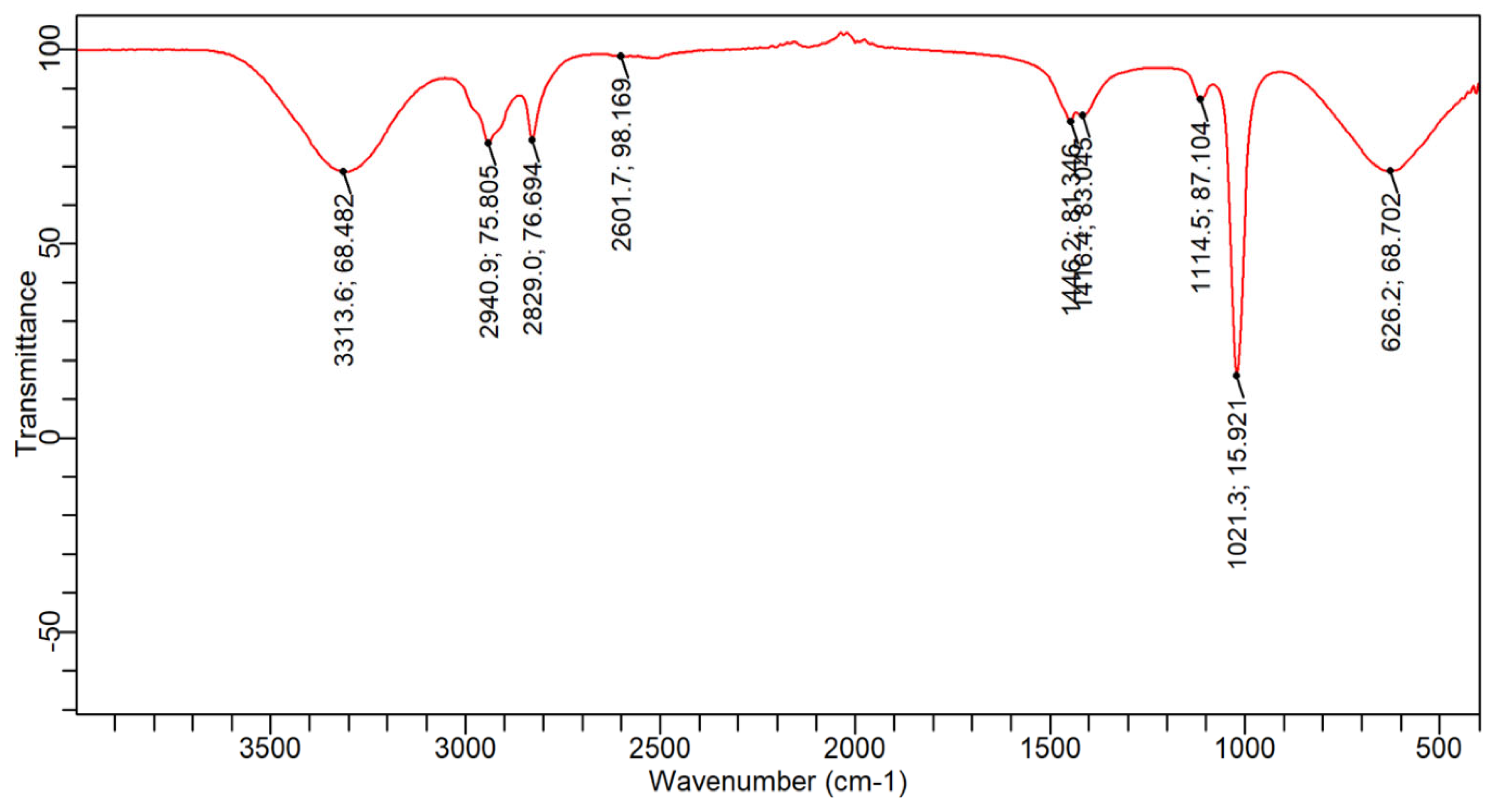
3.6. FTIR Analysis
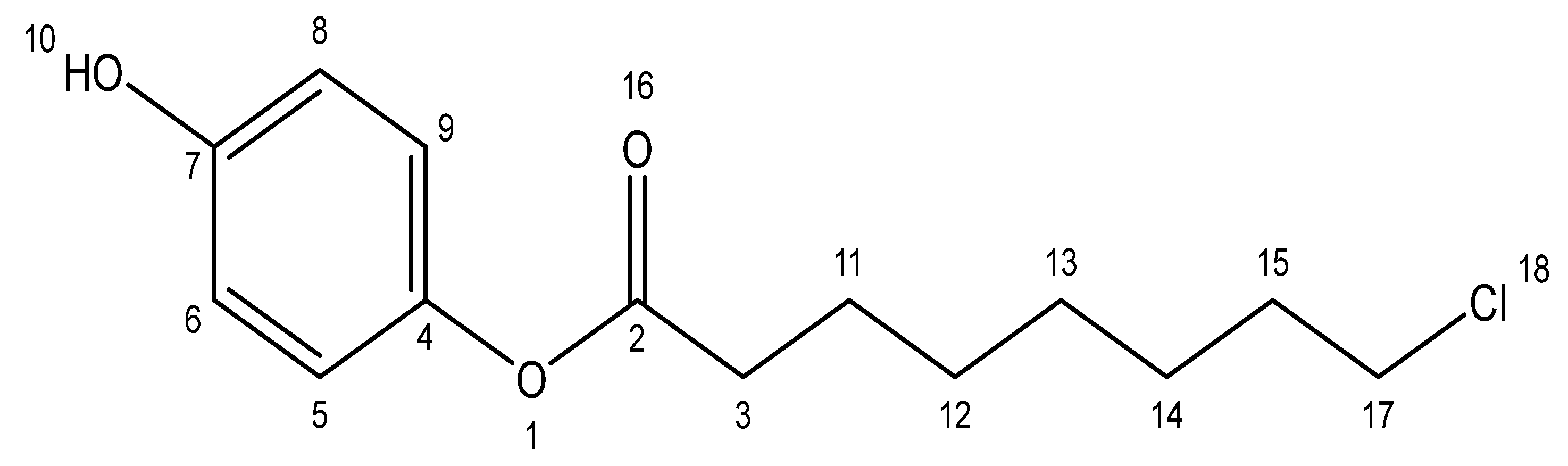
3.7. NMR-Based Structural Elucidation of 4-Hydroxyphenyl 8-Chlorooctanoate
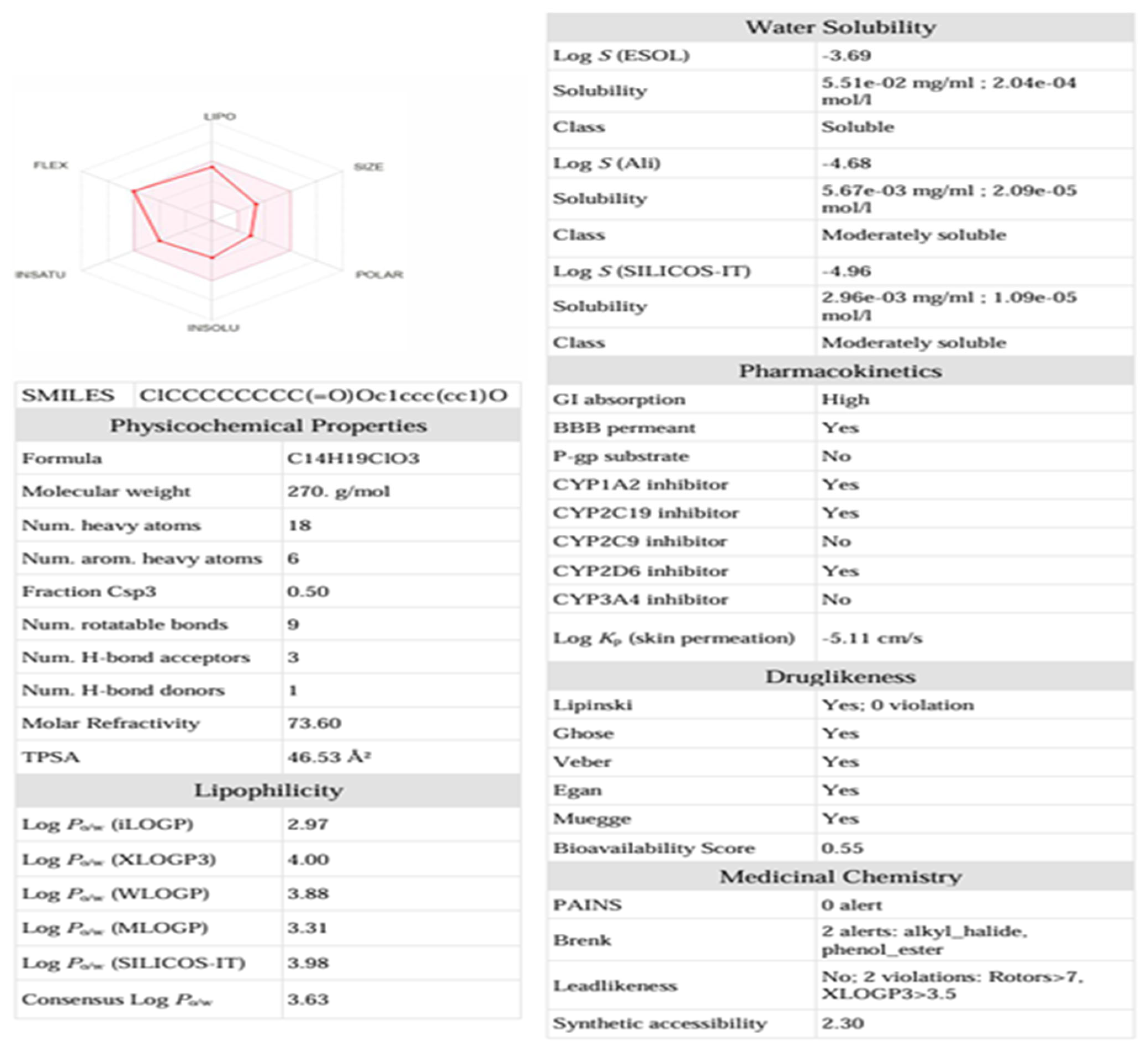
3.8. ADMET Analysis of the Compound-270
3.9. Molecular Docking Studies
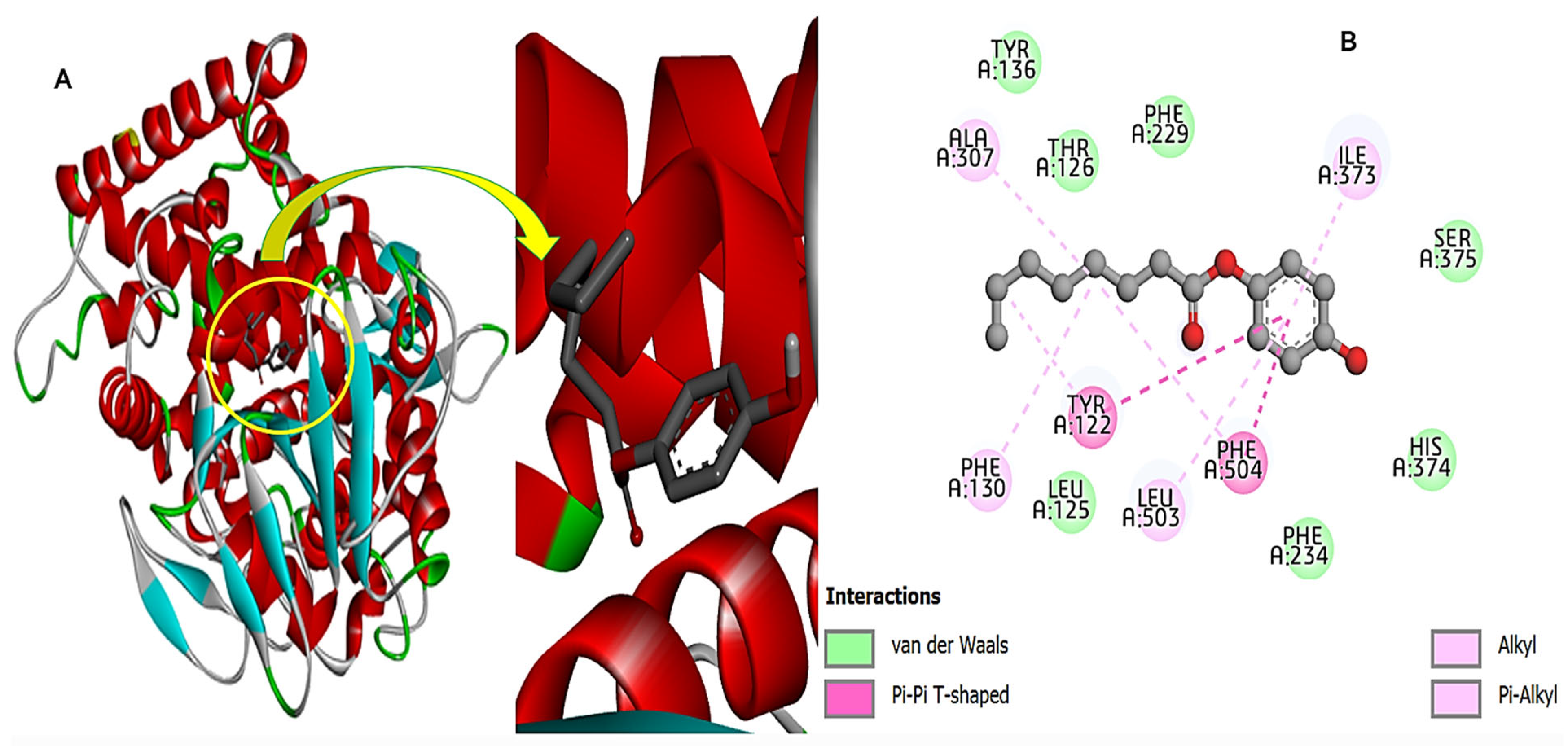
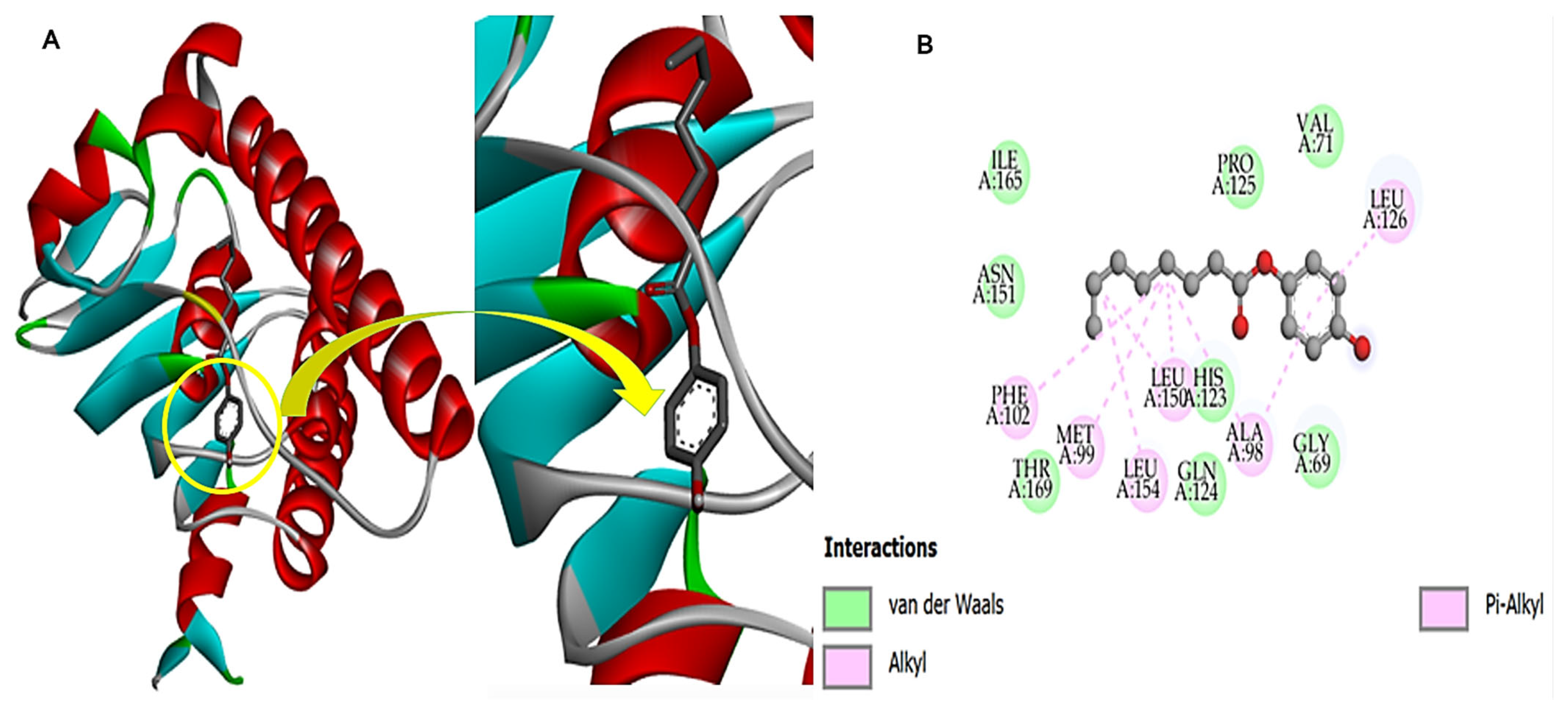
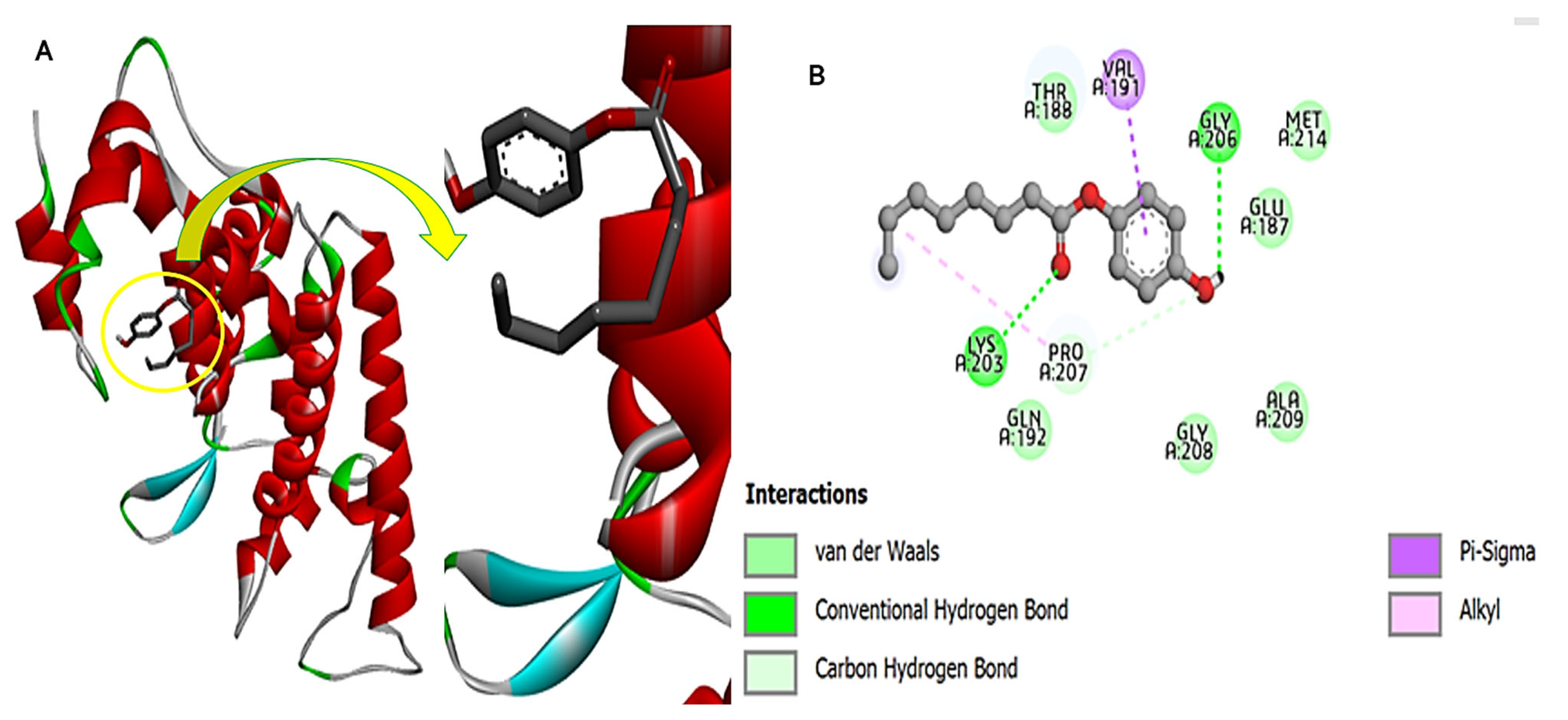
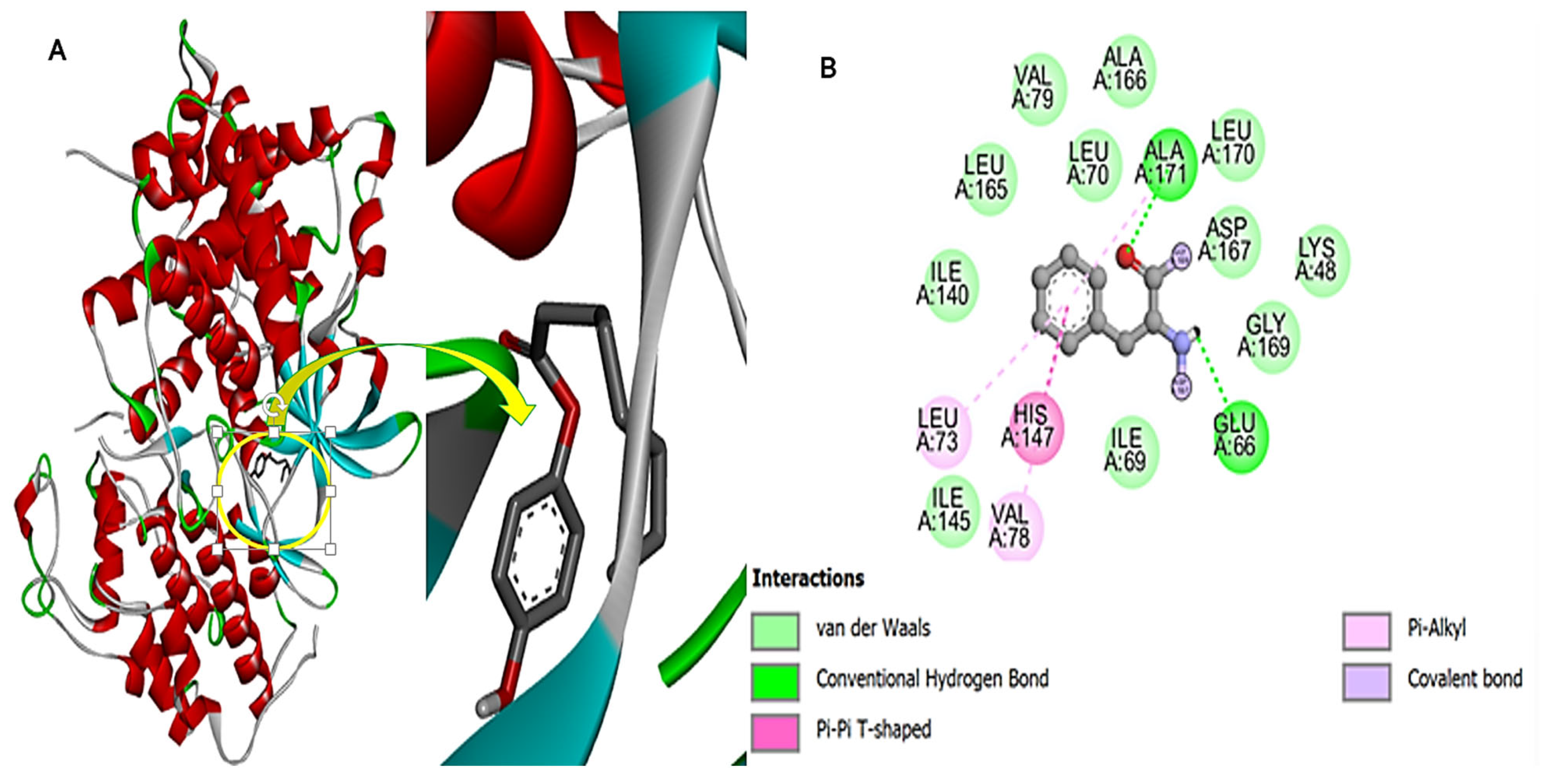
3.10. Fungal Protein and Compound-270 Complex
3.11. Bacterial Protein and Compound-270 Complex
3.12. HIV Protein and Compound-270 Complex
3.13. Cdk-9 Protein and the Compound-270 Complex
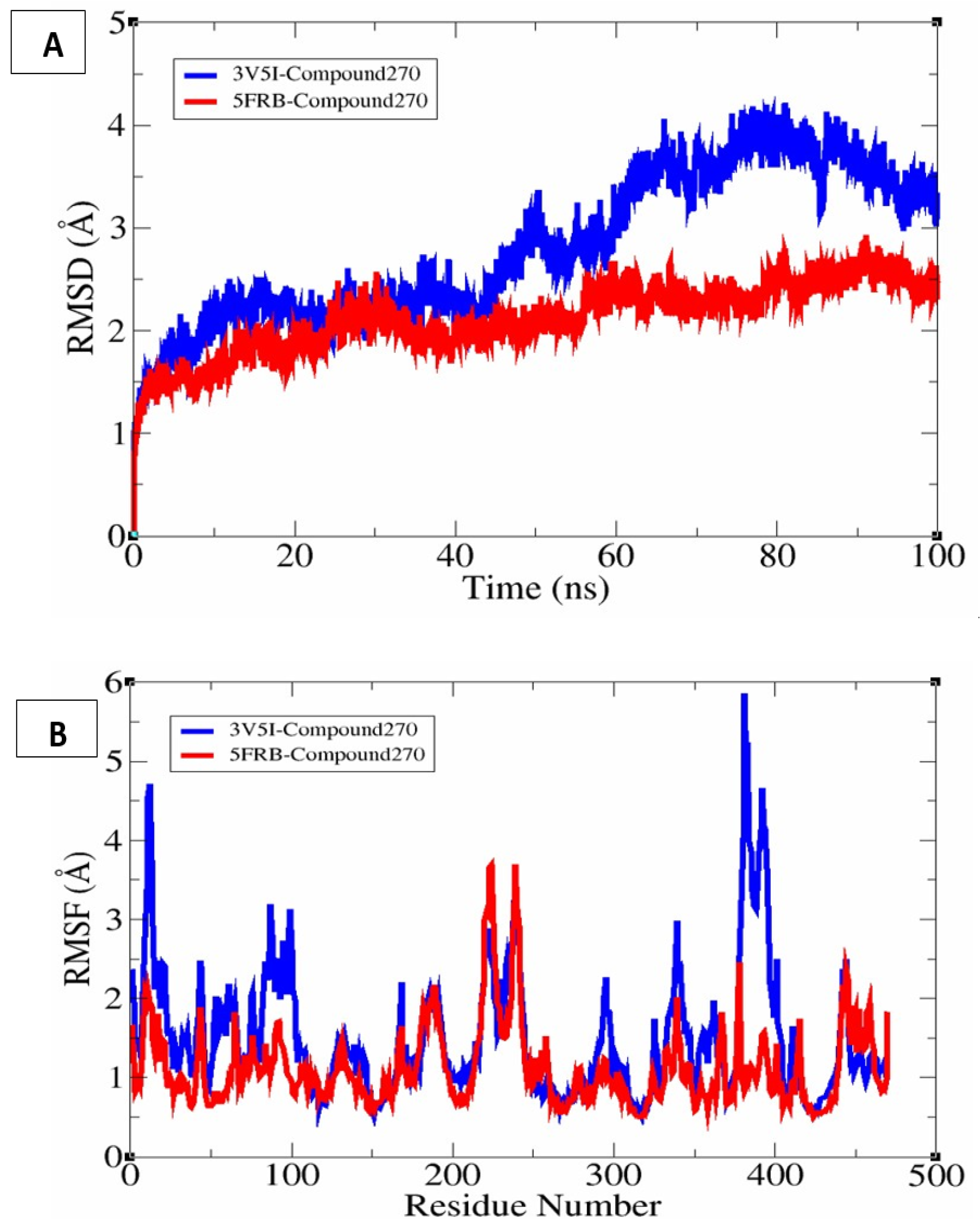
3.14. Molecular Dynamics Simulation
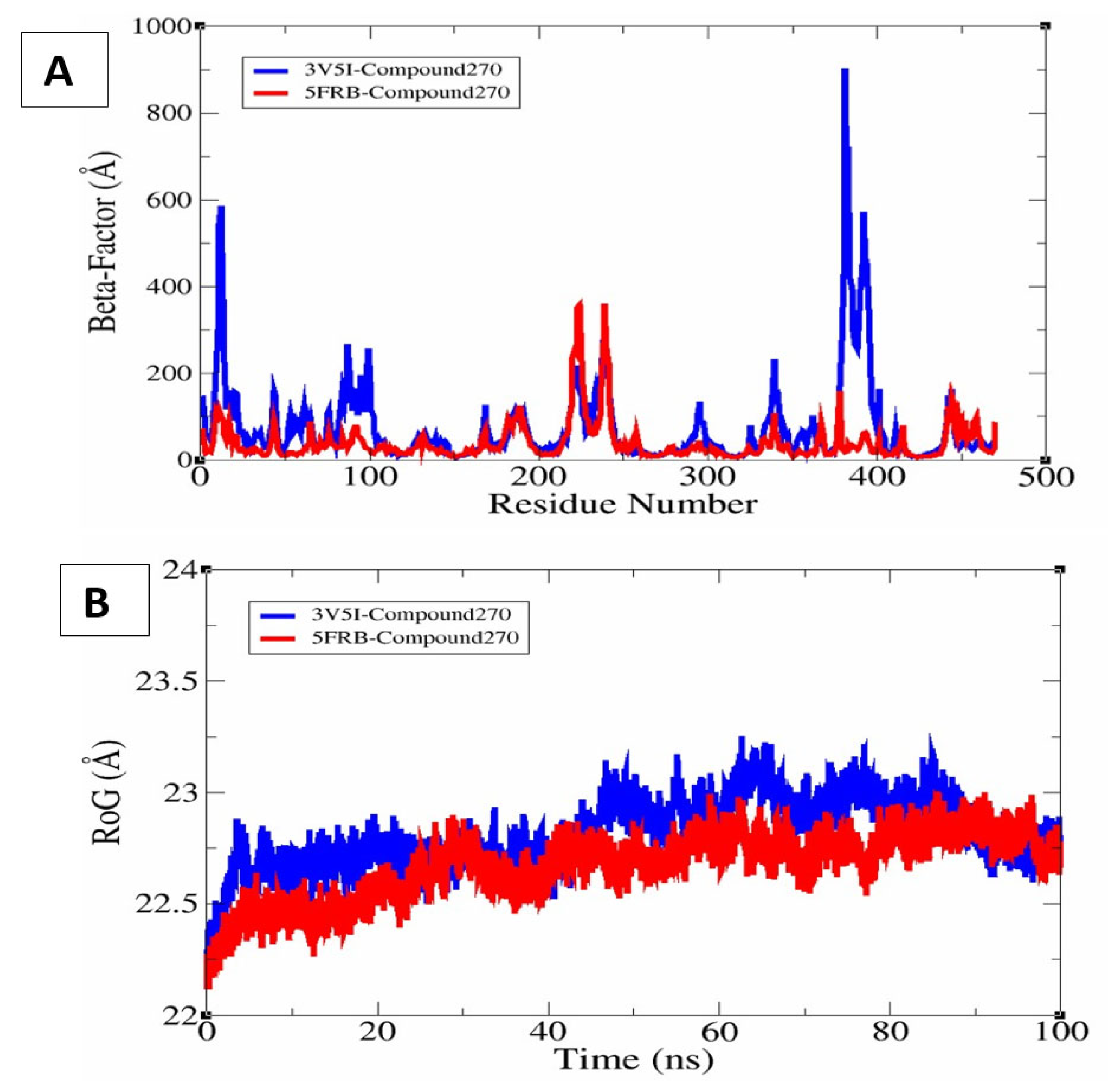
3.15. β-Factor Analysis and Rg
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Data Availability Statement
Conflicts of Interest
References
- Azeem, K.; Fatima, S.; Ali, A.; Ubaid, A.; Husain, F.M.; Abid, M. Biochemistry of bacterial biofilm: Insights into antibiotic resistance mechanisms and therapeutic intervention. Life 2025, 15, 49. [Google Scholar] [CrossRef]
- Niño-Vega, G.A.; Ortiz-Ramírez, J.A.; López-Romero, E. Novel Antibacterial Approaches and Therapeutic Strategies. Antibiotics 2025, 14, 404. [Google Scholar] [CrossRef]
- Correia, J.; Borges, A.; Simões, M.; Simões, L.C. Beyond Penicillin: The potential of filamentous fungi for drug discovery in the age of antibiotic resistance. Antibiotics 2023, 12, 1250. [Google Scholar] [CrossRef]
- Gunatilaka, A.A.L. Natural products from plant-associated microorganisms: Distribution, structural diversity, bioactivity, and implications of their occurrence. J. Nat. Prod. 2006, 69, 509–526. [Google Scholar] [CrossRef] [PubMed]
- De Groot, A.S.; Moise, L.; Terry, F.; Gutierrez, A.H.; Hindocha, P.; Richard, G.; Hoft, D.F.; Ross, T.M.; Noe, A.R.; Takahashi, Y. Better epitope discovery, precision immune engineering, and accelerated vaccine design using immunoinformatics tools. Front. Immunol. 2020, 11, 442. [Google Scholar] [CrossRef] [PubMed]
- Baker, P.; Tiroumalechetty, A.; Mohan, R. Fungal enzymes for bioremediation of xenobiotic compounds. In Recent Advancement in White Biotechnology Through Fungi. Volume 3: Perspective for Sustainable Environments; Springer: Cham, Switzerland, 2019; pp. 463–489. [Google Scholar]
- Tekuri, S.K.; Pasupuleti, S.K.; Konidala, K.K.; Amuru, S.R.; Bassaiahgari, P.; Pabbaraju, N. Phytochemical and pharmacological activities of Solanum surattense Burm. f.–A review. J. Appl. Pharm. Sci. 2019, 9, 126–136. [Google Scholar] [CrossRef]
- Narayanan, M.; Jayashree, T.; Kandasamy, S.; Natarajan, D.; Liu, G.; Elesawy, B.H.; Elfasakhany, A.; Pugazhendhi, A. An in vitro investigation of the antidermatophytic, antioxidant, and nephroprotective activity of Solanum surattense. Process Biochem. 2021, 109, 178–185. [Google Scholar] [CrossRef]
- Parvez, M.K.; Al-Dosari, M.S.; Arbab, A.H.; Alam, P.; Alsaid, M.S.; Khan, A.A. Hepatoprotective effect of Solanum surattense leaf extract against chemical-induced oxidative and apoptotic injury in rats. BMC Complement. Altern. Med. 2019, 19, 154. [Google Scholar] [CrossRef]
- Sashikumar, J.M.; Remya, M.; Janardhanan, K. Antimicrobial activity of ethno medicinal plants of Nilgiri biosphere reserve and Western Ghats. Asian J. Microbiol. Biotechnol. Environ. Sci. 2003, 5, 183–185. [Google Scholar]
- Khan, N.; Afroz, F.; Begum, M.N.; Rony, S.R.; Sharmin, S.; Moni, F.; Hasan, C.M.; Shaha, K.; Sohrab, M.H. Endophytic Fusarium solani: A rich source of cytotoxic and antimicrobial napthaquinone and aza-anthraquinone derivatives. Toxicol. Rep. 2018, 5, 970–976. [Google Scholar] [CrossRef]
- Gupta, C.M.; Tripathi, K.; Tiwari, S.; Rathore, Y.; Nema, S.; Dhanvijay, A.G. Current trends of clinicomycological profile of dermatophytosis in Central India. IOSR-JDMS 2014, 13, 23–26. [Google Scholar]
- James, J.E.; Santhanam, J.; Zakaria, L.; Mamat Rusli, N.; Abu Bakar, M.; Suetrong, S.; Sakayaroj, J.; Abdul Razak, M.F.; Lamping, E.; Cannon, R.D. Morphology, phenotype, and molecular identification of clinical and environmental Fusarium solani species complex isolates from Malaysia. J. Fungi 2022, 8, 845. [Google Scholar] [CrossRef]
- Ortlieb, N.; Klenk, E.; Kulik, A.; Niedermeyer, T.H.J. Development of an agar-plug cultivation system for bioactivity assays of actinomycete strain collections. PLoS ONE 2021, 16, e0258934. [Google Scholar] [CrossRef]
- Tan, W.-N.; Nagarajan, K.; Lim, V.; Azizi, J.; Khaw, K.-Y.; Tong, W.-Y.; Leong, C.-R.; Chear, N.J.-Y. Metabolomics analysis and antioxidant potential of endophytic Diaporthe fraxini ED2 grown in different culture media. J. Fungi 2022, 8, 519. [Google Scholar] [CrossRef]
- Khattak, S.U.; Ahmad, M.; Ahmad, J.; Ikram, S.; Ahmad, S.; Alshabrmi, F.M.; Alatawi, E.A. Purification of Potential Antimicrobial Metabolites from Endophytic Fusarium oxysporum Isolated from Myrtus communis. Appl. Biochem. Biotechnol. 2024, 196, 8940–8964. [Google Scholar] [CrossRef]
- Felisbino, J.K.R.P.; Vieira, B.S.; de Oliveira, A.; da Silva, N.A.; Martins, C.H.G.; Santiago, M.B.; Munoz, R.A.A.; Cunha, L.C.S.; Sousa, R.M.F. Identification of substances produced by Cercospora brachiata in absence of light and evaluation of antibacterial activity. J. Fungi 2021, 7, 680. [Google Scholar] [CrossRef] [PubMed]
- Adams, R.P. Identification of Essential Oil Components by Gas Chromatography/Mass Spectrometry, 5th ed.; Texensis Publishing: Gruver, TX, USA, 2017; pp. 46–52. [Google Scholar]
- Devi, N.N.; Singh, M.S. GC-MS Analysis of metabolites from endophytic fungus Colletotrichum gloeosporioides isolated from Phlogacanthus thyrsiflorus Nees. Int. J. Pharm. Sci. 2013, 23, 392–395. [Google Scholar]
