Abstract
Aptamers are artificial oligonucleotides with excellent molecule-targeting ability. Compared with monoclonal antibodies, aptamers have the advantages of low cost, no batch effect, and negligible immunogenicity, making them promising candidates for cancer immunotherapy. To date, a series of aptamer agonists/antagonists have been discovered and directly used to activate immune response, such as immune checkpoint blockade, immune costimulation, and cytokine regulation. By incorporating both tumor- and immune cell-targeting aptamers, multivalent bispecific aptamers were designed to pursue high tumor affinity and enhanced immune efficacy. More importantly, benefiting from feasible chemical modification and programmability, aptamers can be engineered with diverse nanomaterials (e.g., liposomes, hydrogels) and even living immune cells (e.g., NK cells, T cells). These aptamer-based assemblies exhibit powerful capabilities in targeted cargo delivery, regulation of cell–cell interactions, tumor immunogenicity activation, tumor microenvironment remodeling, etc., holding huge potential in boosting immunotherapeutic efficacy. In this review, we focus on the recent advances in aptamer-based immune drug systems (AptIDCs) and highlight their advantages in cancer immunotherapy. The current challenges and future prospects of this field are also pointed out in this paper.
1. Introduction
Over the past decade, immunotherapy has been recognized as an efficacious treatment for cancers, and sometimes can be an alternative to chemotherapy and radiotherapy in cases where they are ineffective [1]. There are mainly three immunotherapy strategies, i.e., adoptive cell therapy, tumor vaccines, and antibodies. Currently, antibodies are the most popular antitumor immune agents and have yielded encouraging results in advanced cancers [2,3]. However, some limitations still exist. First, the tedious preparation and purification process leads to the high cost of antibodies (especially monoclonal) in practice [4]. Moreover, due to the high immunogenicity of antibodies, patients usually suffer from severe immune side effects clinically [5]. In addition, tumor tissues are characterized with an immunosuppressive microenvironment (ISM), which often causes low immune response and, subsequently, a suboptimal outcome [6]. Thereby, seeking new immunotherapeutic agents that overcome the weaknesses of traditional antibodies is crucial to improve the current status of tumor immunotherapy.
Aptamers are short single-stranded DNA/RNA molecules (20~100 nt) with high binding affinity toward targets, selected by systematic evolution of ligands by exponential enrichment (SELEX) technology [7,8]. Unlike animal-derived antibodies, aptamers can be prepared in large quantities simply by chemical synthesis, greatly reducing the cost. In addition, aptamers also have the advantages of low immunogenicity, superior chemical and physical stabilities, convenient storage, and deep tumor penetration [9,10,11]. All these virtues make aptamers more attractive biomedical reagents for cancer immunotherapy than traditional antibodies. At present, increasing aptamers have been identified to serve as agonists or antagonists in directly regulating immune responses for tumor therapy. For instance, the screened anti-PD1/PDL1 aptamers can specifically block immune checkpoints and inhibit tumor immune escape, and have shown comparable efficacies to the corresponding antibodies [12]. In addition, aptamers targeting immune costimulation (e.g., CD28) [13] and cytokine regulation (e.g., IL-8) [14] are also reported to exhibit favorable immunomodulatory activities. As nucleic acid molecules, the flexible conformation change, easy chemical modification, and accurate base complementary pairing laws impart well-engineered feasibility to aptamers [15]. Therefore, aptamers can be designed in multivalent conformations or can be conjugated to different materials, including small molecules, biomacromolecules, nanoparticles, and even cells [16,17,18,19,20]. Theses multifunctional AptIDCs have been used for targeted immune agent delivery, regulation of intercellular interactions, tumor immunogenicity activation, and tumor microenvironment remodeling, exhibiting high potency in cancer immunotherapy.
In this review, we briefly introduce the classic SELEX technology and new approaches for aptamer selection. Then, we summarize and classify the monomeric aptamer agonists/antagonists that can directly regulate immune response for cancer therapy. Strategies for engineering aptamer-functionalized assemblies, including bivalent aptamers, aptamer–nanoparticle conjugates, and aptamer–cell assemblies, are systematically described and highlighted for their versatility in enhancing the efficacy of cancer immunotherapy. Lastly, we state the current challenges of AptIDCs and provide an outlook on the future prospects of this field.
2. Aptamer Selection
Aptamers were first reported by Gold [7] and Elligton [8], in 1990, as evolved from an in vitro selection method called SELEX. The classic SELEX consists of a defined procedure: ssDNA/RNA library design, target incubation, positive selection, counter selection, sequence isolation, and PCR amplification. After about 20 rounds of selection, specific target-binding sequences can be sufficiently enriched, followed by sequence analysis and affinity evaluation. For instance, Gilboa’s group reported selecting several CTLA-4-specific RNA aptamers for tumor treatment by means of classic SELEX technology [21]. They chose murine CTLA-4/Fc fusion protein for positive selection, while human IgG1 was chosen for counter selection. After successive rounds of selection, RNA aptamers that selectively bound with murine CTLA-4/Fc were obtained. So far, SELEX remains the most popular and reliable approach to discover nucleic acid aptamers for various target molecules.
Aptamers that specifically bind with cell surface receptors play an important role in accurate disease diagnostics [22]. In conventional SELEX, cell surface proteins are isolated and purified from living cells, which can be lethal to their naive conformations and functions. As a result, the screened aptamers often failed in practical applications. To address this issue, cell-SELEX technology was established to generate aptamers against known or unknown target proteins on cell membranes [23]. Living cells, instead of isolated proteins, are used in cell-SELEX to maintain the original state of the displayed membrane proteins. The naïve proteins thereby enable selection of robust aptamers for biomedical applications [24]. One example is the selection of the CTLA-4 receptor-specific aptamer by Huang et al. [25]. In their study, CTLA-4 expressing HEK293T cell line was employed for positive selection. After 12 rounds of selection, the enriched DNA sequence information was analyzed and processed (Figure 1A). The identified aptamer showed high binding affinity toward CTLA-4 and could efficiently inhibit tumor growth.
However, conventional SELEX is a time-consuming and laborious job. Normally, selecting an ideal aptamer can take the server weeks or even months with a low success rate, severely limiting the development and application of aptamers [26]. Over the past decades, with the assistance of high-throughput bioinformatic analysis techniques and material sciences, multiple new methods have been reported, which have made the selection process more convenient, rapid, and efficient. Microfluidic SELEX [27], capillary electrophoresis–SELEX [28], and magnetic bead-based SELEX [29] were established to vastly reduce the selection time and cost. For example, Yang and Bowser successfully screened high affinity DNA aptamers after only three selection cycles, based on CE-SELEX [30]. Very recently, Chang et al. devised a high-dimensional microfluidic method to screen aptamers with quantitatively defined binding affinities in only a single selection round, called Pro-SELEX (Figure 1B) [31]. After particle display, magnetic particle sorting, high-dimensional feature selection and high-content bioinformatics, a large quantity of human myeloperoxidase-specific aptamers, with predefined binding affinities, were successfully identified. Recently, Saito’s team also obtained PC-9 cell-specific aptamer candidates via single-round selection (Figure 1C) [32]. Further, for the purpose of enhancing the binding interactions between oligonucleotides and targets, researchers are experimenting with selection of chemically-modified nucleotides. By replacing nature thymine with modified uridine at the 5′-end, Janjic’s team have developed slow off-rate modified aptamers (SOMAmers), which exhibit superior binding affinity against target proteins. SOMAmers are now commercially available and serve as a powerful and versatile platform for the analysis of thousands of protein biomarkers for disease diagnosis [33].
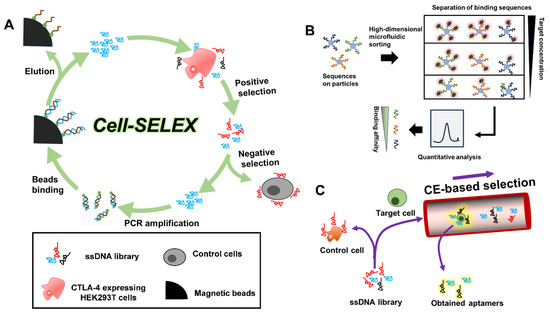
Figure 1.
(A) Isolation of CTLA-4-specific aptamers by cell-SELEX [25]. (B) Pro-SELEX enabled rapid selection of aptamers with programmable affinity [31]. (C) Single-round selection of PC-9 DNA aptamer by polymer-enhanced capillary transient isotachophoresis [32].
3. AptIDCs for Enhanced Cancer Immunotherapy
Nowadays, thanks to the constant efforts of researchers, there is increasing identification of aptamers to serve as effective immunomodulatory tools in boosting the efficacy of cancer immunotherapy. Based on structural and compositional differences, AptIDCs can be classified into the following four types: (1) aptamer agonists/antagonists, (2) multivalent bispecific aptamers, (3) aptamer–nanoparticle conjugates, and (4) aptamer-engineered immune cells. This section elaborates on the relevant engineering techniques associated with AptIDCs, and details their unique physiological functions and application advantages in cancer immunotherapy.
3.1. Aptamer Agonists/Antagonists
Escape from immune surveillance is an important cause of malignant tumor progression, which is associated with upregulated immune checkpoint molecules (e.g., PD-L1, PD-1, CTLA-4), downregulated costimulatory molecules (e.g., B7, CD28, 4-1BB), and inappropriate expression of anti-inflammatory cytokines (e.g., IL-10/-8, TNF-β, IFN-γ) in the tumor microenvironment [34,35]. Attempts have been made clinically to target specific molecules and reverse the ISM of cancers using monoclonal antibodies. As a superior alternative, increasing studies have reported that selected aptamers could be directly employed as agonists or antagonists to achieve equivalent, or even better, anti-tumor immunity.
3.1.1. Immune Checkpoints
Immune checkpoints (ICs) are inhibitory pathways that prevent any excessive immune response of the body and play important roles in maintaining self-tolerance, averting autoimmune reactions and minimizing tissue damage caused by continued immune cell activation. Unfortunately, the overexpressed ICs of tumors create an immunosuppressed microenvironment, helping the tumor to evade immune surveillance and gain a survival advantage [36]. Practical evidence reveals that immune checkpoint blockade (ICB) is one of the most valuable methods for treating tumors, especially in late stages. Among a range of identified ICs, the most successful ICB targets are two pathways, PD-1/PD-L1 and CTLA-4/CD80 (CD86) [37]. There is an urgent clinical need to develop inhibitors that can effectively block these pathways.
In 2015, Jean Gariépy et al. reported developing DNA aptamers to block the PD-1/PD-L1 pathway via specific binding with the murine extracellular domain of PD-1, obtained from an in vitro SELEX (Figure 2A) [12]. One of the sequences, named MP7, could inhibit PD-L1 by suppressing interleukin-2 (IL-2) secretion in primary T cells. In vivo data implied that PEGylated MP7 had a prolonged half-life, preserved the ability of PD-1/PD-L1 interaction and inhibited the growth of PD-L1+ colon carcinoma cells. Recently, Gao et al. isolated a PD4S aptamer targeting PD-1 with excellent affinity (dissociation constant, Kd = 10.3 nM) using cell-SELEX technology, which exerted excellent anti-tumor effects in the CT26 carcinoma model [38]. Accordingly, aptamers against PD-L1 have also continuously emerged. Lai and colleagues obtained an aptamer (AptPD-L1) that can block PD-1/PD-L1 interaction and promote in vitro lymphocyte proliferation [39]. Murine assays demonstrated that aptPD-L1 could enhance cytotoxic T cell infiltration and upregulated inflammatory cytokine secretion, thereby reversing the tumor immunosuppressive environment and inhibiting tumor growth. Furthermore, unnatural nucleic acids were explored to select anti-PD-L1 or PD-1 aptamers, with the aim of obtaining high-quality candidates. For example, employing a bead-based X-aptamer library with different amino acid functional groups modified at 5′-dU, Wang et al. obtained two aptamers, XA-PD1 and XA-PDL1, that could bind with PD-1 and PD-L1, respectively [40]. The Kd of XA-PD1 and XA-PDL1 aptamers were measured to be a nanomolar order of magnitude. In another study, Li and co-workers developed a threose nucleic acid (TNA) aptamer, N5, that can specifically bind to the PD-L1 of cancer cells with nanomolar affinity (Figure 2B). As a result of the unique 2′-3′ phosphodiester bond in the backbone structures of threose nucleic acids, TNA aptamer N5 has superior resistance to nucleases. Animal experiments showed that TNA aptamer N5 could selectively accumulate in tumors and have favorable anti-tumor immune effects [41].
