Construction of an ATP-Activated Y-Shape DNA Probe for Smart miRNA Imaging in Living Cells
Abstract
1. Introduction
2. Materials and Methods
2.1. Materials
2.2. Instruments
2.3. Preparation of the Y-DNA
2.4. Gel Electrophoresis Experiments
2.5. Fluorescence Monitoring
2.6. Biostability of Y-DNA
2.7. Material Cell Culture and MTT Assays
2.8. In Vitro miRNA Imaging
3. Results
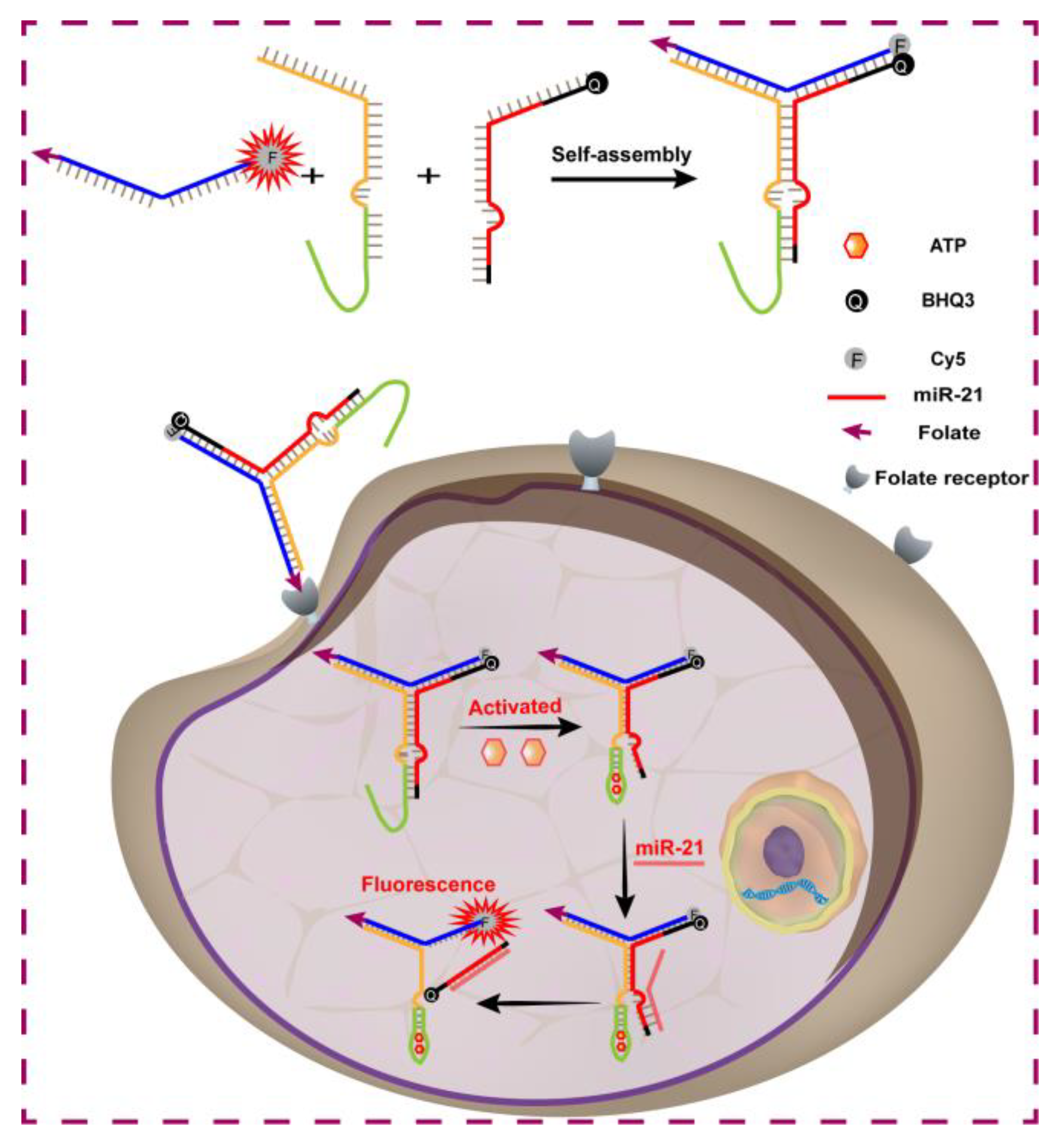
3.1. Principle of the Assay
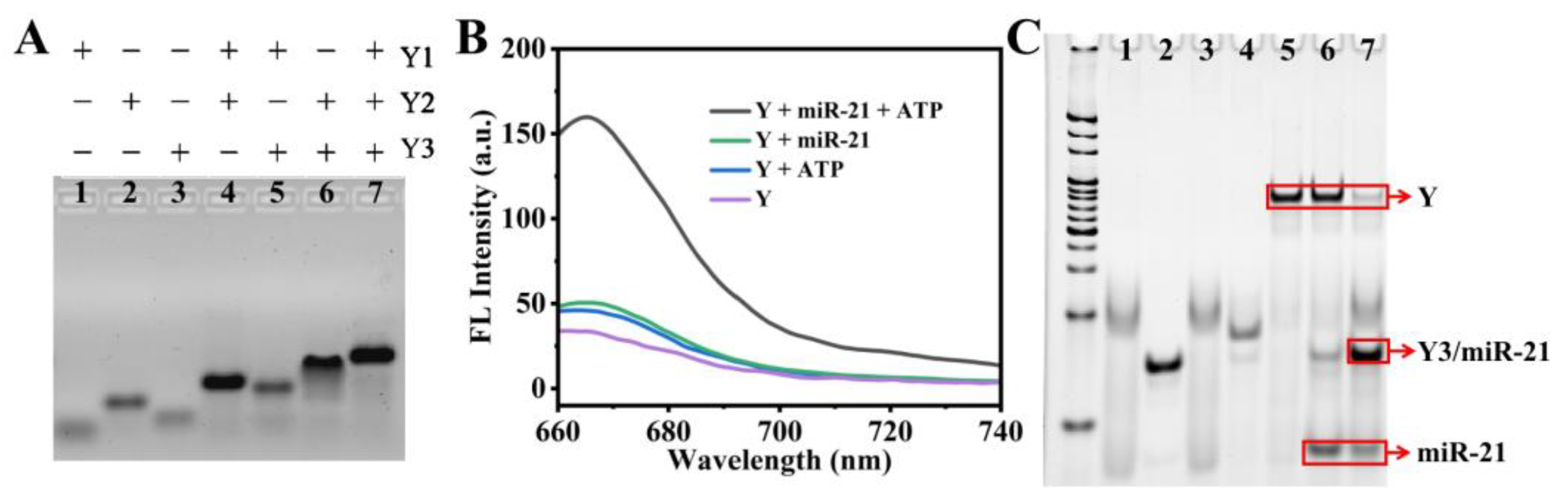
