Responsive DNA Nanostructures for Bioanalysis and Therapy
Abstract
:1. Introduction
2. Responsive DNA Nanostructures for Biosensing and Bioimaging
2.1. DNA Nanostructures for Detection Signal Amplification
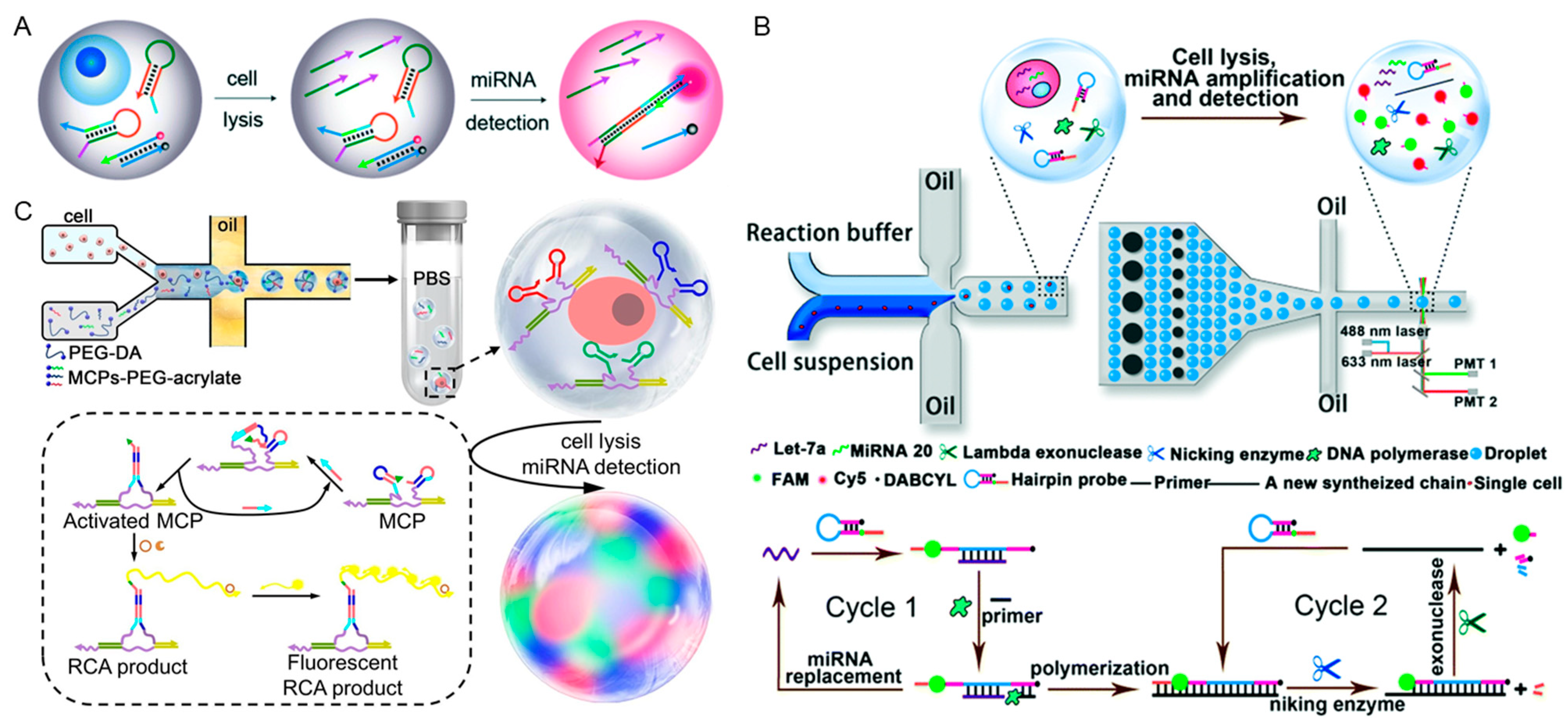
2.1.1. In Vivo miRNAs Detection
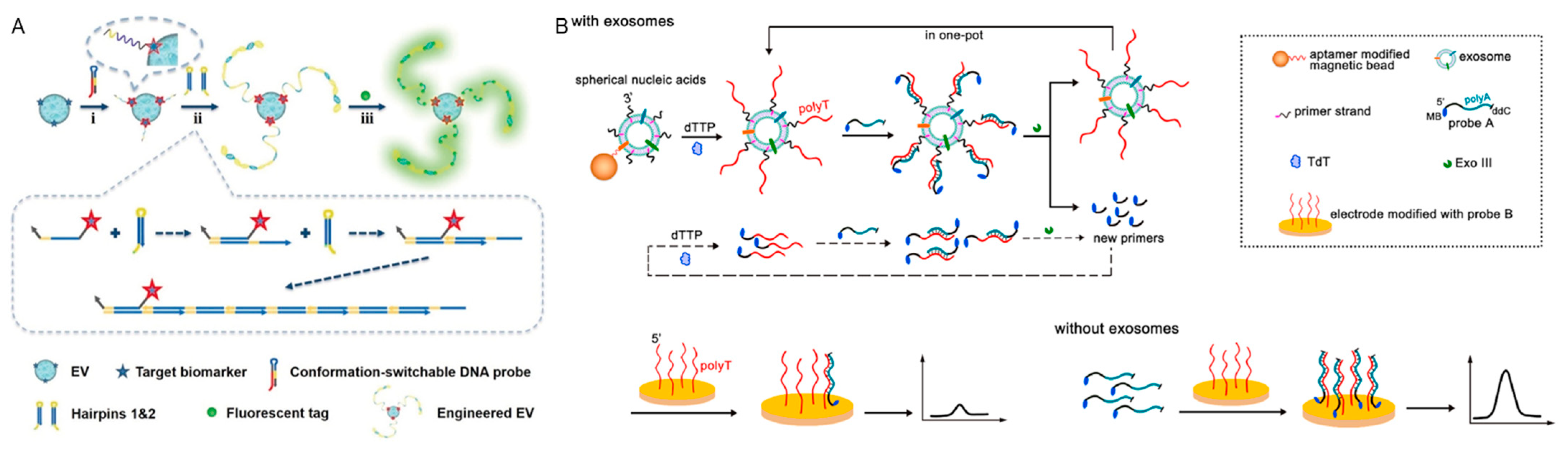
2.1.2. Extracellular Vesicles Detection
2.1.3. Circulating Tumor Cells Detection
2.2. DNA Nanostructures against False-Positive Detection Signal
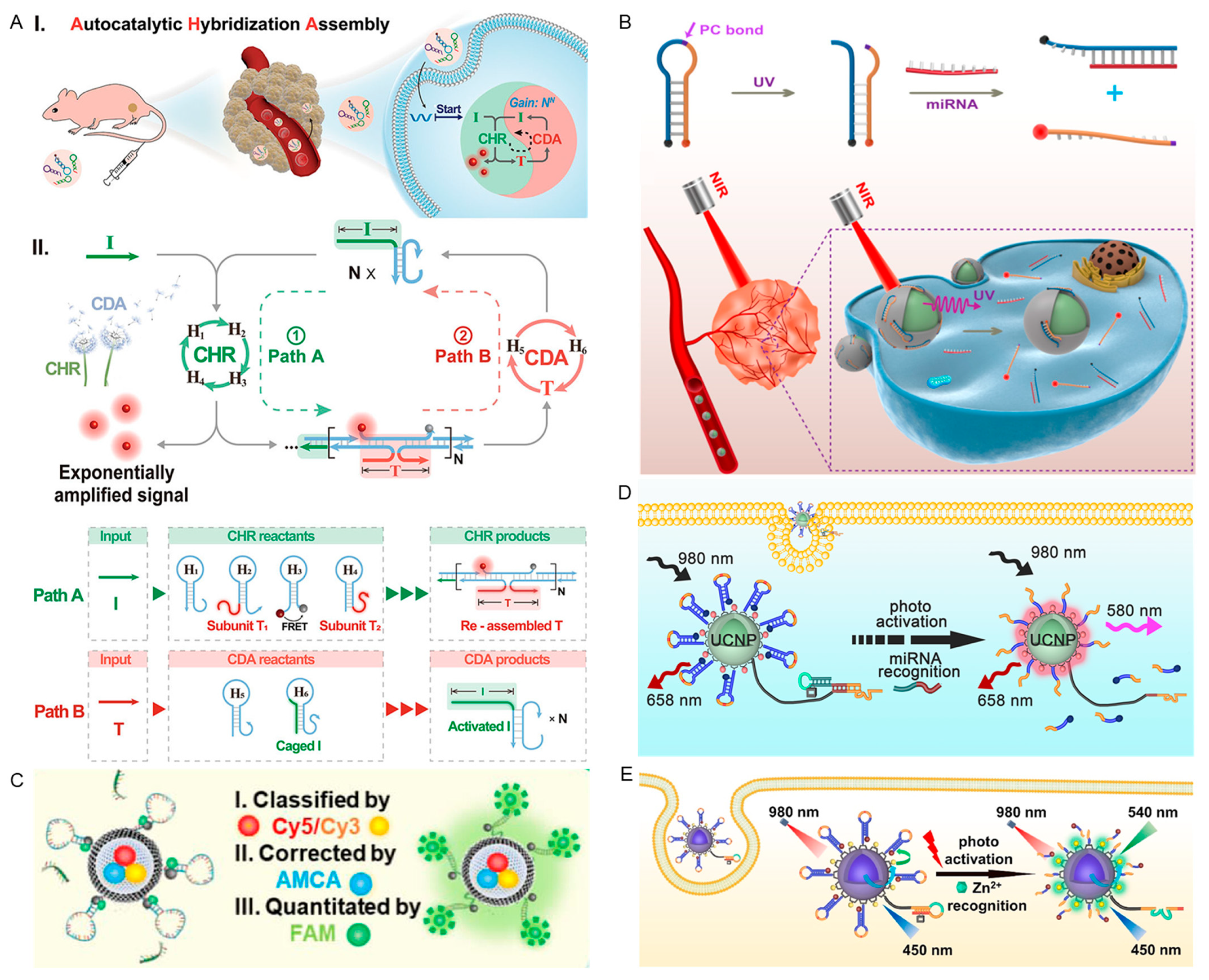
3. Smart Responsive DNA Nanostructures for Therapy
3.1. Response to Small Molecules
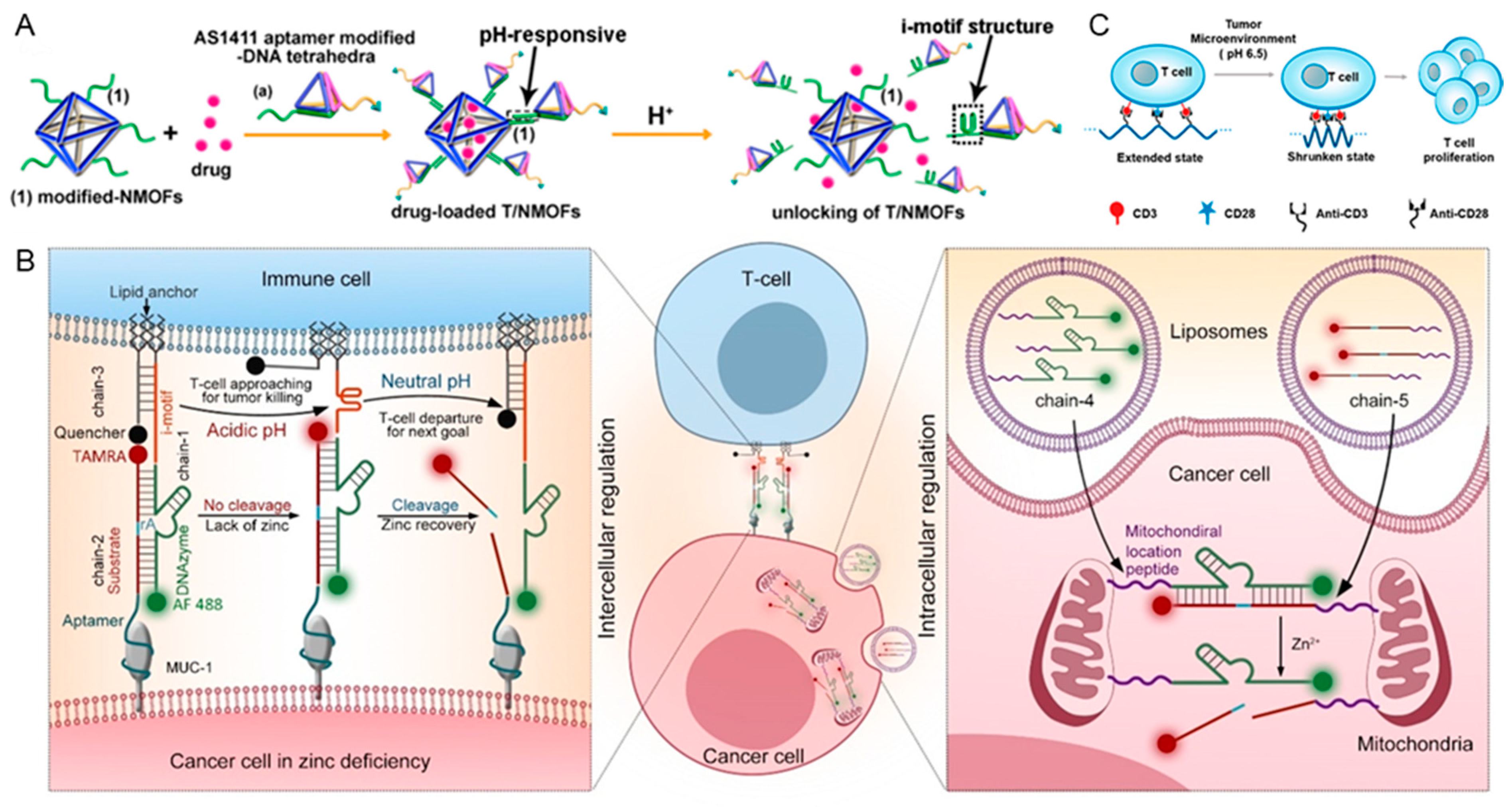
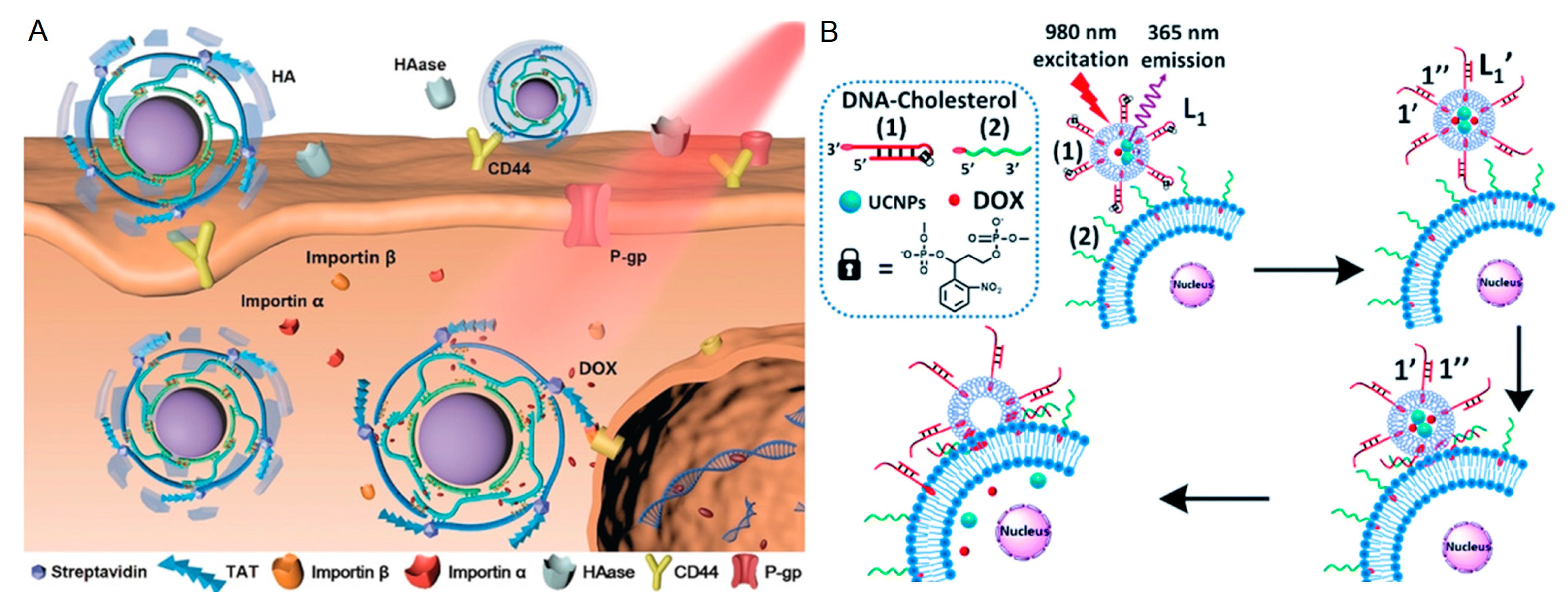
3.1.1. pH
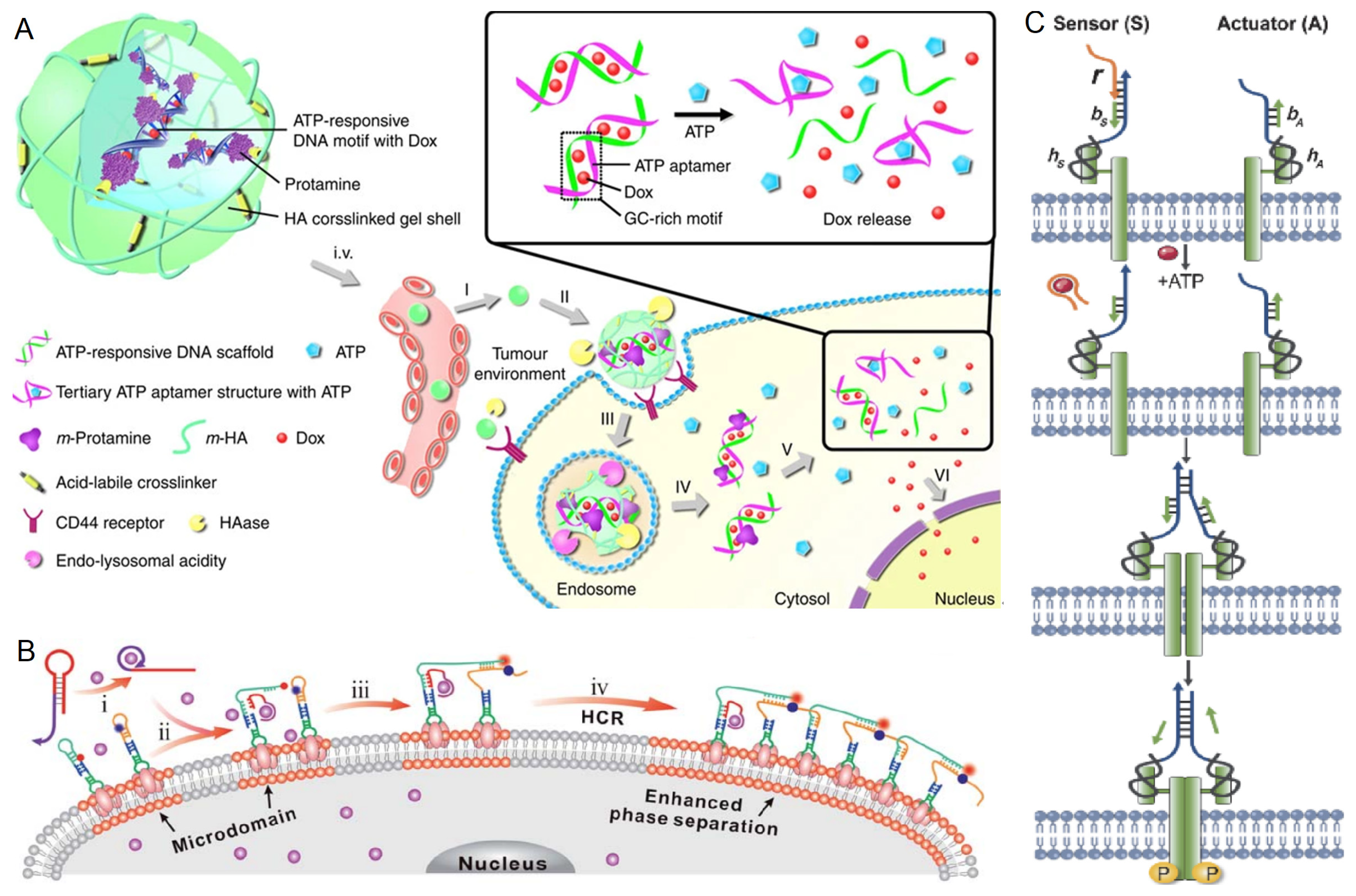
3.1.2. ATP
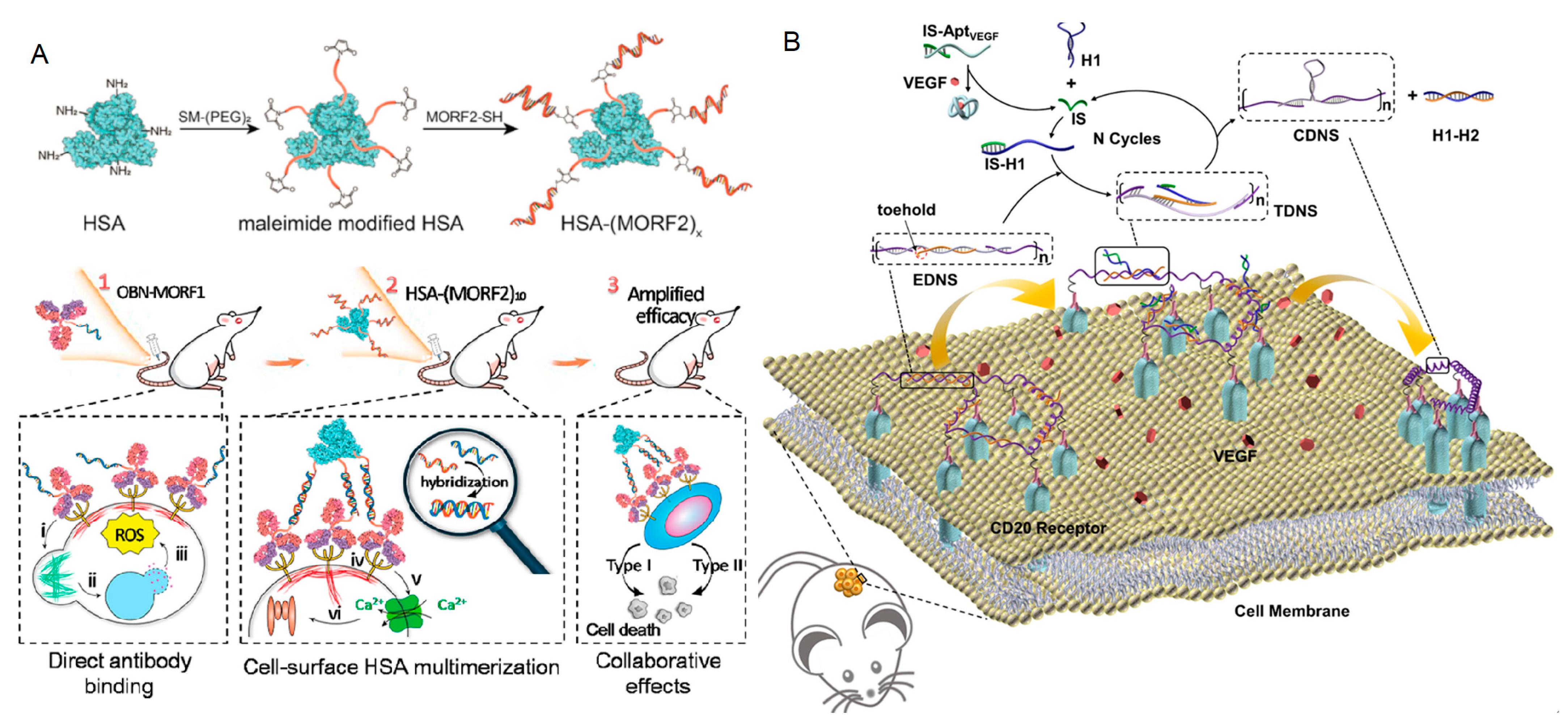
3.2. Response to Biomacromolecules
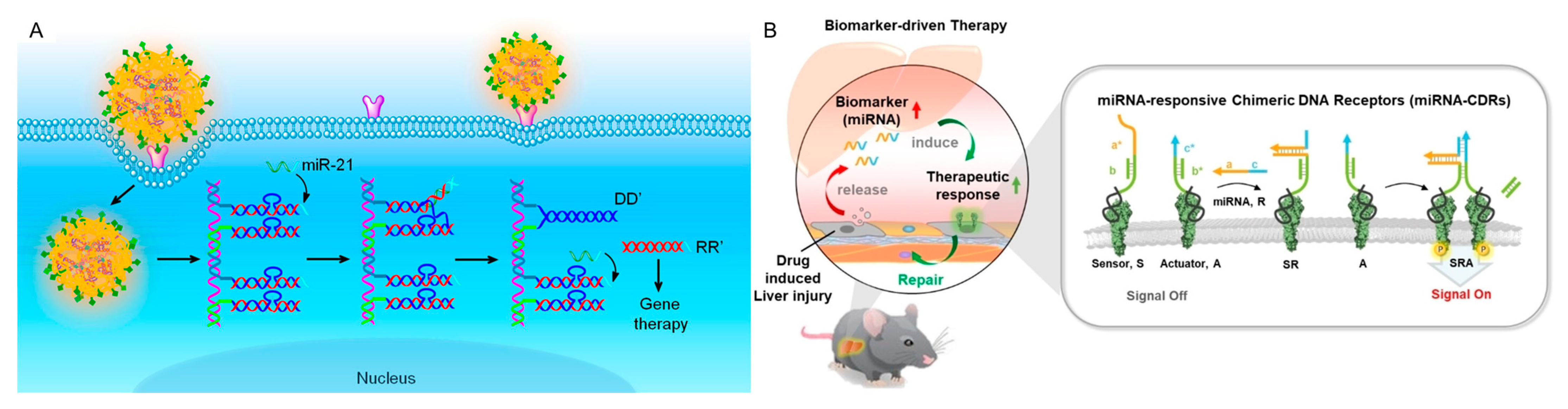
3.2.1. Nucleic Acids
3.2.2. Proteins
3.3. Response to Light Irradiation
3.4. Dual-Responsive DNA Nanostructures
4. Conclusions and Perspectives
Funding
Data Availability Statement
Conflicts of Interest
Abbreviations
| miRNA | microRNA |
| HCR | Hybridization chain reaction |
| CHA | Catalytic hairpin assembly |
| RCA | Rolling circle amplification |
| MCP | miRNA capture probe |
| EVs | Extracellular vesicles |
| MB | Methylene blue |
| Exo III | Exonuclease III |
| SNAs | Spherical nucleic acids |
| CTCs | Circulating tumor cells |
| AP | aptamer |
| FluoELs | Fluorophores-encoded error-corrected labels |
| MSNs | Mesoporous silica nanoparticles |
| PC | Photo-cleavable |
| DQAsome | Dequalinium-based Liposome-like vesicle |
| UCNPs | Upconversion nanoparticles |
| NIR | Near-infra-red |
| AHA | Autocatalytic hybridization assembly |
| Dox | Doxorubicin |
| iDNS | Interlocked DNA nano-spring |
| ATP | 5’-adenosine triphosphate |
| HA | Hyaluronic acid |
| D-CID | DNA-mediated chemically induced dimerization |
| GSH | Glutathione |
| mRNA | messenger RNA |
| UTR | 3’ untranslated region |
| DR/D’R’ | DNA/RNA hybrids |
| SMARC | Selective receptor aggregation |
| PLGA | Poly (D, L-lactide glycolic acid |
| ROS | Reactive oxygen species |
| PDT | Photo-dynamic therapy |
References
- Mo, L.; Liang, D.; Qin, R.; Mo, M.; Yang, C.; Lin, W. Three-Dimensional CHA-HCR System Using DNA Nanospheres for Sensitive and Rapid Imaging of miRNA in Live Cells and Tissues. Anal. Chem. 2023, 95, 11777–11784. [Google Scholar] [CrossRef] [PubMed]
- Ai, L.; Zuo, Q.; Li, Y.; Wu, S.; Sima, Y.; Zhang, Y.; Xie, S.; Zhao, Z.; Tan, W. Programming Affinity for Precise Tumor Recognition with Allosteric Nanosensing-Circles. ACS Nano 2023, 17, 13430–13440. [Google Scholar] [CrossRef] [PubMed]
- Zhi, S.; Zhang, X.; Zhang, J.; Wang, X.Y.; Bi, S. Functional Nucleic Acids-Engineered Bio-Barcode Nanoplatforms for Targeted Synergistic Therapy of Multidrug-Resistant Cancer. ACS Nano 2023, 17, 13533–13544. [Google Scholar] [CrossRef]
- Li, M.; Yang, G.; Zheng, Y.; Lv, J.; Zhou, W.; Zhang, H.; You, F.; Wu, C.; Yang, H.; Liu, Y. NIR/pH-triggered aptamer-functionalized DNA origami nanovehicle for imaging-guided chemo-phototherapy. J. Nanobiotechnol. 2023, 21, 186. [Google Scholar] [CrossRef]
- Rothemund, P.W. Folding DNA to create nanoscale shapes and patterns. Nature 2006, 440, 297–302. [Google Scholar] [CrossRef]
- Ma, S.; Ren, Q.; Jiang, L.; Liu, Z.; Zhu, Y.; Zhu, J.; Zhang, Y.; Zhang, M. A triple-aptamer tetrahedral DNA nanostructures based carbon-nanotube-array transistor biosensor for rapid virus detection. Talanta 2023, 266, 124973. [Google Scholar] [CrossRef]
- Zhang, H.; Zheng, X.; Zhao, T.; Chen, Y.; Luo, Y.; Dong, Y.; Tang, H.; Jiang, J. Real-Time Monitoring of Exosomes Secretion from Single Cell Using Dual-Nanopore Biosensors. ACS Sens. 2023, 8, 2583–2590. [Google Scholar] [CrossRef]