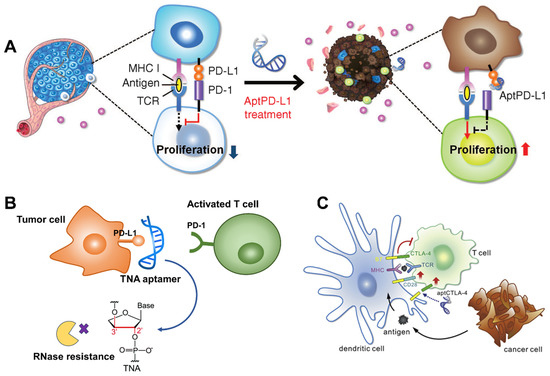
Figure 2.
(A) Anti-PD-L1 aptamer blocking PD-1/PD-L1 axis for ICB immunotherapy. Copyright 2015, Elsevier [12]. (B) Nuclease-resistant threose nucleic acid (TNA) aptamer targeting PD-L1 to avoid immune escape [41]. (C) CTLA-4 antagonizing DNA Aptamer for cancer immunotherapy. Copyright 2017, Elsevier [25].
As for the CTLA-4/CD80 (CD86) pathway, the first anti-CTLA-4 aptamers were screened by Gilboa’s group in 2003, using 2′-fluoropyrimidine modified RNA library against murine CTLA-4/Fc fusion protein in vitro [21]. The identified RNA aptamers had high binding affinity (Kd = 10 nM) and specificity toward CTLA-4 and inhibited its function in vitro. Moreover, the aptamers could self-assemble into tetrameric forms, which significantly enhanced the in vivo anti-tumor bioactivity. However, a high dosage of RNA aptamers (3–5 nmol/injection) was required in vivo, compared to αCTLA-4 antibodies (0.667 nmol/injection). This is due to the lower bioavailability of RNA aptamers in vivo. In 2017, Huang et al. reported identifying a CTLA-4-antagonizing DNA aptamer (aptCTLA-4, Kd = 11.8 nM) via the cell-based SELEX method (Figure 2C) [25]. Studies showed that aptCTLA-4 had good serum stability and could elicit similar tumor-suppress ability to the CTLA-4 monoclonal antibody (mAb), but with less side effects. Notably, the dosage of aptCTLA-4 (2 mg/kg) was five times smaller than that of CTLA-4 mAb (10 mg/kg), which might suggest stronger therapeutic efficacy for the aptamers. Although it was claimed that the aptCTLA-4 was stable after 24 h serum incubation, its circulation half-life in vivo was not investigated. In addition, both of these two CTLA-4-specific aptamers were delivered via intraperitoneal injection, which is not an ideal mode of administration. The reason might be that the RNA aptamers and unmodified DNA aptamers were vulnerable to nucleases in the blood. Therefore, developing CTLA-4-specific aptamers with high stability in blood is vital to clinical ICB therapy.
Besides PD-1, PD-L1 and CTLA-4, other IC molecules were also selected as targets to obtain aptamers to augment the efficacy of cancer immunotherapy. For instance, Gilboa’s group isolated the first antagonist TIM3 aptamer to block TIM3, another receptor expressed in exhausted T cells. After injection of TIM3 aptamers, T cells were activated, which, in turn, inhibited tumor growth in mice [42]. Similarly, in 2007, Pastor’s team reported a high-affinity aptamer (LAG3 apt 1) that was able to bind with LAG3-expressing lymphocytes and induce T cell activation [43].
3.1.2. Costimulatory Molecules
The full activation of T cells involves two signals. The engagement of costimulatory molecules (CMs) by costimulatory molecule receptors (CMR) on T cells is an indispensable “second signal”. However, cancer cells are characterized as having low expression of CMs on their surfaces, so T cells are usually in a state of anergy in tumor sites [44]. CMs are divided to two major families: the B7/CD28 family and the tumor necrosis factor (TNF)/tumor necrosis factor receptor (TNFR) family. Providing artificial costimulatory ligands to target costimulatory pathways is a valid method to activate T cell immunity. The first costimulatory aptamer agonist identification was pioneered by Gilboa’s group in 2008 [45]. They isolated an aptamer (M12-23), that could specifically bind with 4-1BB receptors on T cells, after 12 rounds of selection against 4-1BB–Fc. The 4-1BB receptor is a major CMR that belongs to the TNFR family and facilitates the proliferation and activation of T cells. Interestingly, it was found that only the multivalent form of the 4-1BB aptamer enhanced the proliferation of CD8+ T cells. The reason is that the initiation of costimulatory effects requires not only binding but also cross-linking to 4-1BB receptors. Inspired by dimeric mAbs, they elaborately designed a dimeric 4-1BB aptamer with an appropriate distance and orientation to adapt the cross-link with 4-1BB receptors. The in vitro results showed that the constructed dimeric 4-1BB aptamers could specifically bind to 4-1BB+ cell lines and successfully co-stimulate CD8+ T cells. Remarkable tumor regression was also observed in P815 tumor-bearing mice models, comparable to that of anti-4-1BB Abs. Thereafter, the Gilboa’s group continued to use this 4-1BB agonistic aptamer to combine with other treatments and achieved better synergic anti-tumor activity [46,47]. Subsequently, dimeric or multimeric aptamer agonists targeting other T cell CMRs were reported, such as OX40 [48,49], CD28 [13], and CD40 [50], which manifested comparable tumor immunity to rival Abs, both in vitro and in vivo.
It is noteworthily that all reported CM aptamers can only activate T cells in multivalent form, whereas monomers are inactive. This is because CM receptors require not only specific binding, but also cross-linking to initiate the downstream signaling cascade. Therefore, CM aptamers must be elaborately designed as multivalent structures, which should possess suitable distance and orientation to enable the cross-linking of CM receptors. Compared with other aptamer agonists/antagonists, developing effective CM aptamers is a more complicated job.
3.1.3. Immune Cytokines
Cytokines, as information molecules for immune cell communication, play a crucial role in the oncogenesis, development, and treatment of tumors [51]. In tumor microenvironments, tumor cells and tumor-associated immune cells secrete a series of immunosuppressive cytokines that weaken the body’s anti-tumor immunity. Among them, interleukin-10 (IL-10) is the most representative anti-inflammatory cytokine that mediates pathogenic infection and tumor immunosuppression [52,53,54]. Developing an effective IL-10 antagonist is a promising way to improve the efficacy of tumor immunotherapy. After 12 rounds of selection and high-throughput sequencing, Berezhnoy et al. isolated a 2′-fluoropyrimidines modified A1.2 aptamer that could target IL-10 receptors on the cell membrane and block IL-10 functions in vitro (Figure 3A(a)) [55]. Intravenous injection of A1.2 aptamers showed favorable suppression of CT26 tumor growth in animal models, and 20% tumor rejection was observed after repeated injections of either aptamer or Abs (Figure 3A(b)). CXCL12 (also known as stromal cell-derived factor, SDF-1) and its receptors, CXCR4 and CXCR7, are upregulated in multiple cancers and play important roles in promoting tumor growth, invasion, and metastasis, which is also a representative example [56,57,58]. Interfering with the CXCL12/CXCR4/CXCR7 pathways with inhibitors holds high potential to sensitize conventional cancer therapies. NOX-A12 is a mirror-image aptamer, called Spiegelmer, that binds to CXCL12 with high affinity and specificity [59]. Contrary to conventional nucleic acids, NOX-A12 is screened from the mirror-image (L)-oligonucleotide library to avoid nuclease degradation and hybridization with native nucleic acids (Figure 3B). Hoellenriegel et al. claimed that NOX-A12 was a potent CXCL12 inhibitor that interferes with the migration of chronic lymphocytic leukemia (CLL). Additionally, NOX-A12 can sensitize CLL cells to chemotherapy, as a promising complementary approach for treating CLL [60]. At present, NOX-A12 is in an ongoing phase II-a clinical trial against relapsed CLL [61].
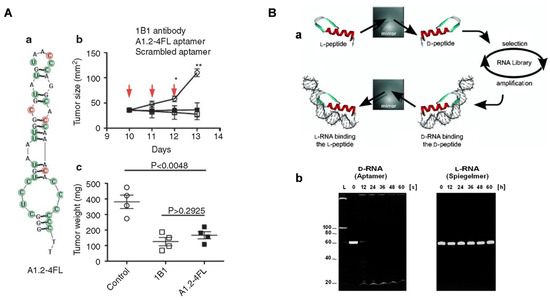
Figure 3.
(A) (a) The predicted secondary structure of IL-10 receptor blocking aptamer and its anti-cancer efficacy reflected by (b) tumor volume and (c) weight [55]. (B) (a) General protocol to produce Spiegelmer mirror-image aptamers (L-RNA) with (b) strong nuclease-resistance ability verified by gel electrophoresis assays. Copyright 2003, Wiley [59].
Aptamers screened as agonists or antagonists for immunotherapy are summarized in Table 1. Despite achieving encouraging results from aptamer agonist/antagonist-mediated cancer immunotherapy in the previous literature, there are still some drawbacks that hamper further clinical applications [22,62]. First, aptamers are generally unstable in vivo and have a shorter half-life (minutes to hours) than Abs, owing to the ubiquitous nuclease. Site-specific modification improves the resistance of aptamers to nuclease degradation to some extent, but increase the cost and may impair binding affinity toward targets. In addition, monospecific aptamer agonists/antagonists have no tumor-targeting ability, and the non-specific distribution might lead to severe immune adverse effects. Third, aptamer agonists/antagonists are readily excreted by the kidney due to their small size (6–30 kDa), which also leads to a short half-life in vivo. All these challenges are calling for novel robust AptIDCs with more powerful antitumor immunity.

Table 1.
Aptamers as agonists or antagonists for immunotherapy.
3.2. Multivalent Bispecific Aptamers
Bispecific aptamers (bsApts) comprise two functional domains: (1) One domain targets tumor antigens for specific tumor-targeting; (2) the other domain binds to immune-related proteins for immune activation. This version ensures specific drug delivery to tumor cells and reduced side effects. According to the valence and specificity of the two domains, bsApts can be categorized by the [m+n] rule, where m and n refer to the valence of the tumor-targeting aptamer domain and immune cell-targeting aptamer domain, respectively [65]. There are mainly three methods to synthesize bsApts: single strand folding, complementary chain hybridization, and DNA ligase-mediated circularization. Currently, several typical types of [m+n] (e.g., [1+1], [1+2], and [2+2]) bsApts have been reported [66].
3.2.1. The [1+1] bsApts
The first [1+1] immunoregulating bsApts were prepared by Boltz et al. [67]. In the research, CD16α-specific aptamers were selected to target natural killer (NK) cells (Figure 4A). Two [1+1] type bsApts, named bsA17 and bsA2, were constructed via designed ssDNA sequences, each of which contained one optimized CD16α-specific aptamer and an anti-c-Met tumor-specific aptamer. Both bsApts could specifically bind with tumor cells and NK cells. In addition, tumor cell lysis was achieved by co-incubating cancer cells and peripheral blood mononuclear cells with bsApts, which were as powerful as Abs. The authors synthesized a range of bsApts with different linker lengths (~0–217 Å) and tested their tumor killing ability. It is noteworthy that only linkers of 49–154 Å were suitable for effective NK cell-mediated tumor lysis, as a consequence of their similarity to the distance from the Fc-binding domain to the complementary determining region in the Abs (~65 Å), while longer linkers (~200 Å) decreased cytotoxicity. This finding emphasizes the need to carefully consider linker distances when designing bio-effective bsApts immunoregulators.
Alternatively, designing bsApts to recruit activated T cells is also an effective strategy to kill malignant cells. In 2020, Tan’s team developed a circular bispecific aptamer (cb-aptamer) that hybridized additional complementary sequences by linking the 3′- and 5′-ends between a T cell targeting aptamer (LD201t1) and a tumor-targeting aptamer (sgc 8) (Figure 4B) [68]. Due to the removal of free ends, that are susceptible to nuclease attack, circular bsApts showed remarkably increased resistance to serum exonucleases, compared to mono-aptamers [69]. The constructed cb-aptamers could bridge the proximity of T cells to tumor cells, but the tumor ablation effects were only initiated by adding extra commercial CD3/CD28 T cell activators. Very recently, Liu’s team engineered two cb-aptamers (PD-L1/CTLA-4 bsApt, CD16/PD-L1 bsApt) for tumor-targeting and cancer ICB (Figure 4C) [70,71]]. In another study, Sun et al. isolated an anti-PD-L1 aptamer, Ap3, that could selectively target the PD-L1 protein on tumor cells and block the PD-1/PD-L1 immune checkpoint [72]. Then, they integrated Ap3 with an anti-PD1 aptamer to prepare Ap3–7c bsApts, which possessed strong binding affinities towards CD8+ T cells and B16F10 tumor cells and could recruit T cells around tumor cells. The Ap3–7c bsApts were further chemically modified with a dibenzocyclooctyne (DBCO) moiety, termed D-Ap3-7c. After specific recognition of target molecules, D-Ap3-7c, bio-orthogonally link with target proteins, called “recognition-then-conjugation”. Consequently, bsApts were proved to increase the stability of target binding and induce effective tumor ICB therapy (Figure 4D). However, there was no comparison of the in vivo ICB efficacy of Ap3–7c and D-Ap3-7c in this study, as well as a lack of in vitro control cells.
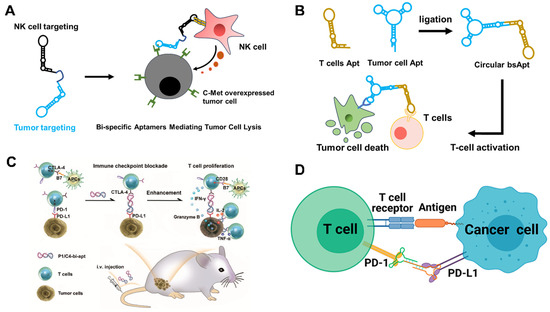
Figure 4.
(A) BsApt-associated tumor apoptosis by NK cell [67]. (B) Circular bsApt-activated T cell immunotherapy via PD-1/PD-L1 blockage [68]. (C) Targeted tumor immunotherapy by multifunctional CTLA-4/PD-L1 bsApt. Copyright 2021, Royal Society of Chemistry [70]. (D) BsApt-based “recognition-then-conjugation” strategy for enhanced PD-1/PD-L1 blockage immunotherapy. Copyright 2022, American Chemical Society [72].
3.2.2. The [1+2] bsApts
So far, all reported [1+2] bsApts were designed to target CMR. Obviously, valence “2” means the required dimeric aptamer domain for CMR binding. Pastor et al. proposed a [1+2] bsApts for immunotherapy in 2011. Specifically, one PMSA-specific tumor-targeting aptamer was hybridized with two 4-1BB agonistic aptamers (dimeric forms) (Figure 5A). Such [1+2] bsApts could selectively accumulate in human PSMA-expressed CT26 cells, and the dimeric 4-1BB agonistic aptamer module indued CD8+ T cell proliferation, thereby triggering anti-tumor immunity [73]. Similarly, another two [1+2] bsApts, [1 (anti-VEGF aptamer) + 2 (4-1BB agonistic aptamers)] [74] and [1 (anti-OPN tumor targeting aptamer) + 2 [4-1BB agonistic aptamers] [75], were also reported as activating the anti-tumor immune response. Another representative example, CD28, has now been recognized as an important CMR for promoting proliferation of effector T cells. Soldevilla et al. engineered a [1+2] MRP1-CD28 bsApts that could target MRP1-expressing tumors and deliver CD28 costimulatory ligands to T cells (Figure 5B(a)) [76]. Then, the MRP1-CD28 bsApts were immobilized on the surface of irradiated melanoma tumor cells, via aptamer-facilitated specific binding with MRP1 proteins. Accordingly, a whole-cell vaccine, named CD28Aptvax, was generated (Figure 5B(b)). After subcutaneous injection of CD28Aptvax, both T cell proliferation and IFN-γ levels were significantly enhanced in tumor-bearing mice, and the tumor growth was noticeably inhibited.
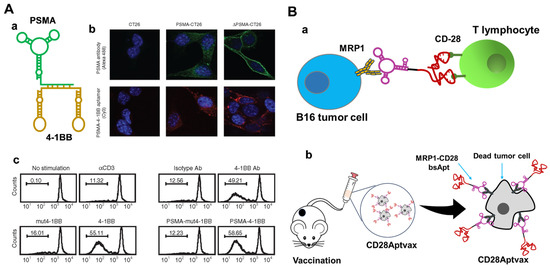
Figure 5.
(A) PSMA-41BB [1+2] bsApt for cancer immunotherapy. (a) Structure diagram of [1+2] bsApt, (b) specific tumor targeting and (c) indued CD8+ T cell proliferation by [1+2] bsApt. Copyright 2011, Cell Press [73]. (B) MRP1-CD28 [1+2] bsApt against drug-resistant melanoma cancer stem cells. (a) MRP1-CD28 [1+2] bsApt-mediated specific cancer immunotherapy, (b) irradiated B16-MRP1 coated with MRP1-CD28 bsApts to generate CD28Aptvax for improved anti-tumor immunity [76].
3.2.3. The [2+2] bsApts
In [2+2] bsApts, each functional domain has two aptamers, which tend to enhance the binding affinity for both tumor cells and immune cells. For instance, Li et al. recently described a tetravalent [2+2] bsApts, termed BBiApt, which contained two anti-MUC1 aptamers and two anti-CD16 aptamers [77]. MUC1 is a high-potential tumor cell surface biomarker that assists in specific drug delivery, whereas CD16 is expressed on several immune cells (e.g., NK cells). The BBiApt is a 420 nt-long ssDNA with three 60 nt ssDNA spacers. Flow cytometry assays demonstrated that BBiApt showed stronger binding affinity to target cells than single MUC1 and CD16 aptamers. BBiApts could mediate effective recruitment of CD16+ immune cells to MUC1+ tumor cells and, subsequently, exert cytotoxicity. Theoretically, [2+2] bsApts are more efficient immunomodulators than the [1+1] and [1+2] types, because of the increased valence of both the tumor-targeting domain and the immune cell-binding domain. Contradictorily, the application of [2+2] bsApts for cancer therapy is rarely reported. This is due to the fact that in [2+2] bsApts, four aptamers need to be assembled into a single structure. Meanwhile, the biological function of each aptamer should also be maintained, thus making sequence engineering a tough job.
Engineered bsApts overcome some serious problems that are often encountered in traditional monovalent aptamer agonists/antagonists. First, benefiting from structural advantages, bsApts, especially circular bsApts, have improved nuclease resistance and prolonged serum half-life time (hours to days). Moreover, the tumor-targeting and immunoregulation abilities of bsApts are enhanced by multivalent assembly, which is conducive to alleviating side effects and improving anti-tumor immunity. However, some points need to be further studied. Linkers/spaces are used to connect aptamer functional domains of bsApts. Through adjusting the distance and positions, linkers are also determinative factors affecting the biological functions for each aptamer. Yet, the understanding of how the composition, flexibility and length of linkers/spacers affect the quality of bsApts is still insufficient. In addition, whether increasing aptamer valency in bsApts can always positively affect the anti-tumor immunity is doubtful. For example, Liu et al. found that increasing the multivalency of bsApts (>4) decreased the cell–cell interactions, implying that blindly increasing the valency of bsApts might be detrimental to binding affinity [78]. Furthermore, bsApts with increased avidity also enhance the affinity towards healthy cells with low expression of target proteins, resulting in off-tumor effects and deleterious cytotoxicity to normal tissues. Thus, when designing bsApts, the aptamer valency, binding affinity, and therapeutic index should be fully investigated and compared to obtain the best-of-breed one.
3.3. Aptamer–Nanoparticle Conjugates (AptNCs)
Nanoparticles, such as liposomes and hydrogels, have opened up new opportunities for tumor therapy, due to their unique properties of good biosafety, high reactivity, efficient drug loading, and improved tumor accumulation [79,80]. Aptamers can be used not only as stand-alone immune therapeutic agents, but also in preparing aptamer–nanoparticle conjugates (AptNCs) by coupling with nanomaterials, via easy chemical linkage or nucleic acid-mediated interface interaction strategies, to enhance the efficacy of immunotherapy. In this section, several representative AptNCs are introduced and their superiorities in targeted drug delivery, immunosuppressive tumor microenvironment remodeling, and enhanced immunotherapy efficacy are emphasized.
3.3.1. Aptamer–Liposome Conjugates
Liposomes are lipid vesicles with a hydrophobic bilayer and aqueous core, which can be employed to load a variety of therapeutic agents. With the advantages of high biocompatibility, favorable stability and excellent drug loading capacity, liposomes are currently the most successful drug delivery nanoparticles [81,82]. Nevertheless, conventional liposomal formulations usually lack specific tumor targeting ability, which leads to compromised therapeutic efficacy and unwanted toxicity to healthy cells. Fortunately, aptamer-modified liposomes with targeted drug delivery capabilities help to improve anti-tumor effects and reduce side effects [83,84]. The attachment of aptamers to liposomes is mainly achieved in the following three ways: direction conjugation strategy, pre-conjugation strategy and post-insertion strategy [85]. In this section, aptamer–liposome conjugates for immunotherapy are presented and categorized, based on these three modification methods.
Direct Conjugation Strategy
Owing to the easy chemical modification of both aptamers and lipids, targeted aptamer ligands can be easily conjugated to the surface of liposomes by means of a range of chemical reactions, including amidation, esterification, and sulfhydryl-mediated Michael Addition Reaction, as well as click reaction [86]. For example, Lu’s group developed aptamer-decorated liposome-modified immuno-activated T cells, in order to improve the targeting anti-tumor immunity of CD8+ T cells (Figure 6A) [87]. Human CD8+ T were isolated from peripheral blood and then the PD-1 gene was knocked out to liberate immunosuppression. Afterwards, PD-1-T cells were further activated by DC/tumor fusion cells to obtain proliferation-promoting tumor-specific cytotoxic T cells (FC/PD-1-CTLs). The liposomes were modified by maleimide/sulfhydryl chemistry using anti-endoglin aptamers (hEnd-Apt) to acquire specific binding ability against HepG2 cells, while CD3 mAb was conjugated to specifically bind FC/PD-1-CTLs. In vivo tumor-xenograft mice assays demonstrated that the hEnd-Apt/CD3-Lipo-modified FC/PD-1-CTLs showed improved anti-tumor ability and survival rate, by increasing FC/PD-1-CTL infiltration in tumor tissue and elevating systemic immuno-activated cytokines levels.
Pre-Conjugation Strategy
In the pre-conjugation strategy, aptamer–lipid conjugates are mixed with other lipids during the liposome preparation process, which allows for a one-step formulation procedure. For instance, Hong and coworkers reported an aptamer-integrated bispecific α-Gal liposome, by means of a hydration lipid film in the presence of Apt-cholesterol conjugates [88]. AS1411 aptamers contribute to specifically target MCF-7 breast cancer cells, while α-Gal can be specifically recognized by anti-Gal Abs to induce antibody-dependent cell-mediated cytotoxicity (ADCC). Under simulated tumor environments, AS1411-Gal-modified liposomes resulted in an obvious and specific cell lysis effect compared to plain liposomes. Polymorphonuclear myeloid-derived suppressor cells (PMN-MDSCs) are a class of immunosuppressive immature myeloid cells that inhibit the proliferation and function of CD8+ T cells, which benefits the tumor cell progression [89]. To address this issue, Liu et al. screened a DNA aptamer, named T1, that could specifically bind with both MDA-MB-231 breast cancer cells and MDSCs [90]. T1 aptamers. modified with thiol at 5′-end. reacted with DSPE-PEG-Mal to generate aptamer–lipid conjugates, followed by integration into liposomes via the hydration process. Then, the T1-functionlized dual-targeting liposomes were encapsulated with DOX, which induced enhanced apoptosis of breast cancer cells and PMN-MDSCs, as well as increased amounts of tumor-infiltrating cytotoxic T cells (Figure 6B).
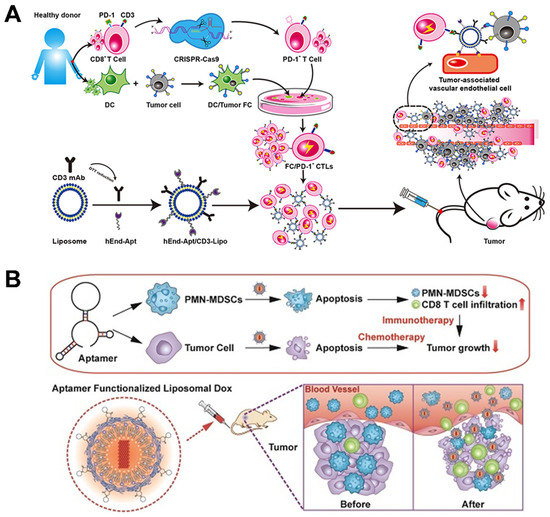
Figure 6.
(A) Endoglin-specific aptamer-modified liposome assembled with PD−1- T cells for improved immunity. Copyright 2021, Dove Medical Press [87]. (B) Dual PMN-MDSC/cancer cell-targeting liposome loaded with DOX to increase infiltration of cytotoxic T cells of breast cancer. Copyright 2018, Ivyspring International Publisher [90].
Post-Insertion Strategy
The post-insertion strategy was originally applied in the preparation of antibody-coupled liposomes and, thereafter, in aptamer modification [91]. Aptamers are first chemically coupled with lipids to synthesize aptamer–lipid conjugates, and are then inserted into bilayers by means of incubation with the as-prepared liposomes. This method is simple, flexible, and free of inorganic solvents that might do harm to aptamers and liposome formulations [92]. Kim et al. used the post-insertion strategy to fabricate a dual-aptamer modified liposome for immunogenic chemotherapy and ICB therapy that reversed tumor immunosuppression (Figure 7) [93]. Firstly, CD44- and PD-L1-specific aptamers were chemically linked to maleimide group micelles. Next, dual aptamer–micelle conjugates were inserted into liposomes via incubation at 60 °C, and loaded with both DOX and indoleamine 2,3-dioxygenase-1 (IDO1) siRNA, simultaneously. Cell binding analysis and fluorescence imaging assays showed that the constructed aptamer-modified liposomes (Aptm[DOX/IDO1]) could be selectively delivered to targeted tumor cells. In addition, the PD-1/PD-L1 axis was blocked to suppress immune inhibition, while the release of DOX and IDO1 siRNA jointly induced immunogenic cell death (ICD) to reverse the immunosuppressive tumor microenvironment. The analysis of tumor tissue extracted from tumor-bearing mice revealed an increase of CD8+ cytotoxic T cells and a decrease of regulatory T cell recruitment. Furthermore, they claimed that Aptm[DOX/IDO1] also inhibited lung metastasis in a mice model, demonstrating the activation of systemic anti-tumor immunity.
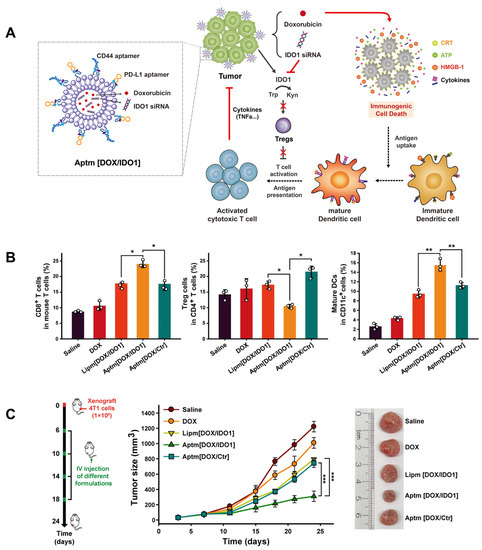
Figure 7.
(A) CD-44/PD-L1 bispecific liposome encapsulating DOX for immunogenic chemotherapy and reversion of immunosuppressive tumor microenvironment. (B) In vivo tumor inhibition effects of dual-specific Aptm[DOX/IDO1]. (C) Tumor-infiltrating immune cell analysis after different drug administrations. (* p < 0.05, ** p < 0.01, *** p < 0.001) Copyright 2022, Elsevier [93].
Direct conjugation of aptamers on pre-formulated liposomes is the most commonly used method to prepare aptamer-functionalized liposomes. Chemical reactions require organic solvents and the catalysts may be harmful to pre-encapsulated drugs. Pre-conjugation and post-insertion approaches, in contrast, are done with pre-synthesized aptamer–lipid conjugates, involving a chemical reaction-free process. Hence, these two methods are safer and have minimal side effects on liposome formulations. However, weaknesses still exist. In pre-conjugation, after co-hydration, aptamers appeared not only on the surface of liposomes, but also on their interiors. The interior aptamers cannot function as cell-targeting ligands, which reduces the efficiency of the aptamers used. The pre-insertion strategy enables all the conjugated aptamers on the surface of liposomes, but needs further purification steps to remove un-anchored aptamer–lipid micelles.
3.3.2. Aptamer–Hydrogel Conjugates
Hydrogels are currently popular soft biomaterials for disease therapy because of having the advantages of good compatibility, tunable mechanical properties, versatile cargo loading ability, and sustained drug release [94]. Natural polymers, like nucleic acids and alginate, are the preferred materials for fabricating because of their environmental friendliness and abundant sources [95]. Through integration with aptamers, natural polymer-based aptamer–hydrogel conjugates exhibit outstanding performance in targeted drug delivery and stimuli-responsive cargo release.
DNA/RNA Hydrogels
DNA/RNA are extremely hydrophilic molecules and can absorb large quantities of water. Together with the characteristics of high programmability, intrinsic biodegradability, and stimuli-responsive structural change (e.g., assembly or disassembly), DNA/RNA are becoming one of the most promising materials for the design of hydrogels [96,97,98]. Since aptamers are essentially nucleic acid molecules, they can be easily integrated with DNA/RNA gels to establish nucleic acid hydrogel-based immune therapies.
Recently, Lee et al. reported a Cas9-responsive DNA polyaptamer hydrogel for the blockage of the PD-1 immune checkpoint (Figure 8A) [99]. The PD-1 DNA hydrogels (PAH) containing PD-1 aptamers and sgRNA-targeting sequences were fabricated using the rolling-circle amplification (RCA) strategy. After the addition of Cas9/sgRNA, the sgRNA-targeting sequences were specifically cleaved, leading to the release of PD-1 aptamers and, thus, the activation of T cells. The levels of tumor-infiltrating cytotoxic CD4+ (12.9%) and CD8+ (5.8%) T cells increased remarkably after intratumoral injection of PAH with Cas9/sgRNA, compared to the control group (CD4+ 0.3%, CD8+ 0.4%). The anti-tumor effects and survival rates improved, in comparison to free PD-1 aptamers. An animal imaging assay indicated that PAH can stay much longer at the tumor site than free aptamers, facilitating long-term immune activation against tumors. In other research, Wei et al. designed a multifunctional DNA nanohydrogel for combined immunochemotherapy by means of a one-step self-assembly strategy (Figure 8B) [100]. This hydrogel construct consists of four key functional elements: (1) anti-MUC1 aptamers for tumor targeting; (2) unmethylated cytosine–phosphate–guanine (CpG) motif as immunoadjuvants; (3) physically intercalated base pairs by DOX; (4) cytosine (C)-rich sequences (I-motif) to initiate pH-response disassembly. Guided by anti-MUC1 aptamers, the multifunctional hydrogels could be specifically internalized into malignant cells and undergo disassembly in late lysosome (pH 5.0–6.0), resulting in efficient release of DOX and the CpG motif. On the one hand, DOX facilitated the apoptosis of tumor cells. On the other hand, the CpG motif was an efficient agonist of toll-like receptor 9 (TLR9), and the exposure of CpG could activate a variety of immune cells and promote the secretion of pro-inflammatory cytokines; thus, eliciting a strong immune response. These aptamer-functionalized CpG-I-DOX hydrogels were capable of inducing high levels of tumor necrosis factor (TNF-α) in serum and obvious tumor regression of MCF-7 tumor xenograft mice. Besides DNA hydrogels, RNA hydrogels are also extensively employed to improve the therapeutical efficacy of cancer. Li’s team developed an RNA hydrogel nano-vaccine against triple-negative breast cancer (Figure 8C) [101]. A single-stranded DNA template, containing antisense miRNA-182/-205, was first designed, and, then, transcribed to poly-RNA copies by rolling circle transcription. Next, CpG–cholesterol and aptamer–cholesterol conjugates were integrated to the RNA transcripts via hydrophobic interactions, and cholesterol also helped to compact the micro-hydrogels into small spaces. Lastly, positively charged MnO2@Ce6 nanoparticles were loaded on the highly negative RNA hydrogel via electrostatic interactions to generate hydrogel/DOX-MnO2@Ce6 (HDMC). Due to the conjugated LXL-1 aptamer, HDMC can be selectively delivered to target MDA-MB-231 cells (human triple-negative breast cancer cell line). The authors claimed that an endogenous Dicer enzyme could cleave HDMC to release therapeutic agents. MnO2@Ce6 could catalyze O2 production from upregulated H2O2 in the tumor microenvironment, which relieves tumor hypoxia and supplies “fuel” for photodynamic therapy. Combined with CpG-induced immune activation and DOX-mediated chemotherapy, HDMC exhibited an outstanding tumor ablation effect.
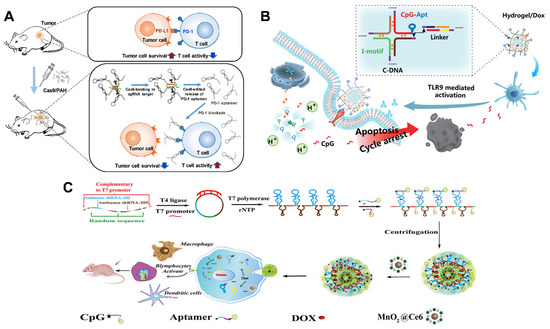
Figure 8.
(A) Cas9-responsive DNA polyaptamer hydrogel of PD-1/PDL-1 immune checkpoint blockage. Copyright 2019, Elsevier [99]. (B) Multifunctional DNA nanohydrogel loading CpG motif for combined immuno-chemo therapy. Copyright 2019, American Chemical Society [100]. (C) RNA hydrogel Nano-vaccine assembled with MnO2 activating immunity for the treatment of triple-negative breast cancers. Copyright 2021, Frontiers [101].
Alginate Hydrogels
Alginate (ALG) is a natural polymer consisting of α-L-Guluronate and β-D-Mannuronate monomers, extracted from brown seaweeds. Due to its low cost, high hydrophilicity, abundant functional groups (-OH, -COOH), and biodegradability, alginate-based hydrogels have received intensive focus in biomedical field [102]. Recently, Liu’s team reported an ATP-responsive ALG hydrogel for enhancing anti-tumor immunity (Figure 9) [103]. Briefly, ALG was linked to ATP-specific aptamers via EDC/NHS chemistry, followed by hybridization with CpG immuno-adjuvant. Immunogenic cell death (ICD) was induced after a low dose of oxaliplatin or X-ray irradiation, which caused the release of intracellular ATP molecules in the tumor microenvironment. The released ATP, consequently, competitively bound to ATP-specific aptamers to trigger CpG release. This smart ALG–aptamer/CPG hydrogel synergistic immunotherapy approach realized dramatic tumor elimination, as well as the inhibition of distant tumor metastases. Additionally, the authors collected the peripheral blood from cured mice to identify memory T cells, which are important immunological cells that suppress tumor recurrence. More memory T cells were observed in peripheral blood compared with untreated groups. Cured mice also proved to acquire immune resistance toward tumor re-challenge. All this evidence indicated that the smart ALG–aptamer/CPG hydrogel holds huge potential for clinical translation.
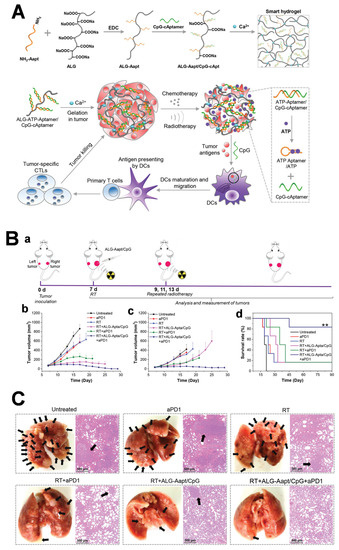
Figure 9.
(A) ATP-responsive alginate hydrogel combined with radiotherapy to achieve immune activation and tumor ablation. (B) Distant tumor elimination via radiotherapy/ATP-responsive CpG release/ICB synergetic therapy. (a) Schematic illustration of the experiment design; (b) Tumor growth curves of (b) the primary and (c) distal tumors (** p < 0.01); (d) Survival rates of CT26-tumor-bearing mice post different treatments. (C) Combined therapy inhibiting lung metastases of 4T1 cell orthotopic tumor model. Copyright 2021, Wiley [103].
3.4. Aptamer-Engineered Immune Cells
Cell-based immunotherapy is considered one of the methods having most potential for cancer therapies, because of the merits of low immunogenicity, prolonged circulation, innate targeting ability, and magic therapeutic outcomes. Precise manipulation of cell–cell interactions during specific immunization can enhance the anti-tumor immunity of cell-based immunotherapy. Aptamers are recognized as powerful cell-surface engineering tools for mediating intercellular interactions. To date, aptamers have been employed to conjugate with different types of cells, such as T cells and NK cells. These aptamer–cell assemblies are reported to have targeted cell–cell recognition, enhanced anti-tumor immunity, and reduced side effects, paving a new way for cell-based immunotherapy.
3.4.1. Aptamer–NK Cell Assemblies
NK cells are cytotoxic lymphocytes that can directly kill malignant cells by secreting cytolytic granules or expressing TNF-related apoptosis inducing ligands without pre-sensitization. Hence, NK cells are the first line to fight the occurrence and progression of cancer. Although having high value in adaptive cancer immunotherapy, the immunotherapeutic potential of NK cells is weakened due to a lack of tumor cell-specific receptors. Similar to chimeric antigen receptor (CAR)-T cell technology, CAR–NK cells are designed to realize specific binding to tumor cells, and showed inspiring results in clinical studies. However, genetic engineering strategies are laborious and costly, not to mention potentially posing long-term safety risks to patients. Fortunately, lipid–aptamer conjugates are emerging as convenient and safe tools for efficient cell membrane labeling, which has already been applied to NK cells for the acquisition of tumor-targeting capability. The first aptamer-modified tumor-specific NK cells were reported by Tan’s group in 2012 (Figure 10A) [104]. The authors used two leukemia cell-specific DNA aptamers, named sgc8 and TD05, respectively, to prepare diacyllipid–DNA aptamer conjugates. These conjugates contained the following three elements: (1) an aptamer head for tumor targeting; (2) a PEG linker for protecting aptamer conformation; (3) a diacyl lipid tail for hydrophobically mediated anchoring on the cell membrane. After membrane insertion, sgc8 aptamer-modified cells could specifically bind CCRF–CEM cells, while the TD05 aptamer modified selectively recognized Ramos cells, and spontaneous cell–cell aggregation was observed. These results indicated that membrane-anchored aptamers still maintained their targeting ability and could facilitate specific cell–cell interactions. Furthermore, the authors fabricated KK1B10 aptamer-modified NK cells to test their specific killing effects on K562 cells (the human chronic myeloid leukemia cell line). In vitro cell viability assays showed that KK1B10 aptamer-modified NK cells caused ~30% death of K562 cells, which was 50% more than in the unmodified NK cells. In addition, the enhanced specific killing capability of KK1B10-NK cells was also preserved in the presence of excess non-targeting Ramos cells. This pioneering work has inspired more scholars to develop aptamer-modified NK cells for targeted immunotherapy. In 2019, Yang et al. selected an efficient aptamer–lipid conjugate to produce aptamer-engineered NK cells (ApEn-NK) for targeted adaptive immunotherapy (Figure 10B(a)) [105]. They conjugated anti-CD-30 aptamers to a series of different lipidic anchors, including single-C18 chain, double-C18 chain, cholesterol, and vitamin E. NK cell-labeling fluorescence imaging and flow cytometry experiments implied that aptamer–double-C18 conjugates (Apt-2xC18) exhibited rapid anchoring (~1 h) and long membrane retention (>10 h), making them the best candidates for membrane anchoring (Figure 10B(b,c)). The fabricated ApEn-NK could specifically bind with CD30+ lymphoma cells and elicit improved anti-tumor immunity, compared to natural NK cells. Finally, they used NK cells obtained from healthy donors to generate ApEn–NK to evaluate the immunotherapeutic potential. In comparison with parental NK cells, ApEn–NK were more efficient in apoptosis of target lymphoma cells, demonstrating their potential for clinical application.
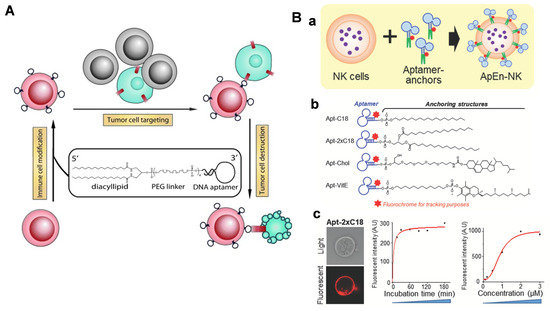
Figure 10.
(A) Aptamer-modified NK cells to realize targeted immune cytotoxicity toward leukemia cells by diacyllipid–DNA aptamer conjugate-mediated membrane insertion. Copyright 2012, Wiley [104]. (B) Optimal Apt- 2xC18 amphiphiles with efficient and stable modification of NK cells for robust tumor apoptosis. (a) Scheme of engineering of ApEn-NK. (b) Aptamer-lipid conjugates with four different lipidic tails: Apt-2xC18, Apt-Chol, Apt-C18, and Apt-VitE. (c) Profile of Apt-2xC18 by confocal imaging and flow cytometry. Copyright 2019, Wiley [105].
3.4.2. Tunable Aptamer–Antibody Nano-Assembly for Precise T Cell Immunotherapy
T cell-based immunotherapies are currently showing tremendous potential in the treatment of malignant cancers, with three approved CAR-T cell therapies having been approved by 2020 [106]. Although CAR-T cell technology renders improved anti-tumor immunity, tumor heterogenicity and ISM usually lead to off-target toxicity and insufficient T cell activation. To address this issue, Wei and Nie’s group recently collaborated to design a chimeric antibody–nucleic acid T cell engager (CAN-TE) for precise triple-antigen tumor recognition and selective T cell activation (Figure 11A) [107]. The constructed CAN-TE consists of two moieties: (1) a DNA assembled “logic circuit” for precise tumor antigen recognition; (2) a conjugated CD3-binding scFv for the binding and activation of TCR co-receptor CD3. Based on the dynamic AND-logic DNA circuit, the CAN-TE could precisely recognize target tumor cells that displayed multiple antigens (EpCAM, MUC1, and Met), avoiding undesired off-target T cell killing effects (Figure 11B). Moreover, to regulate the tumor cell avidity and T cell immune response, the authors modified the DNA nanostructure to produce an avidity-controlled CAN-TE (v2.0β) (Figure 11C). After installing multivalent antigen-targeting aptamer on the CAN-TE, the targeting avidity of T cells increased remarkably; subsequently, achieving effective immune tumor elimination in vivo.
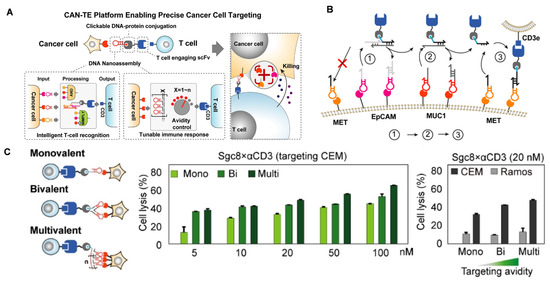
Figure 11.
(A) CAN-TE platform for precise tumor-antigen recognition and tunable T cell activation. (B) Cell-specific recognition by the programmed sequential actuations of the AND-logic CAN-TE. (C) CAN-TE platform (v2.0β) incorporating multivalent tumor-targeting ligands to tune avidity-dependent T cell killing response toward targeted CEM cells. Copyright 2022, Wiley [107].
Increasing studies suggest that aptamers are emerging as versatile tools for manipulating cell–cell communications, offering great promise in improving the efficacy of immune cell-based cancer therapy. Yet, some concerns still need to be taken into consideration. For example, by means of lipid-mediated hydrophobic membrane insertion (a non-covalent strategy), aptamers may readily dissociate from the cell surface with prolonged circulation time in vivo. Secondly, aptamers originally displaying on the cell surface could also be internalized into the cell cytoplasm over time, thereby losing their intended physiological functions. More importantly, nucleases, which are ubiquitous in vivo, may degrade the exposed aptamers on the cell surface and weaken the eventual anti-tumor immunity.
4. Conclusions
Immunotherapy is regarded as an effectual treatment for a variety of malignant cancers, and has achieved great clinical success. However, the immunosuppressive microenvironment, expensive treatment costs and non-negligible adverse effects still hinder the advancement of immunotherapy. Aptamers are chemically synthesized single-stranded DNA or RNA sequences with high target binding affinity, cheapness and low immunogenicity. Due to the development of selection technologies, aptamers targeting immune-related proteins for immunomodulation are being increasingly discovered, and show promising potential in cancer treatment. Moreover, strict complementary base-pairing rules, flexible structure transformation and ready chemical modification endow aptamers with highly engineerable features. On this basis, different types of aptamer assemblies, such as bsApts and aptamer-functionalized nanomaterials, are being exploited as efficient tools for enhanced anti-tumor immunity. Furthermore, aptamers can also be utilized as versatile tools to manipulate cell–cell communications. Aptamer-engineered NK/T cell assembles have opened up new directions for cell-based immunotherapy, owing to the advantages of precise tumor-targeting, tunable immune cell activation, convenient operation and high biosafety. In summary, AptIDCs have tremendous potency in boosting immune efficacy, providing attractive opportunities for effective cancer immunotherapy.
However, despite the numerous merits that AptIDCs have shown in immunotherapy, there are still some challenges ahead for clinical transformation. For example, more comprehensive pharmacokinetics information, like half-life time in blood, organ distribution, metabolism and excretion, should be provided in subsequent studies to elucidate the fate of AptIDCs in vivo. In addition, the possible immunotoxicity also needs to be carefully evaluated, which has been observed in a recent phase-III clinical trial REG1 (anti-IX coagulation factor PEGylated aptamer). The progression of AptIDCs is closely related to real reliable aptamers that can perform stable biofunctions in complex physiological conditions. Although multiple aptamers have been claimed to be effective in the published literature, the publicly accepted ones are still a small minority. Joint efforts by interdisciplinary scholars are indispensable from initial selection to final bio-applications, as a truly robust aptamer needs broad acceptance. Nuclease-mediated degradation in vivo is always a headache for aptamers. Hence, developing artificial nucleic acids with strong resistance to natural nucleases is a hopeful solution. Among them, mirror-image aptamer, NOX-A12, is the candidate with most potential for clinical immunotherapy, due to the abilities of anti-nuclease degradation and non-hybridization with native nucleic acids. Aptamer-engineered NK cells and T cells also show advantages in cell-based tumor therapy, compared with conventional CAR-T/NK cells. However, robust anchors, capable of fast and stable membrane insertion, but avoiding cellular internalization, are still inadequate. In the future, with the rapid development of nanotechnology, nucleic acid chemistry, biotechnology and artificial intelligence, we believe that AptIDCs will embrace a bright future in cancer immunotherapy and, ultimately, benefit patients.
Author Contributions
Conceptualization and writing—original draft preparation, H.X.; writing—review and editing, L.L., X.L., H.J. and X.W. All authors have read and agreed to the published version of the manuscript.
Funding
This work is supported by the National Natural Science Foundation of China (82061148012, 82027806, 91753106, 92061121, 21974019), the National Key Research and Development Program of China (2017YFA0205300) and Primary Research & Development Plan of Jiangsu Province (BE2019716), and the ISF-NSFC Joint Research Program (grant No. 3258/20).
Data Availability Statement
Data is contained within the article.
Conflicts of Interest
The authors declare that they have no known competing financial interest or personal relationships that could have appeared to influence the work reported in this paper.
References
- Varadé, J.; Magadán, S.; González-Fernández, Á. Human immunology and immunotherapy: Main achievements and challenges. Cell. Mol. Immunol. 2021, 18, 805–828. [Google Scholar] [CrossRef]
- Kimiz-Gebologlu, I.; Gulce-Iz, S.; Biray-Avci, C. Monoclonal antibodies in cancer immunotherapy. Mol. Biol. Rep. 2018, 45, 2935–2940. [Google Scholar] [CrossRef]
- Lote, H.; Chau, I. Emerging HER2-directed therapeutic agents for gastric cancer in early phase clinical trials. Expert Opin. Investig. Drugs 2022, 31, 59–78. [Google Scholar] [CrossRef]
- Zhu, G.; Niu, G.; Chen, X. Aptamer-drug conjugates. Bioconjugate Chem. 2015, 26, 2186–2197. [Google Scholar] [CrossRef]
- Pastor, F. Aptamers: A new technological platform in cancer immunotherapy. Pharmaceuticals 2016, 9, 64. [Google Scholar] [CrossRef]
- Kalaora, S.; Nagler, A.; Wargo, J.A.; Samuels, Y. Mechanisms of immune activation and regulation: Lessons from melanoma. Nat. Rev. Cancer 2022, 22, 195–207. [Google Scholar] [CrossRef]
- Tuerk, C.; Gold, L. Systematic evolution of ligands by exponential enrichment: RNA ligands to bacteriophage T4 DNA polymerase. Science 1990, 249, 505–510. [Google Scholar] [CrossRef]
- Ellington, A.D.; Szostak, J.W. In vitro selection of RNA molecules that bind specific ligands. Nature 1990, 346, 818–822. [Google Scholar] [CrossRef]
- Xuan, W.; Peng, Y.; Deng, Z.; Peng, T.; Kuai, H.; Li, Y.; He, J.; Jin, C.; Liu, Y.; Wang, R.; et al. A basic insight into aptamer-drug conjugates (ApDCs). Biomaterials 2018, 182, 216–226. [Google Scholar] [CrossRef]
- Zhao, Z.; Wang, X. Investigations upon the bioconjugation-based construction technologies and applications of aptamer-drug conjugates. Chem. J. Chin. Univ. Chin. 2021, 42, 3367–3378. [Google Scholar]
- Xiang, D.; Zheng, C.; Zhou, S.-F.; Qiao, S.; Tran, P.H.L.; Pu, C.; Li, Y.; Kong, L.; Kouzani, A.Z.; Lin, J.; et al. Superior performance of aptamer in tumor penetration over antibody: Implication of aptamer-based theranostics in solid tumors. Theranostics 2015, 5, 1083–1097. [Google Scholar] [CrossRef]
- Prodeus, A.; Abdul-Wahid, A.; Fischer, N.W.; Huang, E.H.B.; Cydzik, M.; Gariépy, J. Targeting the PD-1/PD-L1 immune evasion axis with DNA aptamers as a novel therapeutic strategy for the treatment of disseminated cancers. Mol. Ther. Nucleic Acids 2015, 4, e237. [Google Scholar] [CrossRef]
- Pastor, F.; Soldevilla, M.M.; Villanueva, H.; Kolonias, D.; Inoges, S.; de Cerio, A.L.; Kandzia, R.; Klimyuk, V.; Gleba, Y.; Gilboa, E.; et al. CD28 aptamers as powerful immune response modulators. Mol. Ther. Nucleic Acids 2013, 2, e98. [Google Scholar] [CrossRef]
- Sung, H.J.; Choi, S.; Lee, J.W.; Ok, C.Y.; Bae, Y.S.; Kim, Y.H.; Lee, W.; Heo, K.; Kim, I.H. Inhibition of human neutrophil activity by an RNA aptamer bound to interleukin-8. Biomaterials 2014, 35, 578–589. [Google Scholar] [CrossRef]
- Xiong, H.; Liu, L.; Wang, Y.; Jiang, H.; Wang, X. Engineered aptamer-organic amphiphile self-assemblies for biomedical applications: Progress and challenges. Small 2022, 18, 2104341. [Google Scholar] [CrossRef]
- Tian, L.; Shao, M.; Gong, Y.; Chao, Y.; Wei, T.; Yang, K.; Chen, Q.; Liu, Z. Albumin-binding lipid-aptamer conjugates for cancer immunoimaging and immunotherapy. Sci. China Chem. 2022, 65, 574–583. [Google Scholar] [CrossRef]
- Gong, H.; Dai, Q.; Peng, P. Cell-Membrane-Anchored DNA Logic-Gated Nanoassemblies for In Situ Extracellular Bioimaging. ACS Appl. Mater. Interfaces 2022, 14, 43026–43034. [Google Scholar] [CrossRef]
- Song, W.; Hu, J.J.; Song, S.J.; Xu, Y.; Yang, H.; Yang, F.; Zhou, Y.; Yu, T.; Qiu, W.X. Aptamer-gold nanocage composite for photoactivated immunotherapy. Acs Appl. Mater. Interfaces 2022, 14, 42931–42939. [Google Scholar] [CrossRef]
- Geng, Z.; Wang, L.; Liu, K.; Liu, J.; Tan, W. Enhancing anti-PD-1 immunotherapy by nanomicelles self-assembled from multivalent aptamer drug conjugates. Angew. Chem. Int. Ed. 2021, 60, 15459–15465. [Google Scholar] [CrossRef]
- Sun, H.; Zhu, X.; Lu, P.Y.; Rosato, R.R.; Tan, W.; Zu, Y. Oligonucleotide aptamers: New tools for targeted cancer therapy. Mol. Ther. Nucleic Acids 2014, 3, e182. [Google Scholar] [CrossRef]
- Santulli-Marotto, S.; Nair, S.K.; Rusconi, C.; Sullenger, B.; Gilboa, E. Multivalent RNA aptamers that inhibit CTLA-4 and enhance tumor immunity. Cancer Res. 2003, 63, 7483–7489. [Google Scholar]
- Zhou, J.; Rossi, J. Aptamers as targeted therapeutics: Current potential and challenges. Nat. Rev. Drug Discov. 2017, 16, 181–202. [Google Scholar] [CrossRef]
- Li, L.; Xu, S.; Yan, H.; Li, X.; Yazd, H.S.; Li, X.; Huang, T.; Cui, C.; Jiang, J.; Tan, W. Nucleic acid aptamers for molecular diagnostics and therapeutics: Advances and perspectives. Angew. Chem. (Int. Ed. Engl.) 2021, 60, 2221–2231. [Google Scholar] [CrossRef]
- Xiong, H.; Yan, J.; Cai, S.; He, Q.; Peng, D.; Liu, Z.; Liu, Y. Cancer protein biomarker discovery based on nucleic acid aptamers. Int. J. Biol. Macromol. 2019, 132, 190–202. [Google Scholar] [CrossRef]
- Huang, B.T.; Lai, W.Y.; Chang, Y.C.; Wang, J.W.; Yeh, S.D.; Lin, E.P.; Yang, P.C. A CTLA-4 antagonizing DNA aptamer with antitumor effect. Mol. Ther. Nucleic Acids 2017, 8, 520–528. [Google Scholar] [CrossRef]
- Yan, J.; Xiong, H.; Cai, S.; Wen, N.; He, Q.; Liu, Y.; Peng, D.; Liu, Z. Advances in aptamer screening technologies. Talanta 2019, 200, 124–144. [Google Scholar] [CrossRef]
- Lai, H.C.; Wang, C.H.; Liou, T.M.; Lee, G.B. Influenza A virus-specific aptamers screened by using an integrated microfluidic system. Lab A Chip 2014, 14, 2002–2013. [Google Scholar] [CrossRef]
- Mosing, R.K.; Bowser, M.T. Isolating aptamers using capillary electrophoresis-SELEX (CE-SELEX). Methods Mol. Biol. 2009, 535, 33–43. [Google Scholar]
- Duan, N.; Gong, W.; Wu, S.; Wang, Z. An ssDNA library immobilized SELEX technique for selection of an aptamer against ractopamine. Anal. Chim. Acta 2017, 961, 100–105. [Google Scholar] [CrossRef]
- Yang, J.; Bowser, M.T. Capillary Electrophoresis–SELEX selection of catalytic DNA aptamers for a small-molecule porphyrin target. Anal. Chem. 2013, 85, 1525–1530. [Google Scholar] [CrossRef]
- Chang, D.; Wang, Z.; Flynn, C.D.; Mahmud, A.; Labib, M.; Wang, H.; Geraili, A.; Li, X.; Zhang, J.; Sargent, E.H.; et al. A high-dimensional microfluidic approach for selection of aptamers with programmable binding affinities. Nat. Chem. 2023, 15, 773–780. [Google Scholar] [CrossRef]
- Hirose, K.; Tsuchida, M.; Asakura, H.; Wakui, K.; Yoshimoto, K.; Iida, K.; Sato, M.; Shibukawa, M.; Suganuma, M.; Saito, S. A single-round selection of selective DNA aptamers for mammalian cells by polymer-enhanced capillary transient isotachophoresis. Analyst 2017, 142, 4030–4038. [Google Scholar] [CrossRef] [PubMed]
- Mehan, M.R.; Ostroff, R.; Wilcox, S.K.; Steele, F.; Schneider, D.; Jarvis, T.C.; Baird, G.S.; Gold, L.; Janjic, N. Highly multiplexed proteomic platform for biomarker discovery, diagnostics, and therapeutics. Adv. Exp. Med. Biol. 2013, 735, 283–300. [Google Scholar]
- Wang, D.R.; Wu, X.L.; Sun, Y.L. Therapeutic targets and biomarkers of tumor immunotherapy: Response versus non-response. Signal Transduct. Target. Ther. 2022, 7, 331. [Google Scholar]
- Lv, B.; Wang, Y.; Ma, D.; Cheng, W.; Liu, J.; Yong, T.; Chen, H.; Wang, C. Immunotherapy: Reshape the tumor immune microenvironment. Front. Immunol. 2022, 13, 844142. [Google Scholar] [CrossRef]
- Darvin, P.; Toor, S.M.; Sasidharan Nair, V.; Elkord, E. Immune checkpoint inhibitors: Recent progress and potential biomarkers. Exp. Mol. Med. 2018, 50, 1–11. [Google Scholar] [CrossRef]
- Kubli, S.P.; Berger, T.; Araujo, D.V.; Siu, L.L.; Mak, T.W. Beyond immune checkpoint blockade: Emerging immunological strategies. Nat. Rev. Drug Discov. 2021, 20, 899–919. [Google Scholar] [CrossRef]
- Gao, T.; Pei, R. Isolation of DNA aptamer targeting PD-1 with an antitumor immunotherapy effect. Acs Appl. Bio Mater. 2020, 3, 7080–7086. [Google Scholar] [CrossRef]
- Lai, W.Y.; Huang, B.T.; Wang, J.W.; Lin, P.Y.; Yang, P.C. A novel PD-L1-targeting antagonistic DNA aptamer with antitumor effects. Mol. Ther. Nucleic Acids 2016, 5, e397. [Google Scholar] [CrossRef]
- Wang, H.; Lam, C.H.; Li, X.; West, D.L.; Yang, X. Selection of PD1/PD-L1 X-aptamers. Biochimie 2018, 145, 125–130. [Google Scholar] [CrossRef]
- Li, X.; Li, Z.; Yu, H. Selection of threose nucleic acid aptamers to block PD-1/PD-L1 interaction for cancer immunotherapy. Chem. Commun. 2020, 56, 14653–14656. [Google Scholar] [CrossRef] [PubMed]
- Hervas-Stubbs, S.; Soldevilla, M.M.; Villanueva, H.; Mancheño, U.; Bendandi, M.; Pastor, F. Identification of TIM3 2’-fluoro oligonucleotide aptamer by HT-SELEX for cancer immunotherapy. Oncotarget 2016, 7, 4522–4530. [Google Scholar] [CrossRef] [PubMed]
- Soldevilla, M.M.; Hervas, S.; Villanueva, H.; Lozano, T.; Rabal, O.; Oyarzabal, J.; Lasarte, J.J.; Bendandi, M.; Inoges, S.; López-Díaz de Cerio, A.; et al. Identification of LAG3 high affinity aptamers by HT-SELEX and Conserved Motif Accumulation (CMA). PLoS ONE 2017, 12, e0185169. [Google Scholar] [CrossRef] [PubMed]
- Capece, D.; Verzella, D.; Fischietti, M.; Zazzeroni, F.; Alesse, E. Targeting costimulatory molecules to improve antitumor immunity. J. Biomed. Biotechnol. 2012, 926321. [Google Scholar] [CrossRef]
- McNamara, J.O., II; Kolonias, D.; Pastor, F.; Mittler, R.S.; Chen, L.; Giangrande, P.H.; Sullenger, B.; Gilboa, E. Multivalent 4-1BB binding aptamers costimulate CD8+ T cells and inhibit tumor growth in mice. J. Clin. Investig. 2008, 118, 376–386. [Google Scholar] [CrossRef]
- Pastor, F.; Kolonias, D.; Giangrande, P.H.; Gilboa, E. Induction of tumour immunity by targeted inhibition of nonsense-mediated mRNA decay. Nature 2010, 465, 227–230. [Google Scholar] [CrossRef]
- Benaduce, A.P.; Brenneman, R.; Schrand, B.; Pollack, A.; Gilboa, E.; Ishkanian, A. 4-1BB aptamer-based immunomodulation enhances the therapeutic index of radiation therapy in murine tumor models. Int. J. Radiat. Oncol. Biol. Phys. 2016, 96, 458–461. [Google Scholar] [CrossRef]
- Dollins, C.M.; Nair, S.; Boczkowski, D.; Lee, J.; Layzer, J.M.; Gilboa, E.; Sullenger, B.A. Assembling OX40 aptamers on a molecular scaffold to create a receptor-activating aptamer. Chem. Biol. 2008, 15, 675–682. [Google Scholar] [CrossRef]
- Pratico, E.D.; Sullenger, B.A.; Nair, S.K. Identification and characterization of an agonistic aptamer against the t cell costimulatory receptor, OX40. Nucleic Acid Ther. 2013, 23, 35–43. [Google Scholar] [CrossRef]
- Soldevilla, M.M.; Villanueva, H.; Bendandi, M.; Inoges, S.; López-Díaz de Cerio, A.; Pastor, F. 2-fluoro-RNA oligonucleotide CD40 targeted aptamers for the control of B lymphoma and bone-marrow aplasia. Biomaterials 2015, 67, 274–285. [Google Scholar] [CrossRef]
- Atallah-Yunes, S.A.; Robertson, M.J. Cytokine based immunotherapy for cancer and lymphoma: Biology, challenges and future perspectives. Front. Immunol. 2022, 13, 872010. [Google Scholar] [CrossRef]
- Rallis, K.S.; Corrigan, A.E.; Dadah, H.; George, A.M.; Keshwara, S.M.; Sideris, M.; Szabados, B. Cytokine-based cancer immunotherapy: Challenges and opportunities for IL-10. Anticancer Res. 2021, 41, 3247–3252. [Google Scholar] [CrossRef] [PubMed]
- Rallis, K.S.; Corrigan, A.E.; Dadah, H.; Stanislovas, J.; Zamani, P.; Makker, S.; Szabados, B.; Sideris, M. IL-10 in cancer: An essential thermostatic regulator between homeostatic immunity and inflammation—A comprehensive review. Future Oncol. 2022, 18, 3349–3365. [Google Scholar] [CrossRef]
- Ni, G.; Zhang, L.; Yang, X.; Li, H.; Ma, B.; Walton, S.; Wu, X.; Yuan, J.; Wang, T.; Liu, X. Targeting interleukin-10 signalling for cancer immunotherapy, a promising and complicated task. Hum. Vaccines Immunother. 2020, 16, 2328–2332. [Google Scholar] [CrossRef]
- Berezhnoy, A.; Stewart, C.A.; McNamara, J.O., 2nd; Thiel, W.; Giangrande, P.; Trinchieri, G.; Gilboa, E. Isolation and optimization of murine IL-10 receptor blocking oligonucleotide aptamers using high-throughput sequencing. Mol. Ther. J. Am. Soc. Gene Ther. 2012, 20, 1242–1250. [Google Scholar] [CrossRef]
- Burns, J.M.; Summers, B.C.; Wang, Y.; Melikian, A.; Berahovich, R.; Miao, Z.; Penfold, M.E.; Sunshine, M.J.; Littman, D.R.; Kuo, C.J.; et al. A novel chemokine receptor for SDF-1 and I-TAC involved in cell survival, cell adhesion, and tumor development. J. Exp. Med. 2006, 203, 2201–2213. [Google Scholar] [CrossRef] [PubMed]
- Miao, Z.; Luker, K.E.; Summers, B.C.; Berahovich, R.; Bhojani, M.S.; Rehemtulla, A.; Kleer, C.G.; Essner, J.J.; Nasevicius, A.; Luker, G.D.; et al. CXCR7 (RDC1) promotes breast and lung tumor growth in vivo and is expressed on tumor-associated vasculature. Proc. Natl. Acad. Sci. USA 2007, 104, 15735–15740. [Google Scholar] [CrossRef] [PubMed]
- Azab, A.K.; Runnels, J.M.; Pitsillides, C.; Moreau, A.S.; Azab, F.; Leleu, X.; Jia, X.; Wright, R.; Ospina, B.; Carlson, A.L.; et al. CXCR4 inhibitor AMD3100 disrupts the interaction of multiple myeloma cells with the bone marrow microenvironment and enhances their sensitivity to therapy. Blood 2009, 113, 4341–4351. [Google Scholar] [CrossRef]
- Eulberg, D.; Klussmann, S. Spiegelmers: Biostable aptamers. ChemBioChem 2003, 4, 979–983. [Google Scholar] [CrossRef]
- Hoellenriegel, J.; Zboralski, D.; Maasch, C.; Rosin, N.Y.; Wierda, W.G.; Keating, M.J.; Kruschinski, A.; Burger, J.A. The Spiegelmer NOX-A12, a novel CXCL12 inhibitor, interferes with chronic lymphocytic leukemia cell motility and causes chemosensitization. Blood 2014, 123, 1032–1039. [Google Scholar] [CrossRef]
- Vater, A.; Sahlmann, J.; Kröger, N.; Zöllner, S.; Lioznov, M.; Maasch, C.; Buchner, K.; Vossmeyer, D.; Schwoebel, F.; Purschke, W.G.; et al. Hematopoietic stem and progenitor cell mobilization in mice and humans by a first-in-class mirror-image oligonucleotide inhibitor of CXCL12. Clin. Pharmacol. Ther. 2013, 94, 150–157. [Google Scholar] [CrossRef]
- Shigdar, S.; Schrand, B.; Giangrande, P.H.; de Franciscis, V. Aptamers: Cutting edge of cancer therapies. Mol. Ther. 2021, 29, 2396–2411. [Google Scholar] [CrossRef] [PubMed]
- Kang, J.; Lee, M.S.; Copland, J.A.; Luxon, B.A.; Gorenstein, D.G. Combinatorial selection of a single stranded DNA thioaptamer targeting TGF-β1 protein. Bioorganic Med. Chem. Lett. 2008, 18, 1835–1839. [Google Scholar] [CrossRef] [PubMed]
- Marro, M.L.; Daniels, D.A.; McNamee, A.; Andrew, D.P.; Chapman, T.D.; Jiang, M.S.; Wu, Z.; Smith, J.L.; Patel, K.K.; Gearing, K.L. Identification of potent and selective RNA antagonists of the IFN-γ-inducible CXCL10 chemokine. Biochemistry 2005, 44, 8449–8460. [Google Scholar] [CrossRef] [PubMed]
- Thomas, B.J.; Porciani, D.; Burke, D.H. Cancer immunomodulation using bispecific aptamers. Mol. Ther. Nucleic Acids 2022, 27, 894–915. [Google Scholar] [CrossRef]
- Vandghanooni, S.; Eskandani, M.; Barar, J.; Omidi, Y. Bispecific therapeutic aptamers for targeted therapy of cancer: A review on cellular perspective. J. Mol. Med. 2018, 96, 885–902. [Google Scholar] [CrossRef] [PubMed]
- Boltz, A.; Piater, B.; Toleikis, L.; Guenther, R.; Kolmar, H.; Hock, B. Bi-specific aptamers mediating tumor cell lysis. J. Biol. Chem. 2011, 286, 21896–21905. [Google Scholar] [CrossRef]
- Yang, Y.; Sun, X.; Xu, J.; Cui, C.; Safari Yazd, H.; Pan, X.; Zhu, Y.; Chen, X.; Li, X.; Li, J.; et al. Circular bispecific aptamer-mediated artificial intercellular recognition for targeted t cell immunotherapy. ACS Nano 2020, 14, 9562–9571. [Google Scholar] [CrossRef]
- Kuai, H.; Zhao, Z.; Mo, L.; Liu, H.; Hu, X.; Fu, T.; Zhang, X.; Tan, W. Circular bivalent aptamers enable in vivo stability and recognition. J. Am. Chem. Soc. 2017, 139, 9128–9131. [Google Scholar] [CrossRef]
- Du, Y.; Zhang, D.; Wang, Y.; Wu, M.; Zhang, C.; Zheng, Y.; Zheng, A.; Liu, X. A highly stable multifunctional aptamer for enhancing antitumor immunity against hepatocellular carcinoma by blocking dual immune checkpoints. Biomater. Sci. 2021, 9, 4159–4168. [Google Scholar] [CrossRef]
- Zheng, A.; Du, Y.; Wang, Y.; Zheng, Y.; Ning, Z.; Wu, M.; Zhang, C.; Zhang, D.; Liu, J.; Liu, X. CD16/PD-L1 bi-specific aptamer for cancer immunotherapy through recruiting NK cells and acting as immunocheckpoint blockade. Mol. Ther. Nucleic Acids 2022, 27, 998–1009. [Google Scholar] [CrossRef]
- Sun, Y.; Mo, L.; Hu, X.; Yu, D.; Xie, S.; Li, J.; Zhao, Z.; Fang, X.; Ye, M.; Qiu, L.; et al. Bispecific aptamer-based recognition-then-conjugation strategy for PD1/PDL1 axis blockade and enhanced immunotherapy. ACS Nano 2022, 16, 21129–21138. [Google Scholar] [CrossRef] [PubMed]
- Pastor, F.; Kolonias, D.; McNamara Ii, J.O.; Gilboa, E. Targeting 4-1BB costimulation to disseminated tumor lesions with bi-specific oligonucleotide aptamers. Mol. Ther. 2011, 19, 1878–1886. [Google Scholar] [CrossRef] [PubMed]
- Schrand, B.; Berezhnoy, A.; Brenneman, R.; Williams, A.; Levay, A.; Kong, L.Y.; Rao, G.; Zhou, S.; Heimberger, A.B.; Gilboa, E. Targeting 4-1BB costimulation to the tumor stroma with bispecific aptamer conjugates enhances the therapeutic index of tumor immunotherapy. Cancer Immunol. Res. 2014, 2, 867–877. [Google Scholar] [CrossRef]
- Wei, C.Y.; Wang, L.; Zhu, M.X.; Deng, X.Y.; Wang, D.H.; Zhang, S.M.; Ying, J.H.; Yuan, X.; Wang, Q.; Xuan, T.F.; et al. TRIM44 activates the AKT/mTOR signal pathway to induce melanoma progression by stabilizing TLR4. J. Exp. Clin. Cancer Res. 2019, 38, 137. [Google Scholar] [CrossRef] [PubMed]
- Soldevilla, M.M.; Villanueva, H.; Casares, N.; Lasarte, J.J.; Bendandi, M.; Inoges, S.; López-Díaz de Cerio, A.; Pastor, F. MRP1-CD28 bi-specific oligonucleotide aptamers: Target costimulation to drug-resistant melanoma cancer stem cells. Oncotarget 2016, 7, 23182–23196. [Google Scholar] [CrossRef] [PubMed]
- Li, Z.; Hu, Y.; An, Y.; Duan, J.; Li, X.; Yang, X.D. Novel bispecific aptamer enhances immune cytotoxicity against MUC1-positive tumor cells by MUC1-CD16 dual targeting. Molecules 2019, 24, 478. [Google Scholar] [CrossRef] [PubMed]
- Liu, X.; Yan, H.; Liu, Y.; Chang, Y. Targeted cell–cell interactions by DNA nanoscaffold-templated multivalent bispecific aptamers. Small 2011, 7, 1673–1682. [Google Scholar] [CrossRef]
- Etter, E.L.; Mei, K.C.; Nguyen, J. Delivering more for less: Nanosized, minimal-carrier and pharmacoactive drug delivery systems. Adv. Drug Deliv. Rev. 2021, 179, 113944. [Google Scholar] [CrossRef]
- Lee, B.K.; Yun, Y.H.; Park, K. Smart nanoparticles for drug delivery: Boundaries and opportunities. Chem. Eng. Sci. 2015, 125, 158–164. [Google Scholar] [CrossRef]
- Filipczak, N.; Pan, J.; Yalamarty, S.S.K.; Torchilin, V.P. Recent advancements in liposome technology. Adv. Drug Deliv. Rev. 2020, 156, 4–22. [Google Scholar] [CrossRef]
- Zhang, Y.; Sun, C.; Wang, C.; Jankovic, K.E.; Dong, Y. Lipids and lipid derivatives for RNA delivery. Chem Rev 2021, 121, 12181–12277. [Google Scholar] [CrossRef]
- Wang, C.; Liu, B.; Lu, J.; Zhang, G.; Lu, A. Strategies for combination of aptamer and targeted drug delivery. J. Nanosci. Nanotechnol. 2014, 14, 501–512. [Google Scholar] [CrossRef] [PubMed]
- Liu, Y.; Castro Bravo, K.M.; Liu, J. Targeted liposomal drug delivery: A nanoscience and biophysical perspective. Nanoscale Horiz. 2021, 6, 78–94. [Google Scholar] [CrossRef]
- Moosavian, S.A.; Sahebkar, A. Aptamer-functionalized liposomes for targeted cancer therapy. Cancer Lett. 2019, 448, 144–154. [Google Scholar] [CrossRef]
- Odeh, F.; Nsairat, H.; Alshaer, W.; Ismail, M.A.; Esawi, E.; Qaqish, B.; Bawab, A.A.; Ismail, S.I. Aptamers chemistry: Chemical modifications and conjugation strategies. Molecules 2019, 25, 3. [Google Scholar] [CrossRef] [PubMed]
- Xie, S.; Hou, X.; Yang, W.; Shi, W.; Yang, X.; Duan, S.; Mo, F.; Liu, A.; Wang, W.; Lu, X. Endoglin-aptamer-functionalized liposome-equipped PD-1-silenced T cells enhance antitumoral immunotherapeutic effects. Int. J. Nanomed. 2021, 16, 6017–6034. [Google Scholar] [CrossRef]
- Hong, S.; Ding, P.; Luo, Y.; Gao, T.; Zhang, Y.; Pei, R. Aptamer-integrated α-Gal liposomes as bispecific agents to trigger immune response for killing tumor cells. J. Biomed. Mater. Res. Part A 2019, 107, 1176–1183. [Google Scholar] [CrossRef] [PubMed]
- Zou, W. Immunosuppressive networks in the tumour environment and their therapeutic relevance. Nat. Rev. Cancer 2005, 5, 263–274. [Google Scholar] [CrossRef] [PubMed]
- Liu, H.; Mai, J.; Shen, J.; Wolfram, J.; Li, Z.; Zhang, G.; Xu, R.; Li, Y.; Mu, C.; Zu, Y.; et al. A novel DNA aptamer for dual targeting of polymorphonuclear myeloid-derived suppressor cells and tumor cells. Theranostics 2018, 8, 31–44. [Google Scholar] [CrossRef] [PubMed]
- Ishida, T.; Iden, D.L.; Allen, T.M. A combinatorial approach to producing sterically stabilized (Stealth) immunoliposomal drugs. FEBS Lett. 1999, 460, 129–133. [Google Scholar] [CrossRef] [PubMed]
- Bertrand, N.; Wu, J.; Xu, X.; Kamaly, N.; Farokhzad, O.C. Cancer nanotechnology: The impact of passive and active targeting in the era of modern cancer biology. Adv. Drug Deliv. Rev. 2014, 66, 2–25. [Google Scholar] [CrossRef] [PubMed]
- Kim, M.; Lee, J.S.; Kim, W.; Lee, J.H.; Jun, B.H.; Kim, K.S.; Kim, D.E. Aptamer-conjugated nano-liposome for immunogenic chemotherapy with reversal of immunosuppression. J. Control. Release Off. J. Control. Release Soc. 2022, 348, 893–910. [Google Scholar] [CrossRef]
- Muir, V.G.; Burdick, J.A. Chemically modified biopolymers for the formation of biomedical hydrogels. Chem. Rev. 2021, 121, 10908–10949. [Google Scholar] [CrossRef]
- Bao, Z.; Xian, C.; Yuan, Q.; Liu, G.; Wu, J. Natural polymer-based hydrogels with enhanced mechanical performances: Preparation, structure, and property. Adv. Healthc. Mater. 2019, 8, 1900670. [Google Scholar] [CrossRef]
- Li, J.; Mo, L.; Lu, C.H.; Fu, T.; Yang, H.H.; Tan, W. Functional nucleic acid-based hydrogels for bioanalytical and biomedical applications. Chem. Soc. Rev. 2016, 45, 1410–1431. [Google Scholar] [CrossRef] [PubMed]
- Wang, Z.; Chen, R.; Yang, S.; Li, S.; Gao, Z. Design and application of stimuli-responsive DNA hydrogels: A review. Mater. Today Bio 2022, 16, 100430. [Google Scholar] [CrossRef]
- Gacanin, J.; Synatschke, C.V.; Weil, T. Biomedical applications of DNA-based hydrogels. Adv. Funct. Mater. 2020, 30, 1906253. [Google Scholar] [CrossRef]
- Lee, J.; Le, Q.V.; Yang, G.; Oh, Y.K. Cas9-edited immune checkpoint blockade PD-1 DNA polyaptamer hydrogel for cancer immunotherapy. Biomaterials 2019, 218, 119359. [Google Scholar] [CrossRef]
- Wei, H.; Zhao, Z.; Wang, Y.; Zou, J.; Lin, Q.; Duan, Y. One-step self-assembly of multifunctional DNA nanohydrogels: An enhanced and harmless strategy for guiding combined antitumor therapy. Acs Appl. Mater. Interfaces 2019, 11, 46479–46489. [Google Scholar] [CrossRef]
- Wang, W.; Liu, X.; Ding, L.; Jin, H.J.; Li, X. RNA hydrogel combined with MnO2 nanoparticles as a nano-vaccine to treat triple negative breast cancer. Front. Chem. 2021, 9, 797094. [Google Scholar] [CrossRef]
- Tan, J.; Luo, Y.; Guo, Y.; Zhou, Y.; Liao, X.; Li, D.; Lai, X.; Liu, Y. Development of alginate-based hydrogels: Crosslinking strategies and biomedical applications. Int. J. Biol. Macromol. 2023, 239, 124275. [Google Scholar] [CrossRef] [PubMed]
- Sun, L.; Shen, F.; Tian, L.; Tao, H.; Xiong, Z.; Xu, J.; Liu, Z. ATP-responsive smart hydrogel releasing immune adjuvant synchronized with repeated chemotherapy or radiotherapy to boost antitumor immunity. Adv. Mater. 2021, 33, 2007910. [Google Scholar] [CrossRef]
- Xiong, X.; Liu, H.; Zhao, Z.; Altman, M.B.; Lopez-Colon, D.; Yang, C.J.; Chang, L.-J.; Liu, C.; Tan, W. DNA aptamer-mediated cell targeting. Angew. Chem. Int. Ed. 2013, 52, 1472–1476. [Google Scholar] [CrossRef] [PubMed]
- Yang, S.; Wen, J.; Li, H.; Xu, L.; Liu, Y.; Zhao, N.; Zeng, Z.; Qi, J.; Jiang, W.; Han, W.; et al. Aptamer-engineered natural killer cells for cell-specific adaptive immunotherapy. Small 2019, 15, e1900903. [Google Scholar] [CrossRef] [PubMed]
- Gong, N.; Sheppard, N.C.; Billingsley, M.M.; June, C.H.; Mitchell, M.J. Nanomaterials for T-cell cancer immunotherapy. Nat. Nanotechnol. 2021, 16, 25–36. [Google Scholar] [CrossRef]
- Tang, R.; Fu, Y.H.; Gong, B.; Fan, Y.Y.; Wang, H.H.; Huang, Y.; Nie, Z.; Wei, P. A chimeric conjugate of antibody and programmable DNA nanoassembly smartly activates T Cells for precise cancer cell targeting. Angew. Chem. (Int. Ed. Engl.) 2022, 61, e202205902. [Google Scholar]
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).