- Coates, J. Interpretation of infrared spectra, a practical approach. Encycl. Anal. Chem. 2000, 12, 10815–10837. [Google Scholar]
- Elyashberg, M. Identification and structure elucidation by NMR spectroscopy. TrAC Trends Anal. Chem. 2015, 69, 88–97. [Google Scholar] [CrossRef]
- Ghosh, S.; Chetia, D.; Gogoi, N.; Rudrapal, M. Design, molecular docking, drug-likeness, and molecular dynamics studies of 1,2,4-trioxane derivatives as novel plasmodium falciparum falcipain-2 (Fp-2) inhibitors. Biotechnologia 2021, 102, 257–275. [Google Scholar] [CrossRef]
- Jia, C.-Y.; Li, J.-Y.; Hao, G.-F.; Yang, G.-F. A drug-likeness toolbox facilitates ADMET study in drug discovery. Drug Discov. Today 2020, 25, 248–258. [Google Scholar] [CrossRef]
- El Aissouq, A.; Bouachrine, M.; Bouayyadi, L.; Ouammou, A.; Khalil, F. Structure-based virtual screening of novel natural products as chalcone derivatives against SARS-CoV-2 Mpro. J. Biomol. Struct. Dyn. 2023, 41, 13235–13249. [Google Scholar] [CrossRef]
- Dallakyan, S.; Olson, A.J. Small-molecule library screening by docking with PyRx. In Chemical Biology; Springer: Berlin/Heidelberg, Germany, 2015; pp. 243–250. [Google Scholar]
- Gersch, M.; List, A.; Groll, M.; Sieber, S.A. Insights into structural network responsible for oligomerization and activity of bacterial virulence regulator caseinolytic protease P (ClpP) protein. J. Biol. Chem. 2012, 287, 9484–9494. [Google Scholar] [CrossRef]
- Bettayeb, K.; Baunbæk, D.; Delehouze, C.; Loaëc, N.; Hole, A.J.; Baumli, S.; Endicott, J.A.; Douc-Rasy, S.; Bénard, J.; Oumata, N. CDK inhibitors roscovitine and CR8 trigger Mcl-1 down-regulation and apoptotic cell death in neuroblastoma cells. Genes Cancer 2010, 1, 369–380. [Google Scholar] [CrossRef] [PubMed]
- Pornillos, O.; Ganser-Pornillos, B.K.; Kelly, B.N.; Hua, Y.; Whitby, F.G.; Stout, C.D.; Sundquist, W.I.; Hill, C.P.; Yeager, M. X-ray structures of the hexameric building block of the HIV capsid. Cell 2009, 137, 1282–1292. [Google Scholar] [CrossRef] [PubMed]
- Pettersen, E.F.; Goddard, T.D.; Huang, C.C.; Meng, E.C.; Couch, G.S.; Croll, T.I.; Morris, J.H.; Ferrin, T.E. UCSF ChimeraX: Structure visualization for researchers, educators, and developers. Protein Sci. 2021, 30, 70–82. [Google Scholar] [CrossRef]
- Raguette, L.E.; Cuomo, A.E.; Belfon, K.A.A.; Tian, C.; Hazoglou, V.; Witek, G.; Telehany, S.M.; Wu, Q.; Simmerling, C. phosaa14SB and phosaa19SB: Updated Amber force field parameters for phosphorylated amino acids. J. Chem. Theory Comput. 2024, 20, 7199–7209. [Google Scholar] [CrossRef]
- BIOVIA, D.S. BIOVIA Discovery Studio 2019. Available online: https://discover.3ds.com/discovery-studio-visualizer-download (accessed on 7 August 2025).
- Bouribab, A.; Errougui, A.; Chtita, S. CADD Methods for Developing Novel Compounds Synthesized to Inhibit Tyrosine Kinase Receptors. Curr. Top. Med. Chem. 2025, 25, 1141–1164. [Google Scholar] [CrossRef]
- Ismail, S.; Ahmad, S.; Azam, S.S. Immunoinformatics characterization of SARS-CoV-2 spike glycoprotein for prioritization of epitope based multivalent peptide vaccine. J. Mol. Liq. 2020, 314, 113612. [Google Scholar] [CrossRef] [PubMed]
- Love, O.; Pacheco Lima, M.C.; Clark, C.; Cornillie, S.; Roalstad, S.; Cheatham III, T.E. Evaluating the accuracy of the AMBER protein force fields in modeling dihydrofolate reductase structures: Misbalance in the conformational arrangements of the flexible loop domains. J. Biomol. Struct. Dyn. 2023, 41, 5946–5960. [Google Scholar] [CrossRef]
- Naz, A.; Yousaf, H.; Zaman, N.; Rauff, B.; Obaid, A.; Awan, F.M. Comprehensive immunoinformatics and structural biology based design for novel peptide vaccines against Epstein-Barr virus. Gene Rep. 2025, 38, 102137. [Google Scholar] [CrossRef]
- Siddiqui, B.; Yadav, C.S.; Akil, M.; Faiyyaz, M.; Khan, A.R.; Ahmad, N.; Hassan, F.; Azad, M.I.; Owais, M.; Nasibullah, M. Artificial intelligence in computer-aided drug design (cadd) tools for the finding of potent biologically active small molecules: Traditional to modern approach. Comb. Chem. High Throughput Screen. 2025; online ahead of print. [Google Scholar]
- Kräutler, V.; Van Gunsteren, W.F.; Hünenberger, P.H. A fast SHAKE algorithm to solve distance constraint equations for small molecules in molecular dynamics simulations. J. Comput. Chem. 2001, 22, 501–508. [Google Scholar] [CrossRef]
- Miandad, K.; Ullah, A.; Bashir, K.; Khan, S.; Abideen, S.A.; Shaker, B.; Alharbi, M.; Alshammari, A.; Ali, M.; Haleem, A. Virtual Screening of Artemisia annua Phytochemicals as Potential Inhibitors of SARS-CoV-2 Main Protease Enzyme. Molecules 2022, 27, 8103. [Google Scholar] [CrossRef] [PubMed]
- Al-Harbi, A.I.; Ullah, A.; Almanaa, T.N.; Gul, F.; Khan, S.; Waheed, Y.; Ul Haq, M.; Muhammad, R.; Khurram, M.; Ullah, A. A chemoinformatic-biophysics based approach to identify novel anti-virulent compounds against Pseudomonas aeruginosa disulfide-bond protein A1. J. Biomol. Struct. Dyn. 2023, 1–10. [Google Scholar] [CrossRef]
- Roe, D.R.; Cheatham III, T.E. PTRAJ and CPPTRAJ: Software for processing and analysis of molecular dynamics trajectory data. J. Chem. Theory Comput. 2013, 9, 3084–3095. [Google Scholar] [CrossRef] [PubMed]
- Bornot, A.; Etchebest, C.; De Brevern, A.G. Predicting protein flexibility through the prediction of local structures. Proteins Struct. Funct. Bioinform. 2011, 79, 839–852. [Google Scholar] [CrossRef] [PubMed]
- Peredo-Lovillo, A.; Romero-Luna, H.E.; Juárez-Trujillo, N.; Jiménez-Fernández, M. Antimicrobial efficiency of chlorine dioxide and its potential use as anti-SARS-CoV-2 agent: Mechanisms of action and interactions with gut microbiota. J. Appl. Microbiol. 2023, 134, lxad133. [Google Scholar] [CrossRef]
- Maillard, J.-Y.; Pascoe, M. Disinfectants and antiseptics: Mechanisms of action and resistance. Nat. Rev. Microbiol. 2024, 22, 4–17. [Google Scholar] [CrossRef]
- Boecker, D.; Zhang, Z.; Breves, R.; Herth, F.; Kramer, A.; Bulitta, C. Antimicrobial efficacy, mode of action and in vivo use of hypochlorous acid (HOCl) for prevention or therapeutic support of infections. GMS Hyg. Infect. Control 2023, 18, Doc07. [Google Scholar]
- López-Arellanes, M.E.; López-Pacheco, L.D.; Elizondo-Luevano, J.H.; González-Meza, G.M. Algae and Cyanobacteria Fatty Acids and Bioactive Metabolites: Natural Antifungal Alternative Against Fusarium sp. Microorganisms 2025, 13, 439. [Google Scholar] [CrossRef]
- Ham, H.N.; Mention, M.M.; Godon, B.; Brunissen, F.; Thomas, M.; Allais, F.; Lopez, M.; Imatoukene, N. Evaluation of the antimicrobial and antioxidant properties of synthetic phenolipids. Process Biochem. 2024, 141, 190–198. [Google Scholar] [CrossRef]
- Durmus, N.; Gulsunoglu-Konuskan, Z.; Kilic-Akyilmaz, M. Recovery, Bioactivity, and Utilization of Bioactive Phenolic Compounds in Citrus Peel. Food Sci. Nutr. 2024, 12, 9974–9997. [Google Scholar] [CrossRef]
- Cheng, C.; Liu, P.; Zhao, P.; Du, G.; Wang, S.; Liu, H.; Cao, X.; Zhao, Q.; Wang, X. Developing novel oenological tannins from 44 plants sources by assessing astringency and color in model wine. J. Sci. Food Agric. 2023, 103, 1499–1513. [Google Scholar] [CrossRef]
- Vavina, A.V.; Seitkalieva, M.M.; Strukova, E.N.; Ananikov, V.P. Fatty acid-derived ionic liquids as soft and sustainable antimicrobial agents. J. Mol. Liq. 2024, 410, 125483. [Google Scholar] [CrossRef]
- Paduszynska, M.A.; Neubauer, D.; Kamysz, W.; Kamysz, E. Anticandidal Activity of Lipopeptides Containing an LL-37-Derived Peptide Fragment KR12. Molecules 2025, 30, 1598. [Google Scholar] [CrossRef]
- Anwer, S.S.; Sdiq, K.H.; Muhammad, K.R.; Aladdin, L.M. Phenolic compound and fatty acid properties of some microalgae species isolated from Erbil City. Brazilian J. Biol. 2022, 82, e256927. [Google Scholar] [CrossRef] [PubMed]
- Balik, M.; Sułkowska-Ziaja, K.J.; Ziaja, M.; Muszyńska, B. Phenolic acids–occurrence and significance in the world of higher fungi. Med. Int. Rev. 2020, 29, 72–81. [Google Scholar]
- Gribble, G.W. Naturally occuring organohalogen compounds—A comprehensive survery. In Progress in the Chemistry of Organic Natural Products; Springer: Berlin/Heidelberg, Germany, 1996; pp. 1–423. [Google Scholar]

Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Salim, M.; Ahmad, S.; Khattak, S.U. Characterization and Computational Insights into the Potential Biological Activity of 4-Hydroxyphenyl 8-Chlorooctanoate Purified from Endophytic Fusarium solani. Chemistry 2025, 7, 130. https://doi.org/10.3390/chemistry7040130
Salim M, Ahmad S, Khattak SU. Characterization and Computational Insights into the Potential Biological Activity of 4-Hydroxyphenyl 8-Chlorooctanoate Purified from Endophytic Fusarium solani. Chemistry. 2025; 7(4):130. https://doi.org/10.3390/chemistry7040130
Chicago/Turabian StyleSalim, Muhammad, Sajjad Ahmad, and Saeed Ullah Khattak. 2025. "Characterization and Computational Insights into the Potential Biological Activity of 4-Hydroxyphenyl 8-Chlorooctanoate Purified from Endophytic Fusarium solani" Chemistry 7, no. 4: 130. https://doi.org/10.3390/chemistry7040130
APA StyleSalim, M., Ahmad, S., & Khattak, S. U. (2025). Characterization and Computational Insights into the Potential Biological Activity of 4-Hydroxyphenyl 8-Chlorooctanoate Purified from Endophytic Fusarium solani. Chemistry, 7(4), 130. https://doi.org/10.3390/chemistry7040130