3.2. Preparation and Characterization of Y-DNA Probe
3.3. Optimization of Experimental Conditions
3.4. Analytical Performance of Y-DNA Probe
3.5. Biostability and Biocompatibility of Y-DNA
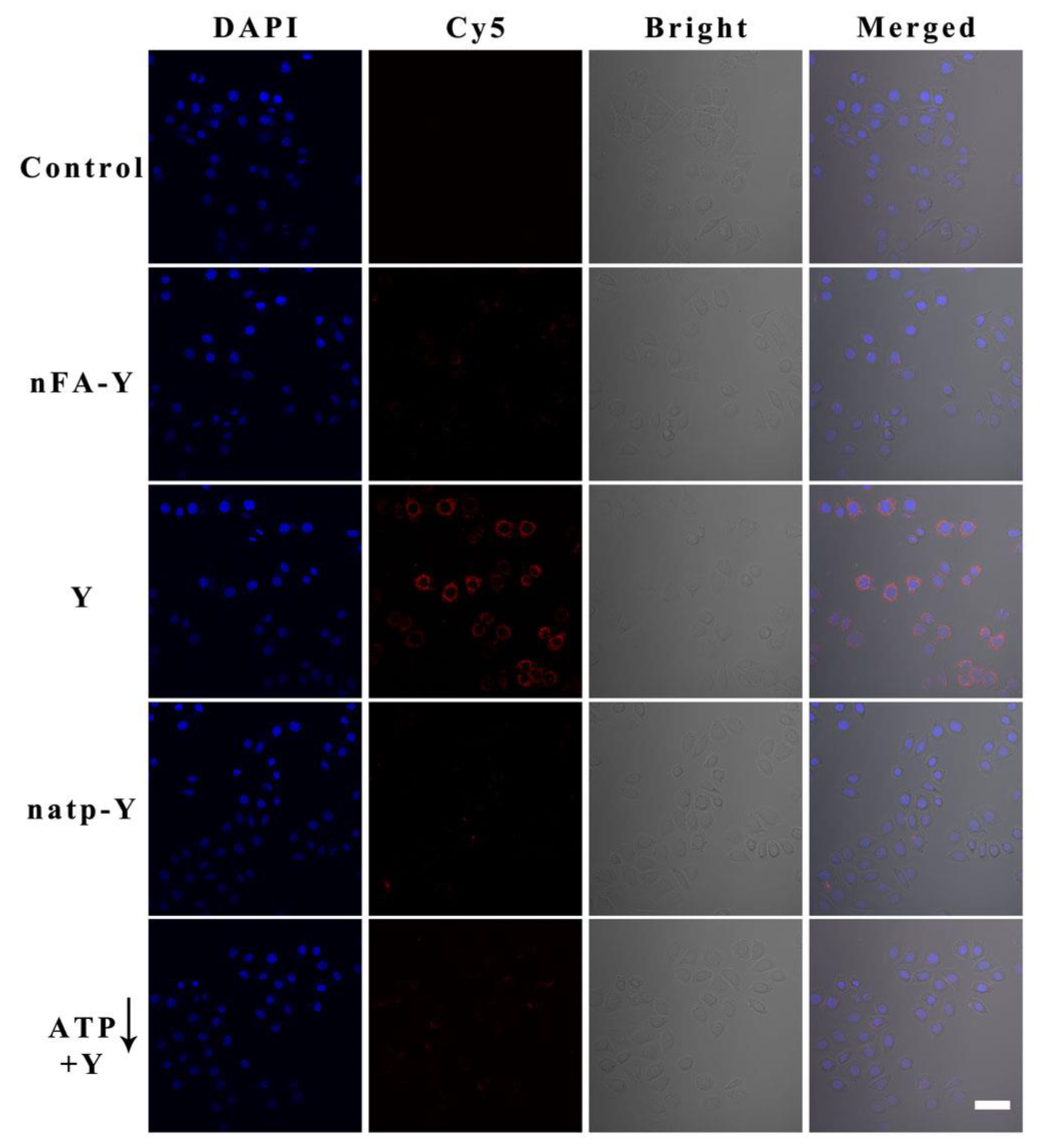
3.6. Sensitivity of miRNA Imaging in Cancer Cells
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Data Availability Statement
Conflicts of Interest
References
- Xi, X.; Li, T.; Huang, Y.; Sun, J.; Zhu, Y.; Yang, Y.; Lu, