Abstract
Mixed-valence complexes contain two metals with different formal oxidation numbers and, therefore, show mixed properties that are influenced by the electronic coupling between the two metals, which is, in turn, regulated by a bridging ligand. This is an attractive point for many researchers. Oxalate is widely used as a bridging ligand for preparing polynuclear complexes. More than 1000 complexes have been reported until now. However, dithiooxamidate, which is an oxalate analog, is less popular as a bridging ligand. Here, a new dithiooxamidate-bridged Ni-diphosphine dinuclear complex with the formula [(μ2-toxa){Ni(dppe)}2](BF4)2 (toxa = dithiooxamidate; dppe = 1,2-bis(diphenylphosphino)ethane) was prepared and characterized via single-crystal X-ray diffraction. When using 1,3-bis(diphenylphosphino)propane (dppp) instead of dppe, dinuclear, trinuclear, and tetranuclear complexes were obtained, i.e., [(μ2-toxa){Ni(dppp)}2](BF4)2, [{μ2-Ni(toxa)2}{Ni(dppp)}2](BF4)2, and [{μ3-Ni(toxa)3}{Ni(dppp)}3](BF4)2, respectively. Bidentate toxa ligands in dinuclear complexes coordinate each Ni atom as κ(S,N). However, the trinuclear and the tetranuclear complexes have the toxa ligands with κ(N,N) and κ(S,S) coordination. The [(μ2-toxa){Ni(dppe)}2](BF4)2 complex undergoes four reversible redox processes, whose analysis via a controlled-potential absorption spectrum reveals the presence of a Ni(II)-Ni(I) mixed-valence state at ∆E1/2 = 0.22 V with a comproportionation constant of 6.1 × 103.
1. Introduction
Mixed-valence complexes have attracted considerable interest since long, leading to numerous reports [1,2,3,4,5]. These complexes contain two metals with different formal oxidation numbers and, therefore, show mixed properties that are influenced by the electronic coupling between the two metals, which is, in turn, regulated by a bridging ligand. In addition, the regulation of the electronic coupling is also achieved by external stimulation (e.g., temperature, pressure, and light), which results in valence tautomerism [6,7].
Oxalate is widely used as a bridging ligand for preparing polynuclear complexes. In contrast, oxamidate and dithiooxamidate, which are oxalate analogs, are less popular as bridging ligands. For example, a search in the CCDC database (updated in November 2022) [8] results in more than 1000 polynuclear complexes bearing oxalate as the bridging ligand, whereas the number of hits for polynuclear complexes with bridging oxamidate and substituted oxamidate ligands, dithiooxamidate and substituted dithiooxamidate ligands is 500 and 30, respectively. Only 12 polynuclear complexes with bridging unsubstituted dithiooxamidate ligands are reported [9,10,11,12,13,14,15,16,17].
To investigate the relationship between the electronic coupling between metal centers and the bridging ligand, several groups studied dinuclear complexes with oxalate analogs. For example, Cotton et al. reported the electronic coupling between Mo2 dimetal units in oxamidate-bridged and dithiooxamidate-bridged bis-(Mo2) complexes [14]. Mikhalyova et al. investigated the exchange coupling in oxalate-bridged, oxamidate-bridged, and dithiooxamidate-bridged dinuclear complexes [15]. Similarly, we aimed to prepare a series of dinuclear Ni complexes bearing oxalate, oxamidate, and dithiooxamidate, respectively. Unfortunately, we only obtained dithiooxamidate–Ni dinuclear complexes.
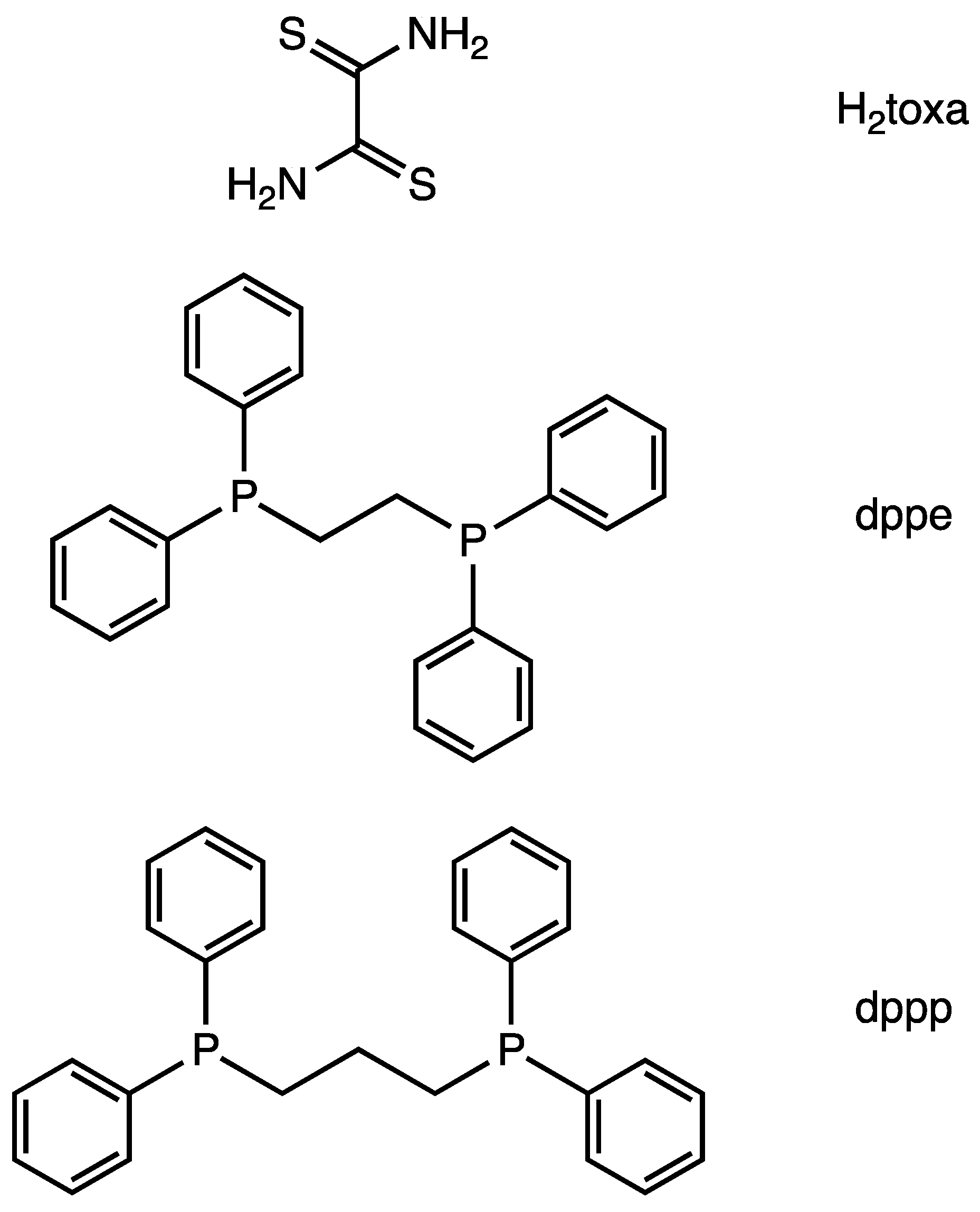
Here, we report the synthesis and X-ray single-crystal structure of a dithiooxamidate-bridged Ni polynuclear complexes, i.e., [(μ2-toxa){Ni(dppe)}2](BF4)2, [(μ2-toxa){Ni(dppp)}2](BF4)2, [{μ2-Ni(toxa)2}{Ni(dppp)}2](BF4)2, and [{μ3-Ni(toxa)3}{Ni(dppp)}3](BF4)2 (H2toxa = dithiooxamide; dppe = 1,2-bis(diphenylphosphino)ethane; dppp = 1,3-bis(diphenylphosphino)propane. Figure 1). The dithiooxamidate shows different coordination between dinuclear complexes and others. We also report electrochemical study of [(μ2-toxa){Ni(dppe)}2](BF4)2.
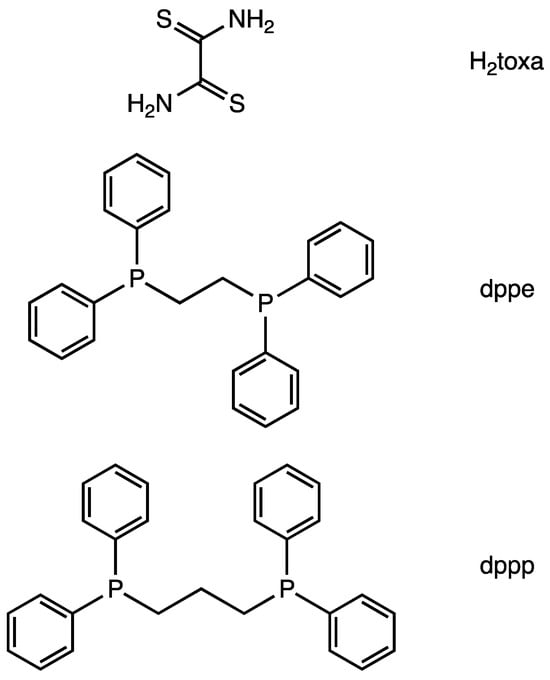
Figure 1.
Molecular structures of ligands.
2. Materials and Methods
All materials were purchased from commercial suppliers (FUJIFILM Wako Pure Chemical Corporation (Osaka, Japan); Kanto Chemical Co., Inc. (Tokyo, Japan); and Tokyo Chemical Industry Co., Ltd. (Tokyo, Japan)) and used as received without further purification.
2.1. Measurements
Absorption spectra were measured using a SHIMADZU UV-3600 UV-vis-NIR spectrophotometer (Kyoto, Tapan). 1H NMR (400 MHz) spectra were measured in CD3CN with a Bruker Avance III HD spectrometer (Rheinstetten, Germany) using CD3CN (δ = 1.94 ppm) as an internal standard. 31P NMR (161 MHz) spectra were measured in CD3CN with a Bruker Avance III HD spectrometer (Rheinstetten, Germany) using 85% H3PO4 in water (δ = 0.00 ppm) as an external standard. Cyclic voltammetry was performed on a BAS BAS100B/W electrochemical workstation (West Lafayette, IN, USA) using a three-electrode cell with a glassy carbon electrode as the working electrode, a Pt coil electrode as the counter electrode, and a homemade Ag+/Ag electrode as the reference electrode. The E1/2 of the ferrocenium/ferrocene couple was 0.11 V vs. Ag+/Ag. The medium was 0.1 mol dm−3 n-Bu4NPF6/CH3CN, and the concentration of the complex was 1 × 10−3 mol dm−3. Controlled-potential absorption spectra were obtained using an optically transparent thin-layer electrode cell and a HOKUTO DENKO HABF501 Potentiostat Galvanostat (Tokyo, Japan). The working electrode was a Pt mesh (80 mesh); the counter electrode was a Pt coil, and the reference electrode was a homemade Ag+/Ag electrode. The optical path length was 1 mm. The medium was 0.1 mol dm−3 n-Bu4NPF6/CH3CN. The concentration of the complex was 3.91 × 10−4 mol dm−3. All electrochemical measurements were performed at room temperature (20 °C) under a nitrogen atmosphere. C, H, and N analyses were conducted by the Service Center for the elementary analysis of organic compounds of Kyushu University.
2.2. Synthesis Procedures
2.2.1. Synthesis of [(μ2-toxa){Ni(dppe)}2](BF4)2 (1)
Dppe (586 mg, 1.47 mmol) was added to a solution of [Ni(H2O)6](BF4)2 (500 mg, 1.47 mmol) in CH3CN (20 mL), and the mixture was stirred for 3 h. To the resulting yellow suspension, dithiooxamide (H2toxa; 88.4 mg, 0.734 mmol) and Et3N (200 μL, 1.47 mmol) were added, and the mixture was stirred overnight. The resulting dark brown solution was evacuated to dryness; the residual solid was dissolved with a small amount of CH3CN, and an excess amount of diethyl ether was added to the solution. The resulting precipitate was collected by filtration and dried in vacuo, affording complex 1 in a 72.5% yield (642 mg). Elemental analysis (%) found: C, 52.45; H, 4.19; N, 2.86. Calc. for C54H50B2F8N2Ni2P4S2·(H2O)2·(CH3CN)0.5: C, 52.32; H, 4.43; N, 2.77. Absorption data (CH3CN) λ/nm(ε/dm3 mol−1 cm−1): 450sh (4800); 380sh (9500); 310sh (29600); 273 (43200) 1H NMR δ 8.23 (s, 2H, N–H); 7.77 (br, 16H, o-phenyl); 7.68 (t, 8H, J = 7.6 Hz, p-phenyl); 7.56 (t, 16H, J = 7.6 Hz, m-phenyl); 2.50 (m, 8H, PCH2CH2P). 31P{1H} NMR δ 62.64 (br). The 1H NMR and 31P NMR spectra are shown in Figures S1 and S2, respectively.
2.2.2. Synthesis of [(μ2-toxa){Ni(dppp)}2](BF4)2 (1a), [{μ2-Ni(toxa)2}{Ni(dppp)}2](BF4)2 (2a), [{μ3-Ni(toxa)3}{Ni(dppp)}3](BF4)2 (3a)
To a solution of [Ni(H2O)6](BF4)2 (200 mg, 0.588 mmol) in CH3CN (10 mL) was added dppp (242 mg, 0.588 mmol), and the mixture was stirred for 3 h. H2toxa (35.0 mg, 0.294 mmol) and Et3N (41 μL, 0.59 mmol) were added to the resulting brown solution, and the mixture was stirred overnight. Subsequent evacuation to dryness and recrystallization of the red-brown crude product via vapor diffusion of diethyl ether into a CH3CN solution afforded brown block crystals along with dark-blue muddy precipitates. The brown block crystals were a mixture of complexes 1a, 2a, and 3a.
2.3. X-ray Crystallography
Single crystals of 1 suitable for single-crystal X-ray analysis were obtained by slow recrystallization via vapor diffusion of diethyl ether into a CH3CN solution of the complex at room temperature. Single crystals of 1a and 2a for X-ray analysis were obtained, following the synthesis method described in Section 2.2.2. Single crystals of 3a for X-ray analysis were obtained via vapor diffusion of diethyl ether into an acetone solution of a mixture of 1a, 2a, and 3a at room temperature. The data were collected on a Rigaku R-AXIS RAPID II diffractometer (Tokyo, Japan) for complexes 1, 1a, and 2a and a Rigaku Saturn 724 CCD area-detector diffractometer for complex 3a. An absorption correction was applied to the intensity data. The structure was solved using a direct method (SHELXT-2014/5) [18] and refined by the full-matrix least-squares method on F2 (SHELXL-2016/6) [18] using the Yadokari-XG software package [19]. All nonhydrogen atoms were refined with anisotropic parameters. H atoms of toxa2− except for complex 1a and water were located in a difference Fourier map, and the coordinate was fixed. Other H atoms were included in the calculated positions and refined using a riding model. Crystallographic diagrams were created using the ORTEP program [20]. Complex 1a seemed to have one diethyl ether and two CH3CN molecules as solvent molecules; however, they were badly disordered and showed large temperature factors. Therefore, they were removed from the final refinement performed using the SQUEEZE program in the PLATON program [21]. The toxa ligands of complex 1a are disordered over two positions (N1a/S1a%/C28/C28%/S1a/N1a% and S2b/N2b%/C28/C28%/N2b/S2b%, N3a/S3a$/C56/C56$/S3a/N3a$ and S4b/N4b$/C56/C56$/N4b/S4b$) and the occupancies are 0.422:0.578 (N1a/S1a%/C28/C28%/S1a/N1a%:S2b/N2b%/C28/C28%/N2b/S2b%) and 0.356:0.644 (N3a/S3a$/C56/C56$/S3a/N3a$:S4b/N4b$/C56/C56$/N4b/S4b$), as a result of optimization. One acetonitrile (N7B-C90B-C91B) and triethylamine in complex 2a are treated as a disorder, and the occupancy is 0.570:0.430 (acetonitrile:triethylamine) as a result of optimization. A phenyl group of a dppp in complex 3a is disordered over two positions (C70A-C71A-C72-C73A-C74A-C75A and C73B-C74B-C75B-C72-C76B-C77B) and the occupancy is 0.604:0.396 (C70A-C71A-C72-C73A-C74A-C75A:C73B-C74B-C75B-C72-C76B-C77B), as a result of optimization. One acetone in complex 3a is also disordered over two positions (O6A-C105-C106-C107 and O6B-C108-C106-C107), and the occupancy is 0.531:0.469 (O6A-C105-C106-C107:O6B-C108-C106-C107), as a result of optimization. Crystallographic data are summarized in Table S1. The CCDC 2259972, 2259973, 2259974, and 2259975 contain the crystallographic data 1, 1a, 2a, and 3a of this paper.
3. Results and Discussion
3.1. Synthesis of μ-Toxa Polynuclear Ni Complexes
The μ2-toxa dinuclear Ni complex [(μ2-toxa){Ni(dppe)}2](BF4)2 (1) was prepared via a one-pot reaction using [Ni(H2O)6](BF4)2, dppe, H2toxa, and Et3N. The reaction afforded a brown crude product, from which the complex was obtained via recrystallization with CH3CN/diethyl ether.
In the case of using dppp instead of dppe, a similar one-pot reaction and evacuation produced similar red-brown crude. However, a treatment of a small amount of CH3CN and the excess amount of diethyl ether produced red-brown sticky solids. To purify the sticky solids, slow vapor recrystallization was carried out. The recrystallization afforded brown block crystals along with dark-blue muddy precipitates. The brown block crystals were mixture of the dinuclear [(μ2-toxa){Ni(dppp)}2](BF4)2 (1a), trinuclear [{μ2-Ni(toxa)2}{Ni(dppp)}2](BF4)2 (2a), and tetranuclear [{μ3-Ni(toxa)3}{Ni(dppp)}3](BF4)2 (3a). In the reaction, the main product was the muddy precipitates, and the yield of the complexes was very low. We speculate that prolonged recrystallization with polar solvent might break complex 1a and might restructure 1a to produce dark-blue muddy precipitates in 2a and 3a. An appropriate synthesis method affording good yields of 1a, 2a, and 3a has not been established yet.
3.2. Structure of μ-Toxa Polynuclear Ni Complexes
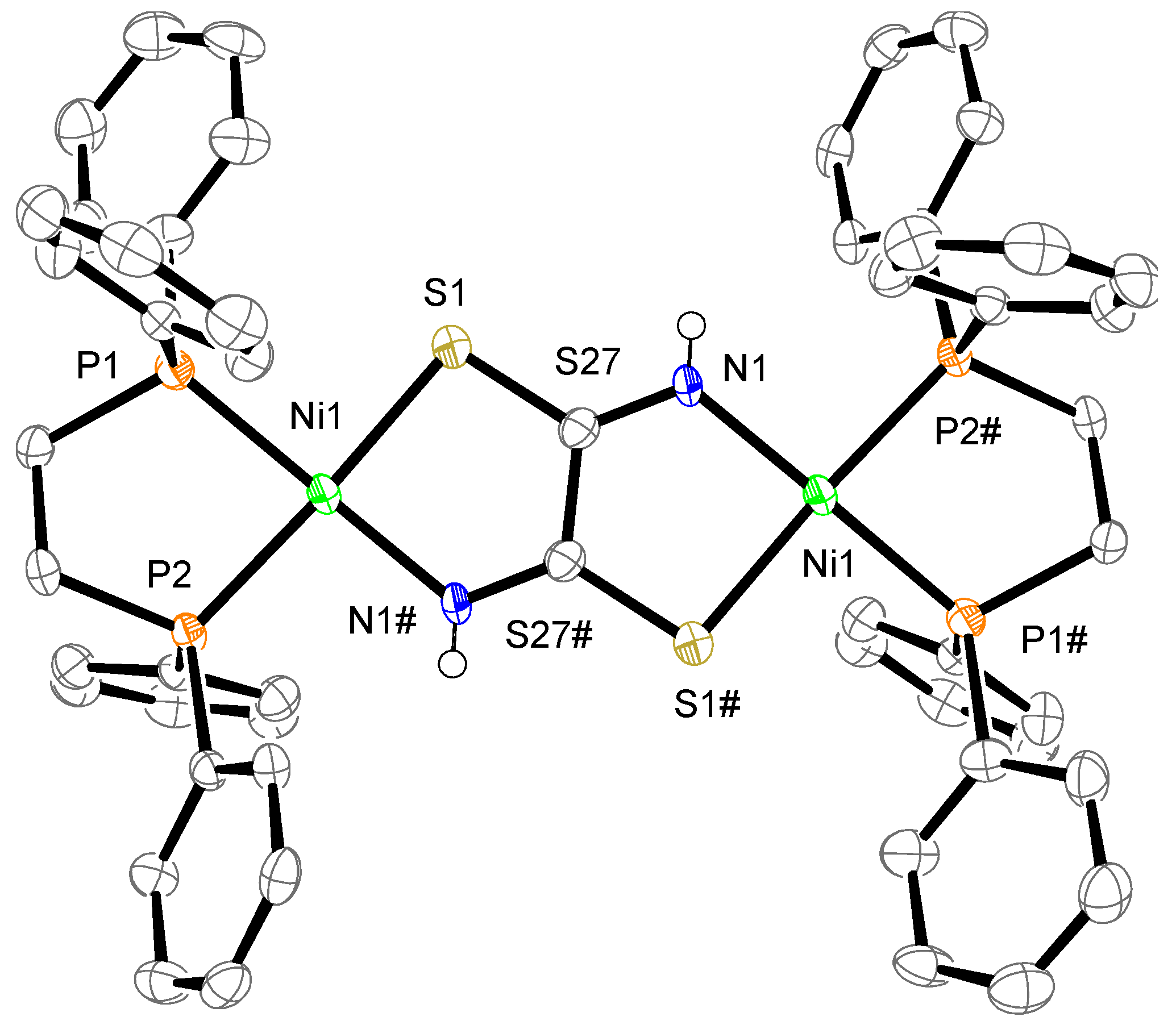
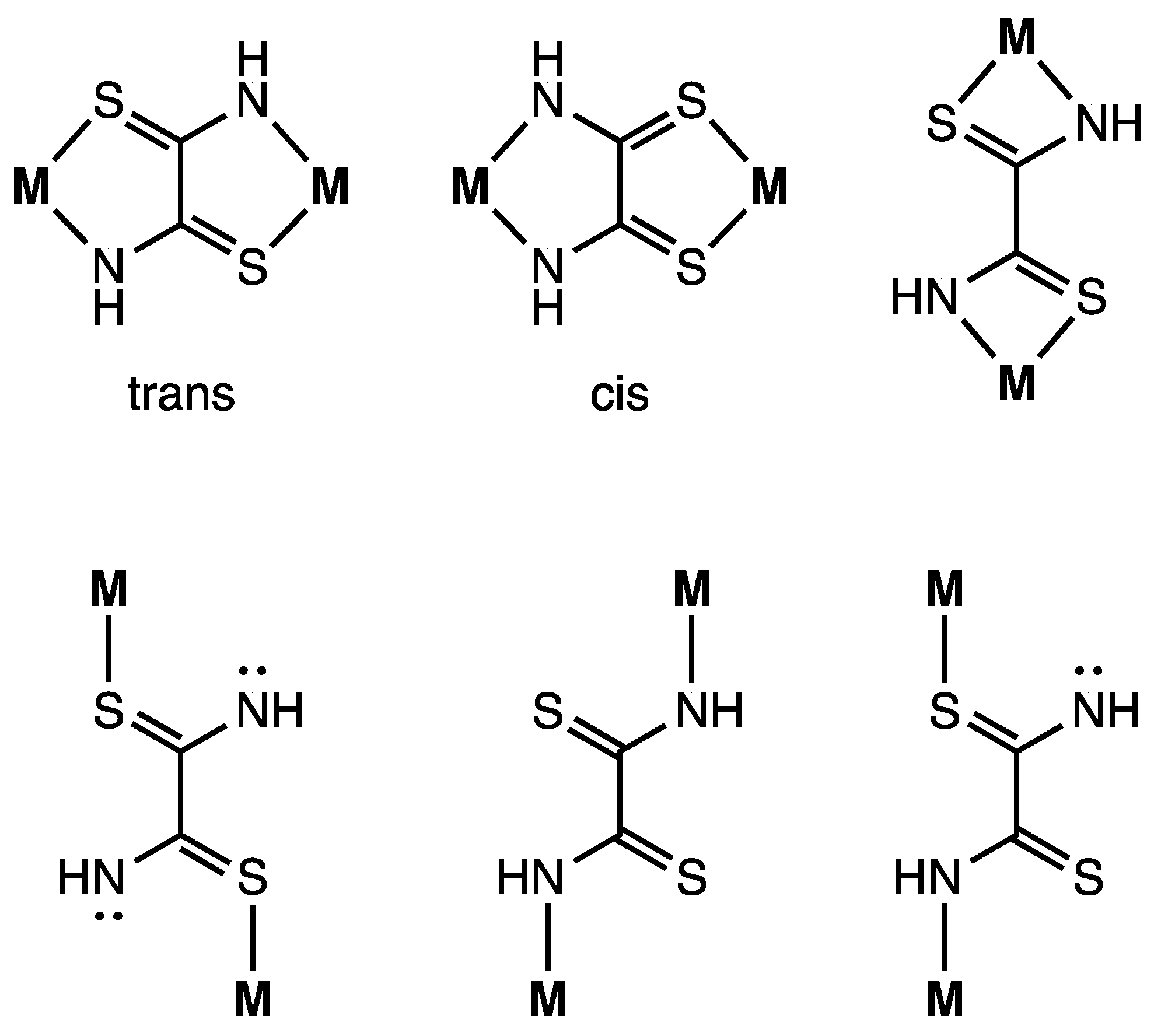
The crystal structure of complex 1 is shown in Figure 2. The complex consists of one complex cation, two BF4− counter anions, two CH3CN, two water molecules, and one dppe dioxide. The cation has two mono-Ni(dppe) units bridged by a toxa ligand, forming the dinuclear Ni complex. The possible bridging modes of the toxa ligand between two metals are shown in Figure 3. The toxa ligand in complex 1 is coordinated in a trans form. Several μ-toxa and μ-substituted-toxa dinuclear structures have been reported [10,13,16,22,23,24]. The difference Fourier map of complex 1 revealed the presence of one proton in each N atom; therefore, deprotonation of dithiooxamide resulted in the loss of one proton from each N atom. Each Ni atom exhibits a P/P/N/S four-coordination environment constructed by the coordination of a bidentate dppe ligand and a toxa-κ(S,N) bridging ligand. The deviation of Ni1 from the least-square mean plane of P1/P2/S1/N1# is 0.03 Å. The dihedral angle between the least-square mean plane of P1/P2/Ni1 and the least-square mean plane of N1#/S1/Ni1 is 9.7(2)°. These data indicate that the Ni(II) ion has a slightly distorted square-planar geometry and two of these structures are arranged in an almost coplanar fashion in complex 1.
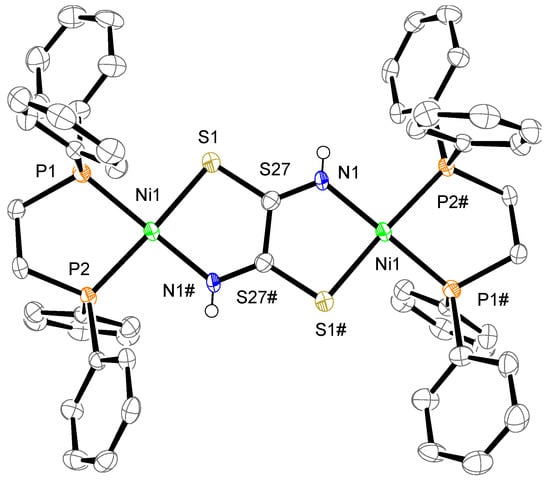
Figure 2.
Crystal structure of the cation of complex 1. The counter anions, solvent molecules, diphosphine dioxide, and hydrogen atoms, except for N–H, are omitted for clarity. The thermal ellipsoids are drawn at the 50% probability level. The subscripts # indicate the equivalent atoms generated by the symmetry operators (1-x, 1-y, -z).
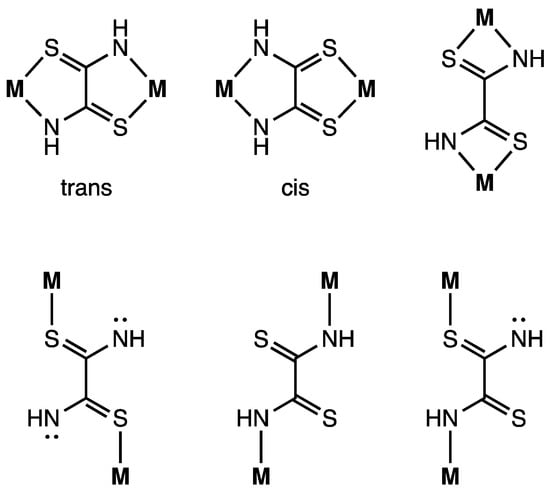
Figure 3.
Bridging modes of the dithiooxamidate ligand.
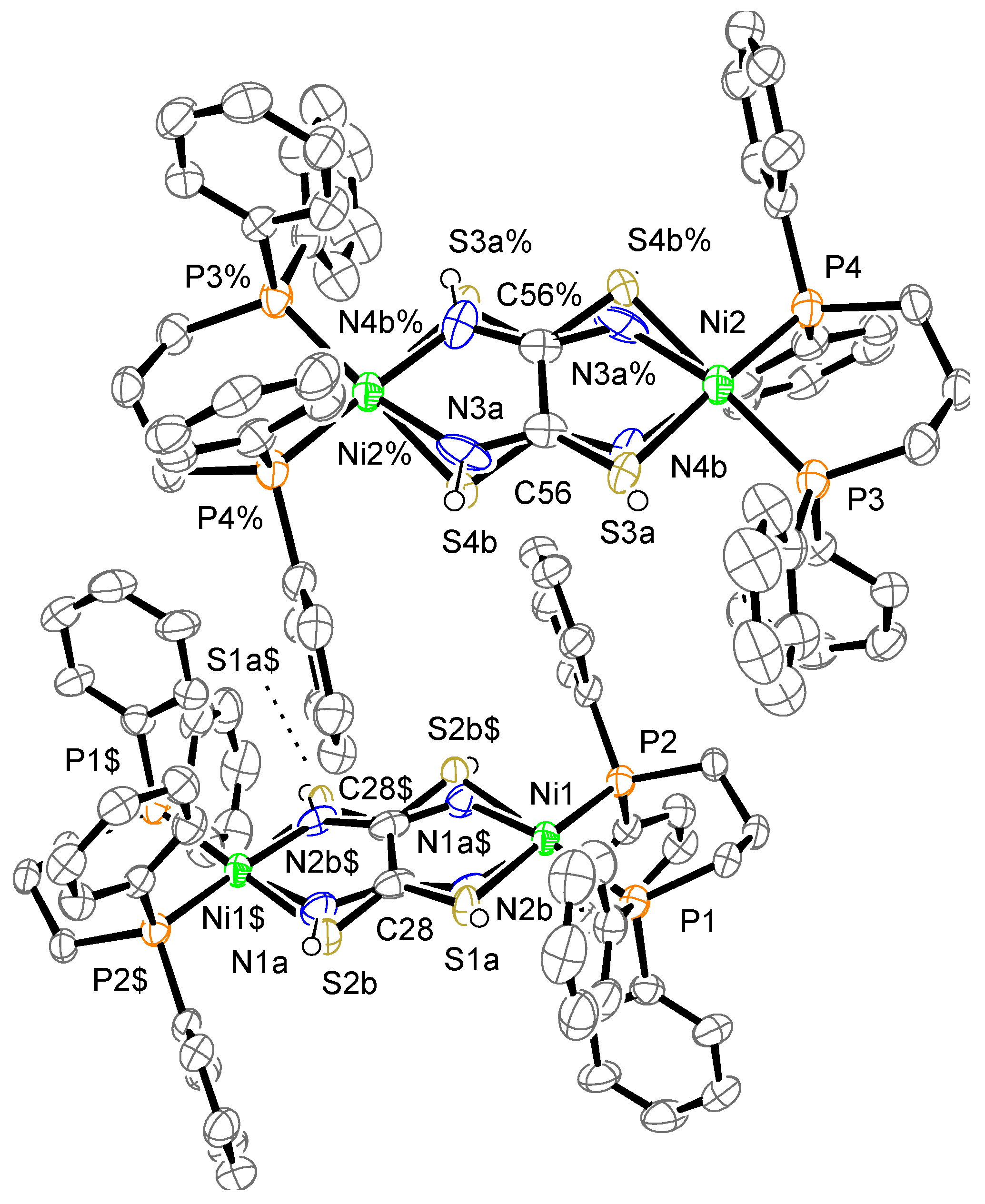
The crystal structure of complex 1a is shown in Figure 4, which displays two crystallographically independent molecules. Complex 1a consists of one complex cation, two BF4− counter anions, and some solvent molecules; however, the solvent molecules were removed from the final refinement performed using the SQUEEZE program. The toxa ligands are disordered over two positions. Protons were not detected over N atoms in the difference Fourier map; therefore, they were treated using a riding model. The cation showed almost the same structure as μ-toxa dinuclear Ni-dppe complex 1. The deviation of Ni from the least-square mean plane of P/P/S/N is 0.08, 0.06, 0.37, and 0.38 Å. The dihedral angle between the least-square mean plane of P/P/Ni and the least-square mean plane of N/S/Ni is 6.5(6)°, 6.2(5)°, 26.0(3)°, and 22.7(2)°. In general, the Ni(II) ion has a slightly distorted square-planar geometry. The dissimilarities between these values can be attributed to the disorder of the toxa ligand. The two four-coordinated square-planar Ni structures are almost coplanar.
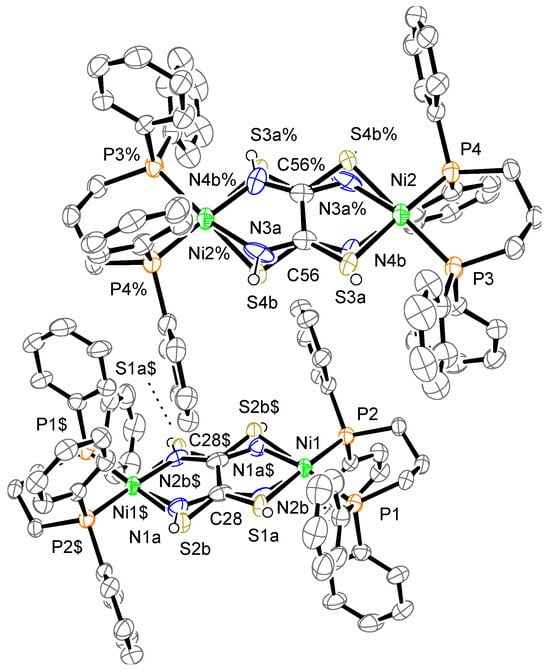
Figure 4.
Crystal structure of the cation of complex 1a. The counter anions and hydrogen atoms, except for N–H, are omitted for clarity. The thermal ellipsoids are drawn at the 50% probability level. The subscripts $ and % indicate the equivalent atoms generated by the symmetry operators (2-x, 2-y, 1-z) and (1-x, 1-y, 2-z), respectively.
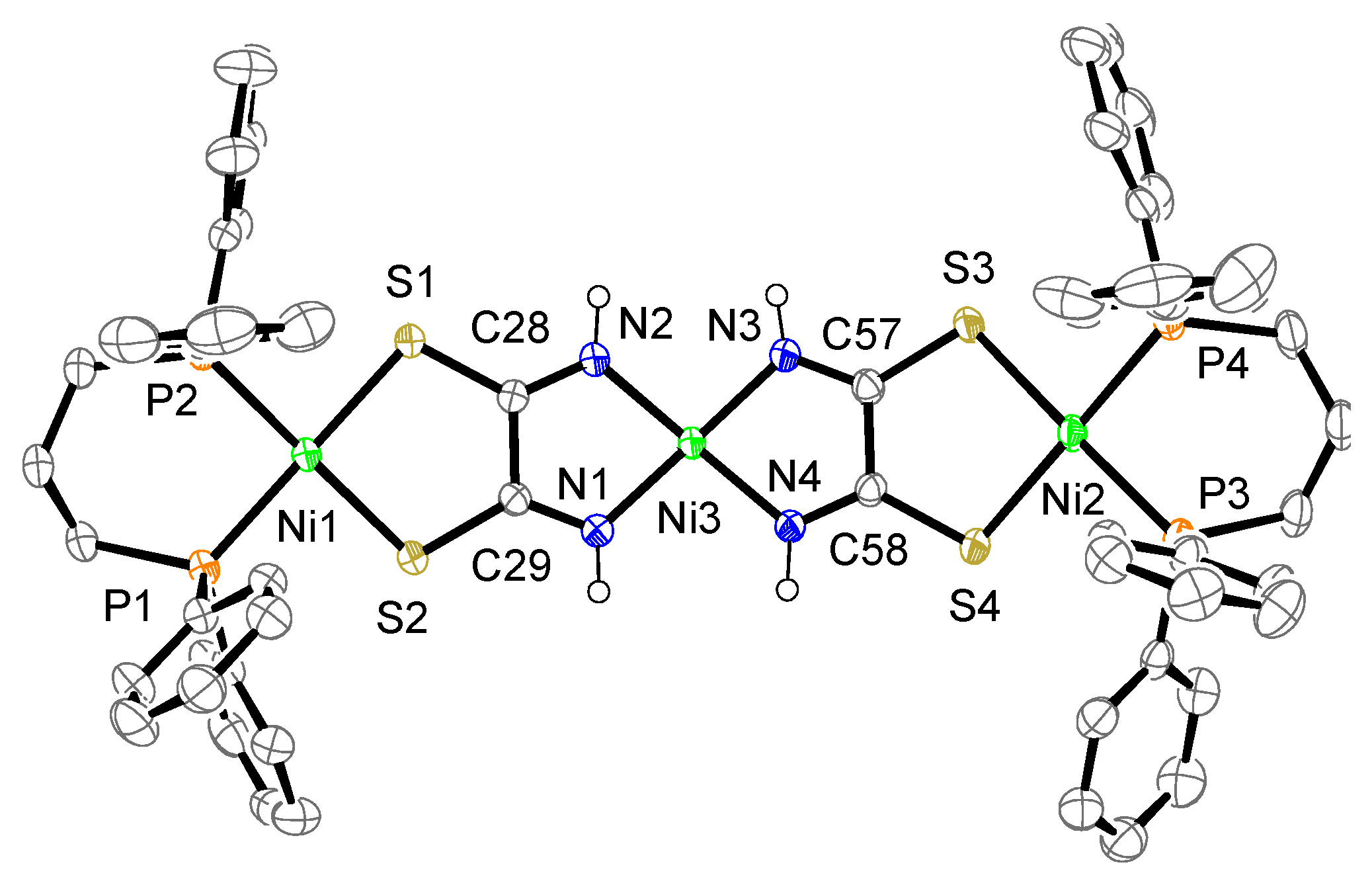
The crystal structure of complex 2a is shown in Figure 5. The complex consists of one complex cation, two BF4− counter anions, one dppp dioxide, and some molecules (2.43 CH3CN·0.57 Et3N). The cation contains two mono-Ni(dppp) units bridged by a [Ni{toxa-κ(N,N)}2]2− moiety instead of a toxa ligand, forming a trinuclear Ni complex. The toxa ligands bridge two Ni atoms in a cis form, which differs from the bridging mode of the toxa ligand in complex 1a. Although similar μ-toxa and μ-substituted-toxa trinuclear structures have been reported [12,25,26,27], our complex shows a unique structure for a homonuclear Ni complex. Two terminal Ni atoms (Ni1, Ni2) are coordinated by the S atoms of toxa and the P atoms of dppp. The deviation of the terminal Ni atoms from the least-square mean plane of P/P/S/S is 0.08 and 0.06 Å. The deviation of the central Ni atom (Ni3) from the least-square mean plane of N/N/N/N is 0.09 Å. The dihedral angles between the least-square mean plane of P/P/terminal Ni and the least-square mean plane of S/S/terminal Ni are 6.2° and 4.3(1)°. The dihedral angle between the least-square mean plane of N1/N2/central Ni and the least-square mean plane of N3/N4/central Ni is 9.1(1)°. As observed for complexes 1 and 1a, the Ni(II) ion exhibits a slightly distorted square-planar geometry.
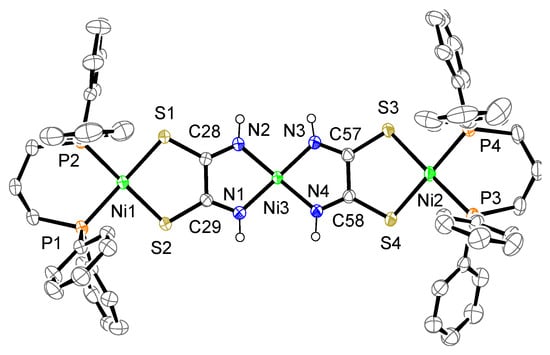
Figure 5.
Crystal structure of the cation of complex 2a. The counter anions, solvent molecules, diphosphine dioxide, triethylamine, and hydrogen atoms, except for N–H, are omitted for clarity. The thermal ellipsoids are drawn at the 50% probability level.
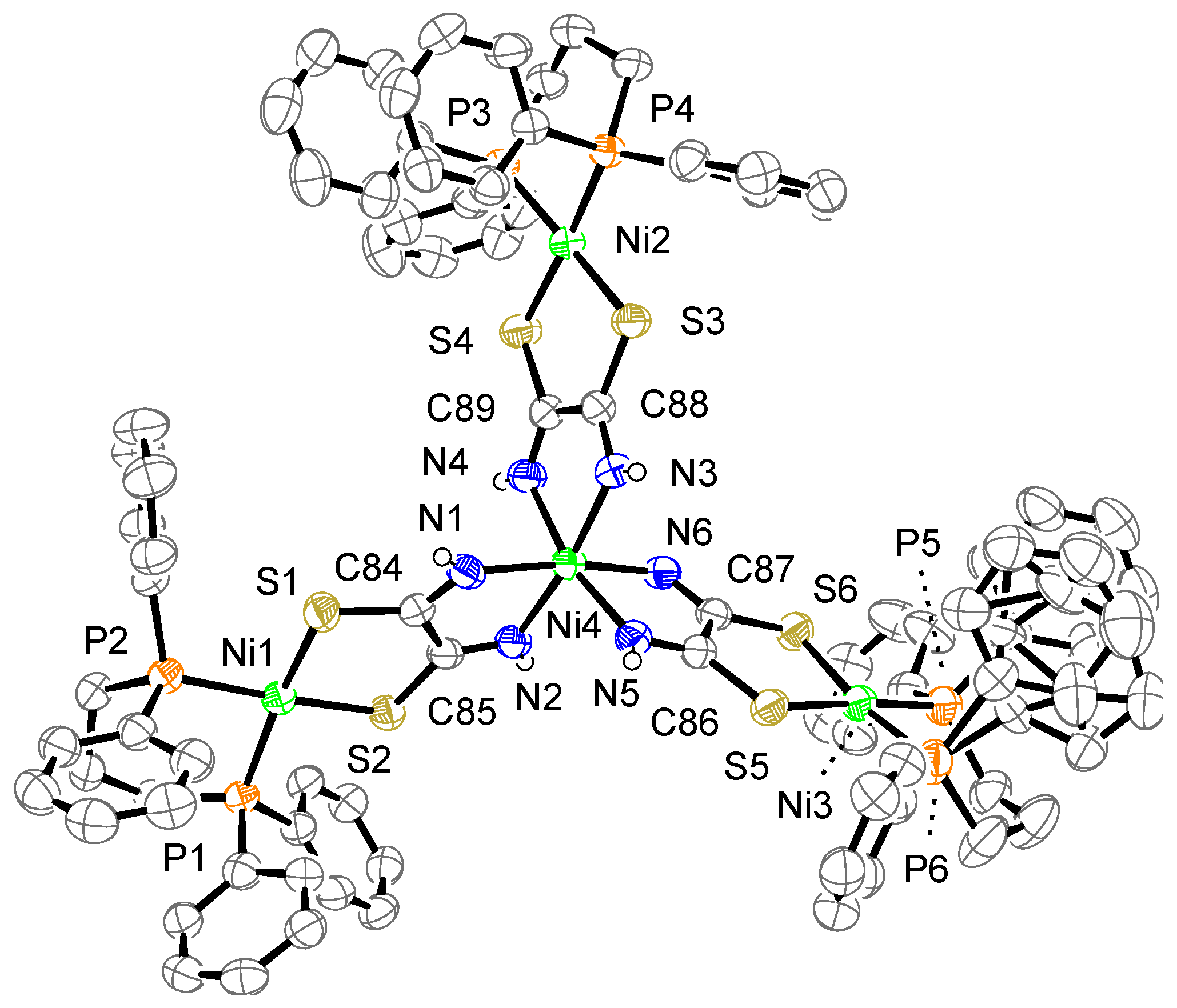
The crystal structure of complex 3a is shown in Figure 6. The complex consists of one complex cation, two BF4− counter anions, and six acetone molecules. One phenyl group bonded to P6 is disordered over two positions. The cation has three mono-Ni (dppp) units bridged by a [Ni{toxa-κ(N,N)}3]4− moiety, which results in a tetranuclear Ni complex. As in the case of complex 2a, the toxa ligands bridge two Ni atoms in a cis form. The central Ni atom (Ni4) is coordinated by six N atoms of toxa ligands, resulting in an octahedral geometry. Three terminal Ni atoms (Ni1, Ni2, and Ni3) are coordinated by the S atoms of toxa and the P atoms of dppp. The deviation of the terminal Ni atoms from the least-square mean plane of P/P/S/S is 0.05, 0.07, and 0.04 Å. The dihedral angles between the least-square mean plane of P/P/terminal Ni and the least-square mean plane of S/S/terminal Ni are 12.9(1)°, 5.1(1)°, and 9.0(1)°. As observed for the other complexes, the terminal Ni(II) ion shows a slightly distorted square-planar geometry.
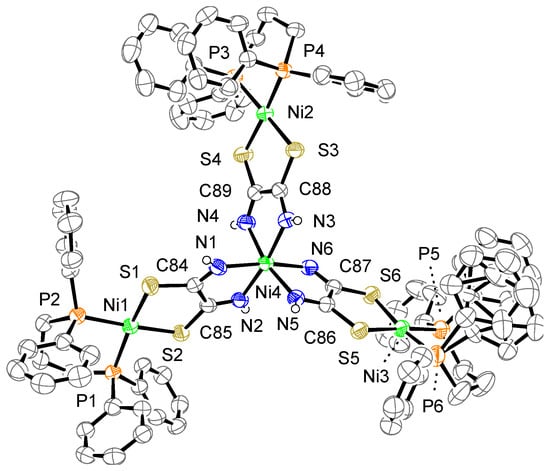
Figure 6.
Crystal structure of the cation of complex 3a. The counter anions, solvent molecules, and hydrogen atoms, except for N–H, are omitted for clarity. The thermal ellipsoids are drawn at the 50% probability level.
Selected bond distances and bond angles are summarized in Table S2. The four complexes show similar Ni–S and Ni–P bond distances but different Ni–N bond distances. Thus, the Ni–N bond distance of Ni-dppp complex 1a is shorter than that of Ni-dppe complex 1, which could be due to the different diphosphine ligands. Comparison of Ni–N distance with complex 1a and complex 2a makes us understand the influence of the coordination mode (trans vs. cis). In this case, there is no significant difference. Meanwhile, complex 3a shows the longest Ni–N distance, which could be due to the different coordination modes (octahedral vs. square-planar). The P–Ni–P bond angle reflects the difference between dppe and dppp. The N–Ni–S angle is almost the same in complexes 1 and 1a. The S–Ni–S angle is almost the same in complexes 2a and 3a; however, the N–Ni–N angle of complex 3a is narrower than that of complex 2a. This difference could be due to the different coordination modes, as observed for the Ni–N bond distance. Bridging toxa ligand is also surveyed by bond distance. The CCDC database shows that the N–C, S–C, and C–C bond lengths of μ-toxa in polynuclear complexes are 1.22 [17]–1.48 [12], 1.33 [17]−1.80 [14], and 1.43 [16]−1.58 [9] Å, respectively, with respective median values of 1.29 [14,28], 1.72 [29,30], and 1.50 [15,30] Å. Therefore, the bond lengths of the toxa ligand in the complexes prepared in this study fall within the range of those previously reported for polynuclear complexes.
3.3. Effect of Ancillary Diphosphine Ligand for the Product
The synthesis of the toxa-bridged Ni complexes involves two steps. [Ni(H2O)6](BF4)2 and diphosphine are mixed in CH3CN in the first step, which produces [Ni(diphosphine)(CH3CN)2]2+. In the second step, H2toxa and Et3N are added to the solution, and toxa2− ligand coordinates Ni atoms of [Ni(diphosphine)(CH3CN)2]2+. The bite angle of the diphosphine ligand is known as an important effect for a property of complexes [31]. The difference between dppe and dppp could invoke the different affinity between the Ni atom and the S/N atoms of toxa. We think that the affinity might produce the difference in the products; however, we could not prove it experimentally in this work.
3.4. Electrochemical Behavior of Dinuclear Ni Complex 1
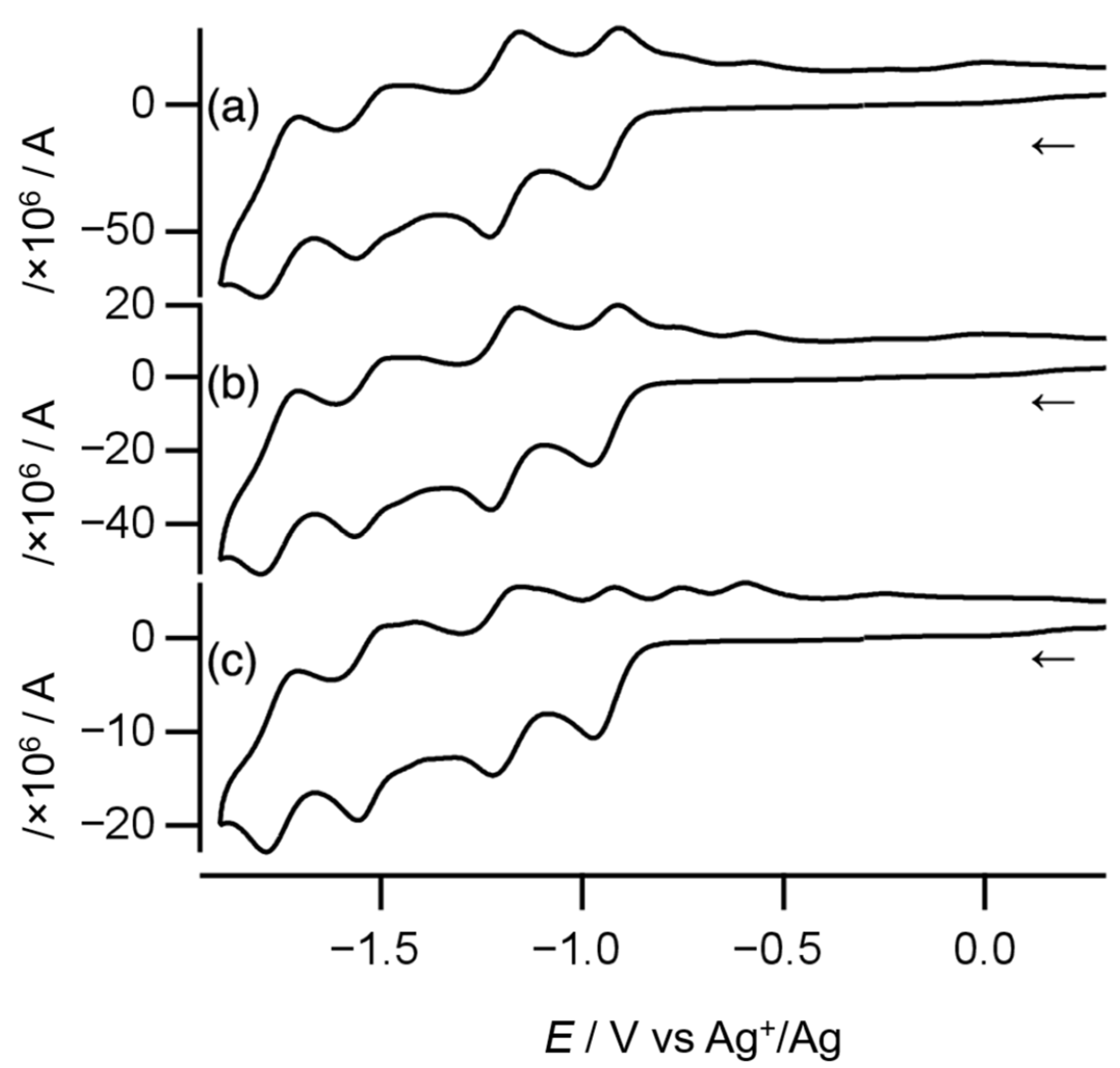
Cyclic voltammetry was conducted to estimate the electronic coupling between Ni atoms through the toxa ligand in complex 1. As shown in Figure 7, the cyclic voltammogram recorded at a scan rate of 100 mV s−1 displays four quasi-reversible one-electron redox couples and two oxidation peaks, as summarized in Table 1. At higher scan rates, the quasi-reversible redox couples turned more reversible, and the oxidation peaks became more obscure. Overall, complex 1 shows four reversible one-electron redox couples.
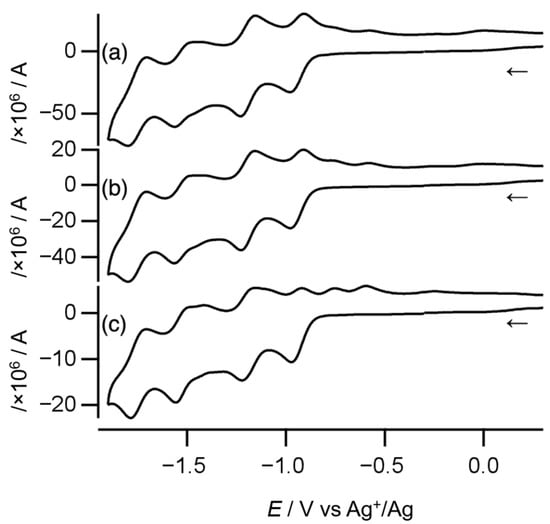
Figure 7.
Cyclic voltammograms of complex 1 recorded at scan rates of (a) 1000 mV s−1, (b) 500 mV s−1, and (c) 100 mV s−1. The arrows indicate the direction of the initial scan.

Table 1.
Electrochemical behavior of complex 1 under various scan rates.
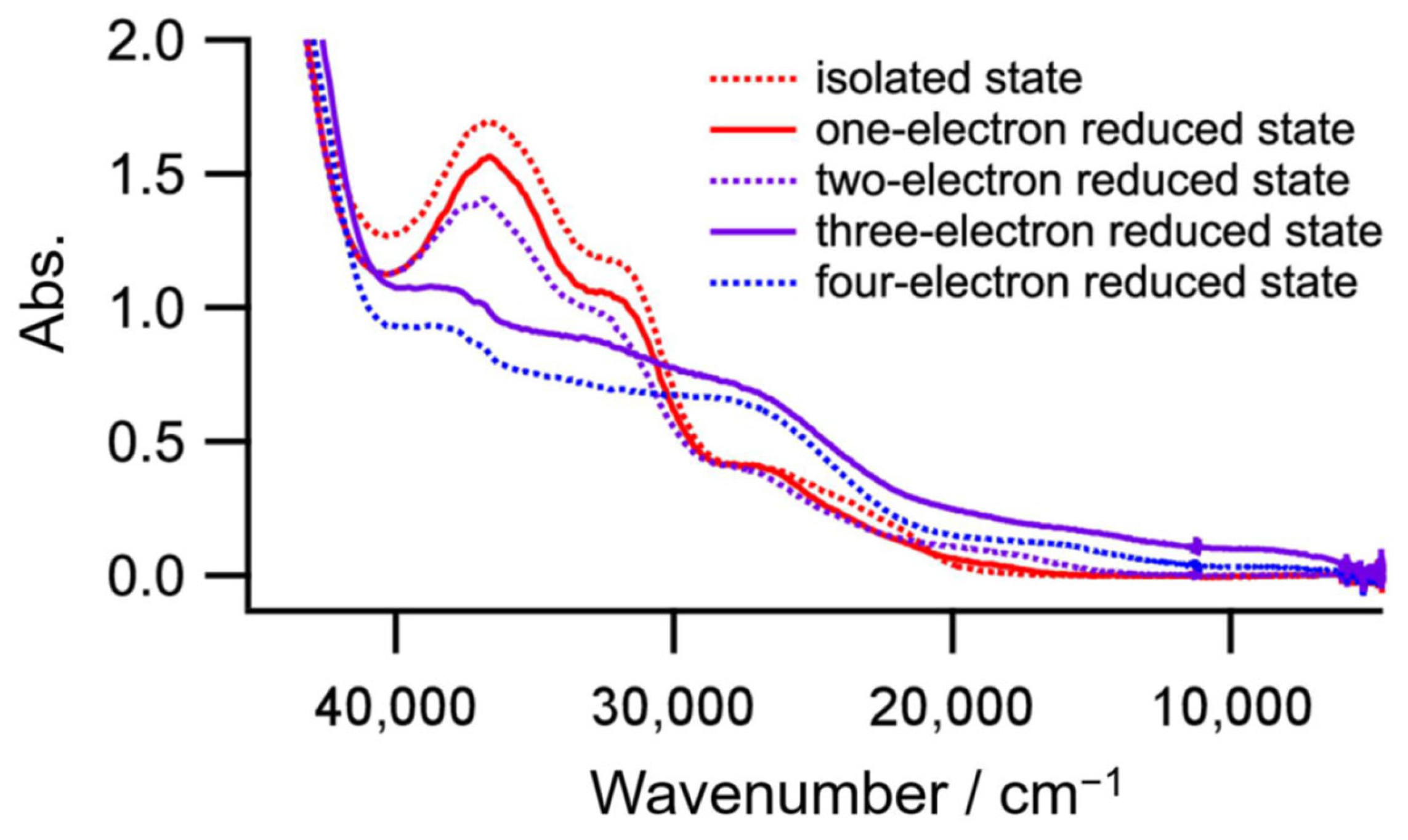
Complex 1 contains two Ni atoms and one toxa ligand, which could act as a redox-active center. To identify the four redox couples where the reduction occurs, a controlled-potential absorption spectrum at each reduced state was measured. As shown in Figure 8, only the three-electron reduced state has a peak (8.9 × 103 cm−1, 2.5 × 103 dm3 mol−1 cm−1) in the near IR region. Furthermore, the peak disappeared in a four-electron reduced state. The peak would be an intervalence charge transfer band; therefore, the third and fourth reduction processes are due to Ni(II)–Ni(II)/Ni(II)–Ni(I) and Ni(II)–Ni(I)/Ni(I)–Ni(I), respectively, and the first and second reduction processes stem from toxa2−/toxa3− and toxa3−/toxa4−, respectively. As far as we know, this is the first report of a mixed valence state of Ni(II)–Ni(I) bridged by toxa3−.
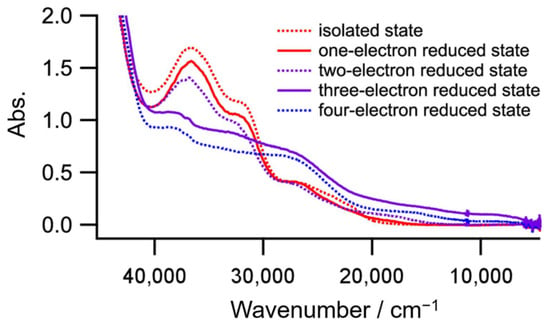
Figure 8.
Absorption spectra of each reduced state of complex 1.
The mixed-valence state Ni(II)–Ni(I) occurs at ∆E1/2 = 0.22 V (E1/2(Ni(II)–Ni(II)/Ni(II)–Ni(I)) = −1.53 V, E1/2(Ni(II)–Ni(I)/Ni(I)–Ni(I)) = −1.75 V). As shown in Equation (1), the separation between the two redox potentials is associated with the comproportionation constant Kc (Equation (1)), which can be calculated using Equation (2) [32]. For complex 1, Kc was determined to be 6.1 × 103.
where E01 and E02 are the standard redox potentials of the first redox reaction and the second redox reaction, respectively; n1 and n2 are the numbers of electrons at the first redox reaction and the second redox reaction, respectively; F is the Faraday constant; R is the gas constant, and T is the temperature.
4. Conclusions
In this study, new toxa-bridged Ni-diphosphine polynuclear complexes were prepared and characterized via X-ray diffraction. Ni-dppe dinuclear complex 1 was obtained from a one-pot reaction with dppe as a diphosphine ligand. When using dppp instead of dppe, dinuclear complex 1a, trinuclear complex 2a, and tetranuclear complex 3a were obtained. The dithiooxamidate shows different coordination between the dinuclear and the trinuclear/tetranuclear complexes. The electrochemical behavior of Ni-dppe dinuclear complex 1 was investigated via cyclic voltammetry and absorption spectra at various potentials, and the electronic coupling between two Ni atoms was discussed.
Further investigation of oxamidate/dithiooxamidate-bridged Ni-diphosphine dinuclear complexes is currently underway in our laboratory, including the optimization of the synthesis method to obtain trinuclear and tetranuclear complexes in good yield to examine the relationship between their molecular structure and their electronic coupling. The synthesis reaction is also explored for the optimization.
Supplementary Materials
The following supporting information can be downloaded at https://www.mdpi.com/article/10.3390/chemistry5040150/s1, Figure S1: 1H NMR spectrum of complex 1 in CD3CN; Figure S2: 31P NMR spectrum of complex 2 in CD3CN (85% H3PO4 aq. shows a singlet peak at δ−1.458 ppm); Table S1: Crystallographic data; Table S2: Selected bond lengths (Å) and angles (°).
Author Contributions
Conceptualization, T.H.; methodology, T.H.; investigation, T.H., R.K., T.Y. and N.T.; resources, I.A. and S.K.; writing—original draft preparation, T.H.; writing—review and editing, I.A. and S.K.; supervision, I.A. and S.K. All authors have read and agreed to the published version of the manuscript.
Funding
This research received no external funding.
Data Availability Statement
CCDC 2259972-2259975 contains the supplementary crystallographic data for this paper. These data can be obtained free of charge from The Cambridge Crystallographic Data Centre via www.ccdc.cam.ac.uk/data_request/cif accessed on 7 September 2023. Supplementary Materials associated with this article can be found, in the online version.
Conflicts of Interest
The authors declare no conflict of interest.
References
- Kaim, W.; Klein, A.; Glöckle, M. Exploration of Mixed-Valence Chemistry: Inventing New Analogues of the Creutz-Taube Ion. Acc. Chem. Res. 2000, 33, 755–763. [Google Scholar] [CrossRef] [PubMed]
- Demadis, K.D.; Hartshorn, C.M.; Meyer, T.J. The Localized-to-Delocalized Transition in Mixed-Valence Chemistry. Chem. Rev. 2001, 101, 2655–2686. [Google Scholar] [CrossRef] [PubMed]
- Brunschwig, B.S.; Creutz, C.; Sutin, N. Optical Transitions of Symmetrical Mixed-Valence Systems in the Class II-III Transition Regime. Chem. Soc. Rev. 2002, 3, 168–184. [Google Scholar] [CrossRef] [PubMed]
- Ceccon, A.; Santi, S.; Orian, L.; Bisello, A. Electronic communication in heterobinuclear organometallic complexes through unsaturated hydrocarbon bridges. Coord. Chem. Rev. 2004, 248, 683–724. [Google Scholar] [CrossRef]
- D’Alessandro, D.M.; Keene, F.R. Current trends and future challenges in the experimental, theoretical and computational analysis of intervalence charge transfer (IVCT) transitions. Chem. Soc. Rev. 2006, 35, 424–440. [Google Scholar] [CrossRef]
- Tezgerevska, T.; Alley, K.G.; Boskovic, C. Valence tautomerism in metal complexes: Stimulated and reversible intramolecular electron transfer between metal centers and organic ligands. Coord. Chem. Rev. 2014, 268, 23–40. [Google Scholar] [CrossRef]
- Sato, O. Dynamic molecular crystals with switchable physical properties. Nat. Chem. 2016, 8, 644–656. [Google Scholar] [CrossRef]
- Groom, C.R.; Bruno, I.J.; Lightfoot, M.P.; Ward, S.C. The Cambridge Structural Database. Acta Crystallogr. Sect. B 2016, 72, 171–179. [Google Scholar] [CrossRef]
- Mosset, A.; Abboudi, M.; Galy, J. Complexes Métalliques De L’acide Rubéa-nique. I.; Synthèse, Structure Moléculaire Et Cristalline D’un Rubéanate De Cuivre(I): CuCl(C2N2S2H4)1.5(H2O)0.45. Z. Für Krist.–Cryst. Mater. 1983, 164, 171–180. [Google Scholar] [CrossRef]
- Veit, R.; Girerd, J.J.; Kahn, O.; Robert, F.; Jeannin, Y.; Murr, N. Amino acid amides of dithiooxalic acid: Spectroscopic, electrochemical, and magnetic properties of copper(II) binuclear complexes and crystal structure of [N,N′-(1,2-dithioxoethane-1,2-diyl)bis(methyl methioninato)]bis(bromocopper(II)). Inorg. Chem. 1984, 23, 4448. [Google Scholar] [CrossRef]
- Draganjac, M.; Minick, D.; Cordes, A.W. Molecular Structures of [CpRu(PPh3)(dtoxa-H2O)]BF4 and {[CpRu(PPh3)2]2(μ-dtoxa)}(BF4)2, dtoxa = dithiooxamide. J. Crystallogr. Spectrosc. Res. 1993, 4, 265–271. [Google Scholar] [CrossRef]
- Kopel, P.; Březina, F.; Trávníček, Z.; Šindelář, Z.; Lasovský, J.; Marek, J. Nickel complexes with sulphur and nitrogen donor ligands, crystal and molecular structure of [(PPh3)2Cu(DTA)Ni(DTA)Cu(PPh3)2] (H2DTA = dithiooxamide). Polyhedron 1995, 14, 991–996. [Google Scholar] [CrossRef]
- Castiñeiras, A.; Vidal, M.C.F.; Romero, J.; Sáez, R.; Matilla, A.; Niclós, J.; Tercero, J.M. Synthesis, Characterization, and Magnetic Behaviour of Dinuclear Nickel(II) Complexes of N,N′-Substituted Dithiooxamides Derived from A-Amino acids. Z. Anorg. Allg. Chem. 2001, 7, 1553–1559. [Google Scholar] [CrossRef]
- Cotton, F.A.; Li, Z.; Liu, C.Y.; Carlos, A. Murillo. Modulating Electronic Coupling Using O– and S–donor Linkers. Inorg. Chem. 2007, 46, 7840–7847. [Google Scholar] [CrossRef]
- Mikhalyova, E.A.; Kolotilov, S.V.; Cador, O.; Zeller, M.; Trofimenko, S.; Ouahab, L.; Addison, A.W.; Pavlishchuk, V.V.; Hunter, A.D. The Role of the Bridging Group in Exchange Coupling in Dinuclear Homo- and Heterometallic Ni(II) and Co(II) Complexes with Oxalate, Oxamidate and Dithiooxamidate Bridges. Dalton Trans 2012, 37, 11319–11329. [Google Scholar] [CrossRef]
- Kee, C.L.; Zhou, F.; Su, H.; Yan, Y.K. Formation of “A-Frame” Dirhenium(I) Hexacarbonyl Complexes by Trans-1,2-Bis(Diphenylphosphino)Ethylene and Bis(Bidentate) Ligands. J. Organomet. Chem. 2015, 792, 211–219. [Google Scholar] [CrossRef]
- Ying; Jun; Gao, N.; Mou, H.; Tian, A. Rare Earth Mono-Substituted Keggin Dimmers: Structures, Electrochemical, Supercapacitor and Magnetic Properties. J. Solid State Chem. 2022, 314, 123343. [Google Scholar]
- Sheldrick, G.M. Crystal Structure Refinement with Shelxl. Acta Crystallogr. Sect. C 2015, 71, 3–8. [Google Scholar] [CrossRef]
- Kabuto, C.; Akine, S.; Nemoto, T.; Kwon, E.; Wakita, K. Release of Software (Yadokari-XG 2009) for Crystal Structure Analyses. J. Cryst. Soc. Jpn. 2009, 51, 218–224. [Google Scholar] [CrossRef]
- Farrugia, L.J. Ortep-3 for Windows–a Version of Ortep-III with a Graphical User Interface (GUI). J. Appl. Crystallogr. 1997, 30, 565. [Google Scholar] [CrossRef]
- Spek, A.L. Single-crystal structure validation with the program PLATON. J. Appl. Crys-Tallogr. 2003, 36, 7–13. [Google Scholar] [CrossRef]
- Lanza, S.; Bruno, G.; Nicolo, F.; Rotondo, A.; Tresoldi, G. Reactions of Secondary Dithioxamides with [(η6-p-cymene)RuCl(μ-Cl)]2: The Role of Steric Hindrance on Amidic Nitrogen in Determining the Reaction Products. Eur. J. Inorg. Chem. 2002, 65–72. [Google Scholar] [CrossRef]
- Bruno, G.; Lanza, S.; Giannetto, A.; Sacca, A.; Rudbari, H. Crystal structure of (μ-N,N′-dibenzyldithiooxamidato-κN,S:N′,S′)bis[(η3-crotyl)palladium(II)]. Acta Crystallogr. Sect. E Cryst. Commun. 2015, 71, 40. [Google Scholar] [CrossRef] [PubMed]
- Askari, B.; Rudbari, H.A.; Micale, N.; Schirmeister, T.; Efferth, T.; Seo, E.J.; Bruno, G.; Schwickert, K. Ruthenium(II) and Palladium(II) Homo- and Heterobimetallic Complexes: Synthesis, Crystal Structures, Theoretical Calculations and Biological Studies. Dalton Trans 2019, 42, 15869–15887. [Google Scholar] [CrossRef] [PubMed]
- Veit, R.; Girerd, J.-J.; Kahn, O.; Robert, F.; Jeannin, Y. Copper(II) and nickel(II) trinuclear species with dithiooxamide derivative ligands: Structural, magnetic, spectroscopic, and electrochemical properties. Inorg. Chem. 1986, 25, 4175. [Google Scholar] [CrossRef]
- Lanza, S.; Bruno, G.; Nicolò, F.; Callipari, G.; Tresoldi, G. Trinuclear Heterobimetallic Complexes with Binucleating Dithioxamides: Stereoselective Synthesis and Solution Behavior Involving Pd−N Bond Rupture. Inorg. Chem. 2003, 42, 4545–4552. [Google Scholar] [CrossRef]
- Askari, B.; Rudbari, H.A.; Micale, N.; Schirmeister, T.; Giannetto, A.; Lanza, S.; Bruno, G.; Mirkhani, V. Synthesis, Solution Be-haviour and Potential Anticancer Activity of New Trinuclear Organometallic Palladium(II) Complex of {S}-1-Phenylethyl Dithiooxamide: Comparison with the Trinuclear Heterobi-metallic Platinum(II) Analogue. Polyhedron 2019, 164, 195–201. [Google Scholar] [CrossRef]
- Bruno, G.; Lanza, S.; Nicolo, F.; Tresoldi, G.; Rosace, G. (η3-Allyl-2κ3C)(chloro-1κcl)(μ-N,N′-diethyldithioxamidato-1:2κ4S,S′:N,N’)[diphenyl(2-pyridyl)phosphine-1κP]palladium(II)platinum(II) chloroform solvate. Acta Crystallographica. Sect. C Cryst. Struct. Commun. 2002, 58, 316–318. [Google Scholar] [CrossRef]
- Chauvel, C.; Girerd, J.; Jeannin, Y.; Kahn, O.; Lavigne, G. Crystal Structure and Magnetic Properties of Di-μ-aqua-bis[μ-[N,N’-bis(2-hydroxyethyl)dithiooxamidato(2–)-N,O,S:N',O',S']]-bis[aquacopper(II)sulfatocopper(II)]. A New Example of Very Strong Antiferromagnetic Coupling Between Copper(II) Ions Far Away from Each Other. Inorg. Chem. 1979, 18, 3015. [Google Scholar] [CrossRef]
- Lanza, S.; Nicolò, F.; Cafeo, G.; Rudbari, H.A.; Bruno, G. The Absolute Configuration of Palladium(II) and Ruthenium(II) Pseudochiral Centers in either Chiral or Achiral Environ-ments. Inorg. Chem. 2010, 49, 9236–9246. [Google Scholar] [CrossRef]
- Miedaner, A.; Haltiwanger, R.C.; DuBois, D.L. Relationship between the bite size of di-phosphine ligands and tetrahedral distortions of “square-planar” nickel(II) complexes: Stabi-lization of nickel(I) and palladium(I) complexes using diphosphine ligands with large bites. Inorg. Chem. 1991, 30, 417–427. [Google Scholar] [CrossRef]
- Richardson, D.E.; Taube, H. Determination of E2° - E1° in multistep charge transfer by stationary-electrode pulse and cyclic voltammetry: Application to binuclear ruthenium ammines. Inorg. Chem. 1981, 20, 1278–1285. [Google Scholar] [CrossRef]
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).