- Yang, C.; Shi, Y.; Zhang, Y.; He, J.; Li, M.; Huang, W.; Yuan, R.; Xu, W. Modular DNA Tetrahedron Nanomachine-Guided Dual-Responsive Hybridization Chain Reactions for Discernible Bivariate Assay and Cell Imaging. Anal. Chem. 2023, 95, 10337–10345. [Google Scholar] [CrossRef]
- Xiao, M.; Zhu, M.; Yuan, R.; Yuan, Y. Dual-sensitized heterojunction PDA/ZnO@MoS(2) QDs combined with multilocus domino-like DNA cascade reaction for ultrasensitive photoelectrochemical biosensor. Biosens. Bioelectron. 2023, 227, 115151. [Google Scholar] [CrossRef]
- Park, J.C.; Na, H.; Choi, S.; Jeon, H.; Nam, Y.S. Target-Catalyzed Self-Assembly of DNA-Streptavidin Nanogel for Enzyme-Free miRNA Assay. Adv. Healthc. Mater. 2023, 12, e2202076. [Google Scholar] [CrossRef]
- Xue, Y.; Xie, H.; Wang, Y.; Feng, S.; Sun, J.; Huang, J.; Yang, X. Novel and sensitive electrochemical/fluorescent dual-mode biosensing platform based on the cascaded cyclic amplification of enzyme-free DDSA and functional nucleic acids. Biosens. Bioelectron. 2022, 218, 114762. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Y.; Chen, W.; Zhang, Y.; Zhang, X.; Liu, Y.; Ju, H. A Near-Infrared Photo-Switched MicroRNA Amplifier for Precise Photodynamic Therapy of Early-Stage Cancers. Angew. Chem. Int. Ed. 2020, 59, 21454–21459. [Google Scholar] [CrossRef] [PubMed]
- Sun, H.; Wang, T.; Ma, W.; Huang, J.; Chen, B.; Cheng, H.; Duan, S.; He, X.; Jian, L.; Wang, K. A stable DNA Tetrahedra-AuNCs nanohybrid: On-site programmed disassembly for tumor imaging and combination therapy. Biomaterials 2022, 288, 121738. [Google Scholar] [CrossRef] [PubMed]
- Yang, L.; Yu, S.; Yan, Y.; Bi, S.; Zhu, J.J. Upconversion Nanoparticle@Au Core-Satellite Assemblies for In Situ Amplified Imaging of MicroRNA in Living Cells and Combined Cancer Phototherapy. Anal. Chem. 2022, 94, 7075–7083. [Google Scholar] [CrossRef]
- Chai, X.; Yi, D.; Sheng, C.; Zhao, J.; Li, L. A Remotely Controlled Nanosystem for Spatiotemporally Specific Gene Regulation and Combinational Tumor Therapy. Angew. Chem. Int. Ed. 2023, 62, e202217702. [Google Scholar] [CrossRef]
- Baig, M.; Ma, J.; Gao, X.; Khan, M.A.; Ali, A.; Farid, A.; Zia, A.W.; Noreen, S.; Wu, H. Exploring the robustness of DNA nanotubes framework for anticancer theranostics toward the 2D/3D clusters of hypopharyngeal respiratory tumor cells. Int. J. Biol. Macromol. 2023, 236, 123988. [Google Scholar] [CrossRef]
- Wang, J.; Li, J.; Chen, Y.; Liu, R.; Wu, Y.; Liu, J.; Yang, X.; Wang, K.; Huang, J. Size-Controllable and Self-Assembled DNA Nanosphere for Amplified MicroRNA Imaging through ATP-Fueled Cyclic Dissociation. Nano Lett. 2022, 22, 8216–8223. [Google Scholar] [CrossRef]
- Yang, Y.; Cai, X.; Shi, M.; Zhang, X.; Pan, Y.; Zhang, Y.; Ju, H.; Cao, P. Biomimetic retractable DNA nanocarrier with sensitive responsivity for efficient drug delivery and enhanced photothermal therapy. J. Nanobiotechnol. 2023, 21, 46. [Google Scholar] [CrossRef]
- Shen, Y.; Hu, M.; Li, W.; Chen, Y.; Xu, Y.; Sun, L.; Liu, D.; Chen, S.; Gu, Y.; Ma, Y.; et al. Delivery of DNA octahedra enhanced by focused ultrasound with microbubbles for glioma therapy. J. Control. Release Off. J. Control. Release Soc. 2022, 350, 158–174. [Google Scholar] [CrossRef]
- Wang, D.; Liu, J.; Duan, J.; Ma, Y.; Gao, H.; Zhang, Z.; Liu, J.; Shi, J.; Zhang, K. Photocontrolled Spatiotemporal Delivery of DNA Immunomodulators for Enhancing Membrane-Targeted Tumor Photodynamic Immunotherapy. ACS Appl. Mater. Interfaces 2022, 14, 44183–44198. [Google Scholar] [CrossRef]
- Chen, Y.; Ke, G.; Ma, Y.; Zhu, Z.; Liu, M.; Liu, Y.; Yan, H.; Yang, C.J. A Synthetic Light-Driven Substrate Channeling System for Precise Regulation of Enzyme Cascade Activity Based on DNA Origami. J. Am. Chem. Soc. 2018, 140, 8990–8996. [Google Scholar] [CrossRef] [PubMed]
- Trinh, T.; Thompson, I.A.P.; Clark, F.; Remington, J.M.; Eisenstein, M.; Li, J.; Soh, H.T. A Photoresponsive Intramolecular Triplex Motif That Enables Rapid and Reversible Control of Aptamer Binding Activity. ACS Nano 2022, 16, 14549–14557. [Google Scholar] [CrossRef] [PubMed]
- Asanuma, H.; Liang, X.; Nishioka, H.; Matsunaga, D.; Liu, M.; Komiyama, M. Synthesis of Azobenzene-Tethered DNA for Reversible Photo-Regulation of DNA Functions: Hybridization and Transcription. Nat. Protoc. 2007, 2, 203–212. [Google Scholar] [CrossRef]
- Ebrahimi, S.; Akhlaghi, Y.; Kompany-Zareh, M.; Rinnan, A. Nucleic acid based fluorescent nanothermometers. ACS Nano 2014, 8, 10372–10382. [Google Scholar] [CrossRef]
- Mariottini, D.; Idili, A.; Ercolani, G.; Ricci, F. Thermo-Programmed Synthetic DNA-Based Receptors. ACS Nano 2023, 17, 1998–2006. [Google Scholar] [CrossRef]
- Du, X.; He, P.-P.; Wang, C.; Wang, X.; Mu, Y.; Guo, W. Fast Transport and Transformation of Biomacromolecular Substances via Thermo-Stimulated Active “Inhalation-Exhalation” Cycles of Hierarchically Structured Smart pNIPAM-DNA Hydrogels. Adv. Mater. 2023, 35, e2206302. [Google Scholar] [CrossRef]
- Maret, G.; Schickfus, M.v.; Mayer, A.; Dransfeld, K. Orientation of Nucleic Acids in High Magnetic Fields. Phys. Rev. Lett. 1975, 35, 397–400. [Google Scholar] [CrossRef]
- Wang, Y.; Yang, Y.; Cao, X.; Liu, Z.; Chen, B.; Du, Q.; Lu, X. Simple and Ultrasensitive Detection of Glioma-Related ctDNAs in Mice Serum by SERS-Based Catalytic Hairpin Assembly Signal Amplification Coupled with Magnetic Aggregation. Int. J. Nanomed. 2023, 18, 3211–3230. [Google Scholar] [CrossRef]
- Hallaj, R.; Ghafary, Z.; Kamal Mohammed, O.; Shakeri, R. Induced ultrasensitive electrochemical biosensor for target MDA-MB-231 cell cytoplasmic protein detection based on RNA-cleavage DNAzyme catalytic reaction. Biosens. Bioelectron. 2023, 227, 115168. [Google Scholar] [CrossRef]
- Serrano-Chacón, I.; Mir, B.; Cupellini, L.; Colizzi, F.; Orozco, M.; Escaja, N.; González, C. pH-Dependent Capping Interactions Induce Large-Scale Structural Transitions in i-Motifs. J. Am. Chem. Soc. 2023, 145, 3696–3705. [Google Scholar] [CrossRef]
- Luo, H.; Wang, Z.; Mo, Q.; Yang, J.; Yang, F.; Tang, Y.; Liu, J.; Li, X. Framework Nucleic Acid-Based Multifunctional Tumor Theranostic Nanosystem for miRNA Fluorescence Imaging and Chemo/Gene Therapy. ACS Appl. Mater. Interfaces 2023, 15, 33223–33238. [Google Scholar] [CrossRef] [PubMed]
- Deng, J.; Walther, A. ATP-Responsive and ATP-Fueled Self-Assembling Systems and Materials. Adv. Mater. 2020, 32, e2002629. [Google Scholar] [CrossRef] [PubMed]
- Kim, N.; Kim, E.; Kim, H.; Thomas, M.R.; Najer, A.; Stevens, M.M. Tumor-Targeting Cholesterol-Decorated DNA Nanoflowers for Intracellular Ratiometric Aptasensing. Adv. Mater. 2021, 33, e2007738. [Google Scholar] [CrossRef] [PubMed]
- Yin, Q.; Zhao, D.; Chang, Y.; Liu, B.; Liu, Y.; Liu, M. Functional DNA Superstructures Exhibit Positive Homotropic Allostery in Ligand Binding. Angew. Chem. Int. Ed. 2023, 135, e202303838. [Google Scholar] [CrossRef]
- Ren, Q.; Jiang, L.; Ma, S.; Li, T.; Zhu, Y.; Qiu, R.; Xing, Y.; Yin, F.; Li, Z.; Ye, X.; et al. Multi-Body Biomarker Entrapment System: An All-Encompassing Tool for Ultrasensitive Disease Diagnosis and Epidemic Screening. Adv. Mater. 2023, e2304119. [Google Scholar] [CrossRef]
- Meng, R.; Zhang, X.; Liu, J.; Zhou, Y.; Zhang, P.; Chai, Y.; Yuan, R. Dual-layer 3D DNA nanostructure: The next generation of ultrafast DNA nanomachine for microRNA sensing and intracellular imaging. Biosens. Bioelectron. 2023, 237, 115517. [Google Scholar] [CrossRef]
- Yu, L.; Peng, Y.; Sheng, M.; Wang, Q.; Huang, J.; Yang, X. Sensitive and Amplification-Free Electrochemiluminescence Biosensor for HPV-16 Detection Based on CRISPR/Cas12a and DNA Tetrahedron Nanostructures. ACS Sens. 2023, 8, 2852–2858. [Google Scholar] [CrossRef]
- Rossi-Gendron, C.; El Fakih, F.; Bourdon, L.; Nakazawa, K.; Finkel, J.; Triomphe, N.; Chocron, L.; Endo, M.; Sugiyama, H.; Bellot, G.; et al. Isothermal self-assembly of multicomponent and evolutive DNA nanostructures. Nat. Nanotechnol. 2023. [Google Scholar] [CrossRef]
- Jin, Y.; Huang, Z.; Xu, B.; Chen, J. Localization of multiple DNAzymes as a branchedzyme-powered nanodevice for the immunoassay of tumor biomarkers. Anal. Chim. Acta 2023, 1274, 341580. [Google Scholar] [CrossRef]
- Xing, Y.; Dorey, A.; Howorka, S. Multi-Stimuli-Responsive and Mechano-Actuated Biomimetic Membrane Nanopores Self-Assembled from DNA. Adv. Mater. 2023, 35, e2300589. [Google Scholar] [CrossRef]
- Xue, Y.Q.; Liao, N.; Li, Y.; Liang, W.B.; Yang, X.; Zhong, X.; Zhuo, Y. Ordered heterogeneity in dual-ligand MOF to enable high electrochemiluminescence efficiency for bioassay with DNA triangular prism as signal switch. Biosens. Bioelectron. 2022, 217, 114713. [Google Scholar] [CrossRef] [PubMed]
- Zhou, C.; Chuai, Y.; Lin, C.; Wang, D.; Wang, Q.; Zou, H. A dual fragment triggered DNA ladder nanostructure based on logic gate and dispersion-to-localization catalytic hairpin assembly for efficient fluorescence assay of SARS-CoV-2 and H1N1. Anal. Chim. Acta 2023, 1275, 341590. [Google Scholar] [CrossRef]
- Deng, W.; Xu, J.Y.; Peng, H.; Huang, C.Z.; Le, X.C.; Zhang, H. DNAzyme motor systems and logic gates facilitated by toehold exchange translators. Biosens. Bioelectron. 2022, 217, 114704. [Google Scholar] [CrossRef] [PubMed]
- Zhou, X.M.; Zhuo, Y.; Yuan, R.; Chai, Y.Q. Target-mediated self-assembly of DNA networks for sensitive detection and intracellular imaging of APE1 in living cells. Chem. Sci. 2023, 14, 2318–2324. [Google Scholar] [CrossRef] [PubMed]
- Shen, M.; Shi, J.; Chen, Z.; Zhang, S.; Zhang, Z. Self-assembly of DNA-hyperbranched aggregates catalyzed by a dual-targets recognition probe for miRNAs SERS detection in single cells. Biosens. Bioelectron. 2023, 222, 114997. [Google Scholar] [CrossRef]
- Du, S.; Xie, B.; Gao, H.; Zhang, J.; Fu, H.; Liao, F.; Liao, Y. Self-Powered DNAzyme Walker Enables Dual-Mode Biosensor Construction for Electrochemiluminescence and Electrochemical Detection of MicroRNA. Anal. Chem. 2023, 95, 7006–7013. [Google Scholar] [CrossRef]
- Garzon, R.; Calin, G.A.; Croce, C.M. MicroRNAs in Cancer. Annu. Rev. Med. 2009, 60, 167–179. [Google Scholar] [CrossRef]
- Bartel, D.P. MicroRNAs: Target recognition and regulatory functions. Cell 2009, 136, 215–233. [Google Scholar] [CrossRef]
- He, L.; Hannon, G.J. MicroRNAs: Small RNAs with a big role in gene regulation. Nat. Rev. Genet. 2004, 5, 522–531. [Google Scholar] [CrossRef]
- Lu, J.; Getz, G.; Miska, E.A.; Alvarez-Saavedra, E.; Lamb, J.; Peck, D.; Sweet-Cordero, A.; Ebert, B.L.; Mak, R.H.; Ferrando, A.A.; et al. MicroRNA expression profiles classify human cancers. Nature 2005, 435, 834–838. [Google Scholar] [CrossRef]
- Chen, X.; Ba, Y.; Ma, L.; Cai, X.; Yin, Y.; Wang, K.; Guo, J.; Zhang, Y.; Chen, J.; Guo, X.; et al. Characterization of microRNAs in serum: A novel class of biomarkers for diagnosis of cancer and other diseases. Cell Res. 2008, 18, 997–1006. [Google Scholar] [CrossRef]
- Kim, T.H.; Lee, Y.; Lee, Y.S.; Gim, J.A.; Ko, E.; Yim, S.Y.; Jung, Y.K.; Kang, S.; Kim, M.Y.; Kim, H.; et al. Circulating miRNA is a useful diagnostic biomarker for nonalcoholic steatohepatitis in nonalcoholic fatty liver disease. Sci. Rep. 2021, 11, 14639. [Google Scholar] [CrossRef] [PubMed]
- Wu, Q.; Li, L.; Jia, Y.; Xu, T.; Zhou, X. Advances in studies of circulating microRNAs: Origination, transportation, and distal target regulation. J. Cell Commun. Signal. 2022, 17, 445–455. [Google Scholar] [CrossRef] [PubMed]
- Chapin, S.C.; Doyle, P.S. Ultrasensitive multiplexed microRNA quantification on encoded gel microparticles using rolling circle amplification. Anal. Chem. 2011, 83, 7179–7185. [Google Scholar] [CrossRef] [PubMed]
- Wu, L.; Wang, Y.; He, R.; Zhang, Y.; He, Y.; Wang, C.; Lu, Z.; Liu, Y.; Ju, H. Fluorescence hydrogel array based on interfacial cation exchange amplification for highly sensitive microRNA detection. Anal. Chim. Acta 2019, 1080, 206–214. [Google Scholar] [CrossRef]
- Guo, S.; Lin, W.N.; Hu, Y.; Sun, G.; Phan, D.T.; Chen, C.H. Ultrahigh-throughput droplet microfluidic device for single-cell miRNA detection with isothermal amplification. Lab Chip 2018, 18, 1914–1920. [Google Scholar] [CrossRef]
- Li, L.; Lu, M.; Fan, Y.; Shui, L.; Xie, S.; Sheng, R.; Si, H.; Li, Q.; Wang, Y.; Tang, B. High-throughput and ultra-sensitive single-cell profiling of multiple microRNAs and identification of human cancer. Chem. Commun. 2019, 55, 10404–10407. [Google Scholar] [CrossRef]
- Wang, Y.; Fang, Y.; Zhu, Y.; Bi, S.; Liu, Y.; Ju, H. Single cell multi-miRNAs quantification with hydrogel microbeads for liver cancer cell subtypes discrimination. Chem. Sci. 2022, 13, 2062–2070. [Google Scholar] [CrossRef]
- Herrmann, I.K.; Wood, M.J.A.; Fuhrmann, G. Extracellular vesicles as a next-generation drug delivery platform. Nat. Nanotechnol. 2021, 16, 748–759. [Google Scholar] [CrossRef]
- Buzas, E.I. The roles of extracellular vesicles in the immune system. Nat. Rev. Immunol. 2023, 23, 236–250. [Google Scholar] [CrossRef]
- Nabet, B.Y.; Qiu, Y.; Shabason, J.E.; Wu, T.J.; Yoon, T.; Kim, B.C.; Benci, J.L.; DeMichele, A.M.; Tchou, J.; Marcotrigiano, J.; et al. Exosome RNA Unshielding Couples Stromal Activation to Pattern Recognition Receptor Signaling in Cancer. Cell 2017, 170, 352–366. [Google Scholar] [CrossRef]
- Shen, W.; Guo, K.; Adkins, G.B.; Jiang, Q.; Liu, Y.; Sedano, S.; Duan, Y.; Yan, W.; Wang, S.E.; Bergersen, K.; et al. A Single Extracellular Vesicle (EV) Flow Cytometry Approach to Reveal EV Heterogeneity. Angew. Chem. Int. Ed. 2018, 57, 15675–15680. [Google Scholar] [CrossRef]
- Wang, L.; Deng, Y.; Wei, J.; Huang, Y.; Wang, Z.; Li, G. Spherical nucleic acids-based cascade signal amplification for highly sensitive detection of exosomes. Biosens. Bioelectron. 2021, 191, 113465. [Google Scholar] [CrossRef] [PubMed]
- Lin, D.; Shen, L.; Luo, M.; Zhang, K.; Li, J.; Yang, Q.; Zhu, F.; Zhou, D.; Zheng, S.; Chen, Y.; et al. Circulating tumor cells: Biology and clinical significance. Signal Transduct. Target. Ther. 2021, 6, 404. [Google Scholar] [CrossRef] [PubMed]
- Eslami, S.Z.; Cortes-Hernandez, L.E.; Thomas, F.; Pantel, K.; Alix-Panabieres, C. Functional analysis of circulating tumour cells: The KEY to understand the biology of the metastatic cascade. Br. J. Cancer 2022, 127, 800–810. [Google Scholar] [CrossRef] [PubMed]
- Peng, Y.; Pan, Y.; Han, Y.; Sun, Z.; Jalalah, M.; Al-Assiri, M.S.; Harraz, F.A.; Yang, J.; Li, G. Direct Analysis of Rare Circulating Tumor Cells in Whole Blood Based on Their Controlled Capture and Release on Electrode Surface. Anal. Chem. 2020, 92, 13478–13484. [Google Scholar] [CrossRef]
- Song, Y.; Shi, Y.; Huang, M.; Wang, W.; Wang, Y.; Cheng, J.; Lei, Z.; Zhu, Z.; Yang, C. Bioinspired Engineering of a Multivalent Aptamer-Functionalized Nanointerface to Enhance the Capture and Release of Circulating Tumor Cells. Angew. Chem. Int. Ed. 2019, 58, 2236–2240. [Google Scholar] [CrossRef]
- Zhang, J.; Lin, B.; Wu, L.; Huang, M.; Li, X.; Zhang, H.; Song, J.; Wang, W.; Zhao, G.; Song, Y.; et al. DNA Nanolithography Enables a Highly Ordered Recognition Interface in a Microfluidic Chip for the Efficient Capture and Release of Circulating Tumor Cells. Angew. Chem. Int. Ed. 2020, 59, 14115–14119. [Google Scholar] [CrossRef]
- Wu, L.; Ding, H.; Qu, X.; Shi, X.; Yang, J.; Huang, M.; Zhang, J.; Zhang, H.; Song, J.; Zhu, L.; et al. Fluidic Multivalent Membrane Nanointerface Enables Synergetic Enrichment of Circulating Tumor Cells with High Efficiency and Viability. J. Am. Chem. Soc. 2020, 142, 4800–4806. [Google Scholar] [CrossRef]
- Wei, J.; Wang, H.; Wu, Q.; Gong, X.; Ma, K.; Liu, X.; Wang, F. A Smart, Autocatalytic, DNAzyme Biocircuit for in Vivo, Amplified, MicroRNA Imaging. Angew. Chem. Int. Ed. 2020, 59, 5965–5971. [Google Scholar] [CrossRef]
- He, S.; Yu, S.; Li, R.; Chen, Y.; Wang, Q.; He, Y.; Liu, X.; Wang, F. On-Site Non-enzymatic Orthogonal Activation of a Catalytic DNA Circuit for Self-Reinforced In Vivo MicroRNA Imaging. Angew. Chem. Int. Ed. 2022, 61, e202206529. [Google Scholar] [CrossRef] [PubMed]
- Wang, H.; He, Y.; Wei, J.; Wang, H.; Ma, K.; Zhou, Y.; Liu, X.; Zhou, X.; Wang, F. Construction of an Autocatalytic Hybridization Assembly Circuit for Amplified In Vivo MicroRNA Imaging. Angew. Chem. Int. Ed. 2022, 61, e202115489. [Google Scholar] [CrossRef] [PubMed]
- Dong, P.; Li, R.; He, S.; Zhang, Q.; Shang, J.; Jiang, Y.; Liu, X.; Wang, F. The compact integration of a cascaded HCR circuit for highly reliable cancer cell discrimination. Chem. Sci. 2023, 14, 2159–2167. [Google Scholar] [CrossRef] [PubMed]
- Li, R.; Zhu, Y.; Gong, X.; Zhang, Y.; Hong, C.; Wan, Y.; Liu, X.; Wang, F. Self-Stacking Autocatalytic Molecular Circuit with Minimal Catalytic DNA Assembly. J. Am. Chem. Soc. 2023, 145, 2999–3007. [Google Scholar] [CrossRef] [PubMed]
- Shang, J.; Yu, S.; Li, R.; He, Y.; Wang, Y.; Wang, F. Bioorthogonal Disassembly of Hierarchical DNAzyme Nanogel for High-Performance Intracellular microRNA Imaging. Nano Lett. 2023, 23, 1386–1394. [Google Scholar] [CrossRef]
- Hong, S.; Zhang, X.; Lake, R.J.; Pawel, G.T.; Guo, Z.; Pei, R.; Lu, Y. A photo-regulated aptamer sensor for spatiotemporally controlled monitoring of ATP in the mitochondria of living cells. Chem. Sci. 2019, 11, 713–720. [Google Scholar] [CrossRef]
- Wen, S.; Zhou, J.; Zheng, K.; Bednarkiewicz, A.; Liu, X.; Jin, D. Advances in highly doped upconversion nanoparticles. Nat. Commun. 2018, 9, 2415. [Google Scholar] [CrossRef]
- Zhao, J.; Chu, H.; Zhao, Y.; Lu, Y.; Li, L. A NIR Light Gated DNA Nanodevice for Spatiotemporally Controlled Imaging of MicroRNA in Cells and Animals. J. Am. Chem. Soc. 2019, 141, 7056–7062. [Google Scholar] [CrossRef]
- Wei, W.; Dai, W.; Yang, F.; Lu, H.; Zhang, K.; Xing, Y.; Meng, X.; Zhou, L.; Zhang, Y.; Yang, Q.; et al. Spatially Resolved, Error-Robust Multiplexed MicroRNA Profiling in Single Living Cells. Angew. Chem. Int. Ed. 2022, 61, e202116909. [Google Scholar] [CrossRef]
- Zhang, Y.; Zhang, Y.; Zhang, X.; Li, Y.; He, Y.; Liu, Y.; Ju, H. A photo zipper locked DNA nanomachine with an internal standard for precise miRNA imaging in living cells. Chem. Sci. 2020, 11, 6289–6296. [Google Scholar] [CrossRef]
- Li, Y.; Xie, Y.; Zhang, Y.; Zhao, H.; Ju, H.; Liu, Y. DNA nanomachine activation and Zn2+ imaging in living cells with single NIR irradiation. Anal. Chim. Acta 2022, 1221, 340149. [Google Scholar] [CrossRef] [PubMed]
- He, C.; Lu, K.; Lin, W. Nanoscale metal-organic frameworks for real-time intracellular pH sensing in live cells. J. Am. Chem. Soc. 2014, 136, 12253–12256. [Google Scholar] [CrossRef] [PubMed]
- Singh, A.K.; Nair, A.V.; Singh, N.D.P. Small Two-Photon Organic Fluorogenic Probes: Sensing and Bioimaging of Cancer Relevant Biomarkers. Anal. Chem. 2021, 94, 177–192. [Google Scholar] [CrossRef] [PubMed]
- Zhang, P.; Ouyang, Y.; Sohn, Y.S.; Nechushtai, R.; Pikarsky, E.; Fan, C.; Willner, I. pH- and miRNA-Responsive DNA-Tetrahedra/Metal-Organic Framework Conjugates: Functional Sense-and-Treat Carriers. ACS Nano 2021, 15, 6645–6657. [Google Scholar] [CrossRef]
- Liu, S.; Jiang, Q.; Zhao, X.; Zhao, R.; Wang, Y.; Wang, Y.; Liu, J.; Shang, Y.; Zhao, S.; Wu, T.; et al. A DNA nanodevice-based vaccine for cancer immunotherapy. Nat. Mater. 2021, 20, 421–430. [Google Scholar] [CrossRef]
- Qian, R.C.; Zhou, Z.R.; Wu, Y.; Yang, Z.; Guo, W.; Li, D.W.; Lu, Y. Combination Cancer Treatment: Using Engineered DNAzyme Molecular Machines for Dynamic Inter- and Intracellular Regulation. Angew. Chem. Int. Ed. 2022, 61, e202210935. [Google Scholar] [CrossRef]
- Zhang, K.; Ma, Y.; Wang, D.; Liu, J.; An, J.; Li, Y.; Ma, C.; Pei, Y.; Zhang, Z.; Liu, J.; et al. In Vivo Activation of T-Cell Proliferation by Regulating Cell Surface Receptor Clustering Using a pH-Driven Interlocked DNA Nano-Spring. Nano Lett. 2022, 22, 1937–1945. [Google Scholar] [CrossRef]
- Mo, R.; Jiang, T.; DiSanto, R.; Tai, W.; Gu, Z. ATP-triggered anticancer drug delivery. Nat. Commun. 2014, 5, 3364. [Google Scholar] [CrossRef]
- Su, Y.; Chen, X.; Wang, H.; Sun, L.; Xu, Y.; Li, D. Enhancing cell membrane phase separation for inhibiting cancer metastasis with a stimuli-responsive DNA nanodevice. Chem. Sci. 2022, 13, 6303–6308. [Google Scholar] [CrossRef]
- Li, H.; Wang, M.; Shi, T.; Yang, S.; Zhang, J.; Wang, H.H.; Nie, Z. A DNA-Mediated Chemically Induced Dimerization (D-CID) Nanodevice for Nongenetic Receptor Engineering To Control Cell Behavior. Angew. Chem. Int. Ed. 2018, 57, 10226–10230. [Google Scholar] [CrossRef]
- Wang, Z.; Song, L.; Liu, Q.; Tian, R.; Shang, Y.; Liu, F.; Liu, S.; Zhao, S.; Han, Z.; Sun, J.; et al. A Tubular DNA Nanodevice as a siRNA/Chemo-Drug Co-delivery Vehicle for Combined Cancer Therapy. Angew. Chem. Int. Ed. 2021, 60, 2594–2598. [Google Scholar] [CrossRef] [PubMed]
- Yoon, A.J.; Wang, S.; Kutler, D.I.; Carvajal, R.D.; Philipone, E.; Wang, T.; Peters, S.M.; LaRoche, D.; Hernandez, B.Y.; McDowell, B.D.; et al. MicroRNA-based risk scoring system to identify early-stage oral squamous cell carcinoma patients at high-risk for cancer-specific mortality. Head Neck 2020, 42, 1699–1712. [Google Scholar] [CrossRef] [PubMed]
- Lakhia, R.; Ramalingam, H.; Chang, C.M.; Cobo-Stark, P.; Biggers, L.; Flaten, A.; Alvarez, J.; Valencia, T.; Wallace, D.P.; Lee, E.C.; et al. PKD1 and PKD2 mRNA cis-inhibition drives polycystic kidney disease progression. Nat. Commun. 2022, 13, 4765. [Google Scholar] [CrossRef] [PubMed]
- Ren, K.; Zhang, Y.; Zhang, X.; Liu, Y.; Yang, M.; Ju, H. In Situ SiRNA Assembly in Living Cells for Gene Therapy with MicroRNA Triggered Cascade Reactions Templated by Nucleic Acids. ACS Nano 2018, 12, 10797–10806. [Google Scholar] [CrossRef]
- He, F.; Wang, M.; Wang, J.; Wang, H.H.; Nie, Z. An Extracellular miRNA-Responsive Artificial Receptor via Dynamic DNA Nano-assembly for Biomarker-Driven Therapy. Angew. Chem. Int. Ed. 2023, 135, e202305227. [Google Scholar] [CrossRef]
- Li, S.; Jiang, Q.; Liu, S.; Zhang, Y.; Tian, Y.; Song, C.; Wang, J.; Zou, Y.; Anderson, G.J.; Han, J.Y.; et al. A DNA nanorobot functions as a cancer therapeutic in response to a molecular trigger in vivo. Nat. Biotechnol. 2018, 36, 258–264. [Google Scholar] [CrossRef]
- Li, L.; Wang, J.; Li, Y.; Radford, D.C.; Yang, J.; Kopeček, J. Broadening and Enhancing Functions of Antibodies by Self-Assembling Multimerization at Cell Surface. ACS Nano 2019, 13, 11422–11432. [Google Scholar] [CrossRef]
- Ueki, R.; Atsuta, S.; Ueki, A.; Sando, S. Nongenetic Reprogramming of the Ligand Specificity of Growth Factor Receptors by Bispecific DNA Aptamers. J. Am. Chem. Soc. 2017, 139, 6554–6557. [Google Scholar] [CrossRef]
- Bi, S.; Chen, W.; Fang, Y.; Wang, Y.; Zhang, Q.; Guo, H.; Ju, H.; Liu, Y. Cancer Cell-Selective Membrane Receptor Clustering Driven by VEGF Secretion for In Vivo Therapy. J. Am. Chem. Soc. 2023, 145, 5041–5052. [Google Scholar] [CrossRef]
- Fang, Y.; Yan, Y.; Bi, S.; Wang, Y.; Chen, Y.; Xu, P.; Ju, H.; Liu, Y. Screening T-Cell Activity via a Photodetachable DNA-Copolymer Nanocage and Its Therapeutic Application. Anal. Chem. 2022, 94, 13205–13214. [Google Scholar] [CrossRef]
- Chen, S.; Xu, Z.; Yang, W.; Lin, X.; Li, J.; Li, J.; Yang, H. Logic-Gate-Actuated DNA-Controlled Receptor Assembly for the Programmable Modulation of Cellular Signal Transduction. Angew. Chem. Int. Ed. 2019, 58, 18186–18190. [Google Scholar] [CrossRef]
- Zhang, Y.; Zhang, Y.; Song, G.; He, Y.; Zhang, X.; Liu, Y.; Ju, H. A DNA-Azobenzene Nanopump Fueled by Upconversion Luminescence for Controllable Intracellular Drug Release. Angew. Chem. Int. Ed. 2019, 58, 18207–18211. [Google Scholar] [CrossRef]
- Huang, F.; Duan, R.; Zhou, Z.; Vázquez-González, M.; Xia, F.; Willner, I. Near-infrared light-activated membrane fusion for cancer cell therapeutic applications. Chem. Sci. 2020, 11, 5592–5600. [Google Scholar] [CrossRef] [PubMed]
- Han, S.; Samanta, A.; Xie, X.; Huang, L.; Peng, J.; Park, S.J.; Teh, D.B.L.; Choi, Y.; Chang, Y.T.; All, A.H.; et al. Gold and Hairpin DNA Functionalization of Upconversion Nanocrystals for Imaging and In Vivo Drug Delivery. Adv. Mater. 2017, 29, 1700244. [Google Scholar] [CrossRef] [PubMed]
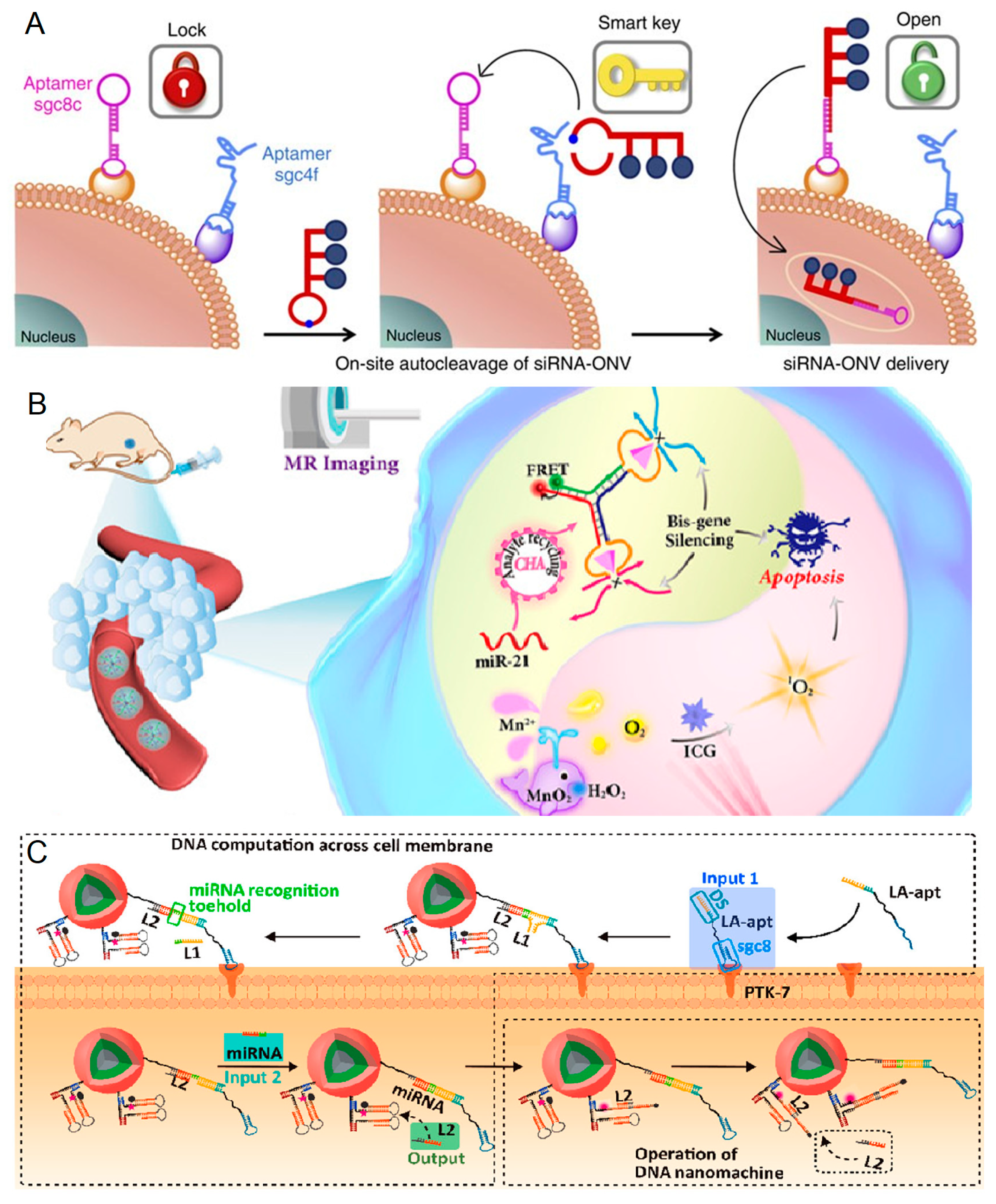
- Ren, K.; Liu, Y.; Wu, J.; Zhang, Y.; Zhu, J.; Yang, M.; Ju, H. A DNA dual lock-and-key strategy for cell-subtype-specific siRNA delivery. Nat. Commun. 2016, 7, 13580. [Google Scholar] [CrossRef] [PubMed]
- Gong, X.; Li, R.; Wang, J.; Wei, J.; Ma, K.; Liu, X.; Wang, F. A Smart Theranostic Nanocapsule for Spatiotemporally Programmable Photo-Gene Therapy. Angew. Chem. Int. Ed. 2020, 59, 21648–21655. [Google Scholar] [CrossRef]
- Zhang, Y.; Chen, W.; Fang, Y.; Zhang, X.; Liu, Y.; Ju, H. Activating a DNA Nanomachine via Computation across Cancer Cell Membranes for Precise Therapy of Solid Tumors. J. Am. Chem. Soc. 2021, 143, 15233–15242. [Google Scholar] [CrossRef]
- Zhou, F.; Wang, P.; Chen, J.; Zhu, Z.; Li, Y.; Wang, S.; Wu, S.; Sima, Y.; Fu, T.; Tan, W.; et al. A photochemically covalent lock stabilizes aptamer conformation and strengthens its performance for biomedicine. Nucleic Acids Res. 2022, 50, 9039–9050. [Google Scholar] [CrossRef]
- Xue, C.; Zhang, S.; Yu, X.; Hu, S.; Lu, Y.; Wu, Z.S. Periodically Ordered, Nuclease-Resistant DNA Nanowires Decorated with Cell-Specific Aptamers as Selective Theranostic Agents. Angew. Chem. Int. Ed. 2020, 59, 17540–17547. [Google Scholar] [CrossRef]



Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Wang, Y.; Zhang, Y.; Ju, H.; Liu, Y. Responsive DNA Nanostructures for Bioanalysis and Therapy. Chemistry 2023, 5, 2182-2204. https://doi.org/10.3390/chemistry5040147
Wang Y, Zhang Y, Ju H, Liu Y. Responsive DNA Nanostructures for Bioanalysis and Therapy. Chemistry. 2023; 5(4):2182-2204. https://doi.org/10.3390/chemistry5040147
Chicago/Turabian StyleWang, Yingfei, Yue Zhang, Huangxian Ju, and Ying Liu. 2023. "Responsive DNA Nanostructures for Bioanalysis and Therapy" Chemistry 5, no. 4: 2182-2204. https://doi.org/10.3390/chemistry5040147
APA StyleWang, Y., Zhang, Y., Ju, H., & Liu, Y. (2023). Responsive DNA Nanostructures for Bioanalysis and Therapy. Chemistry, 5(4), 2182-2204. https://doi.org/10.3390/chemistry5040147







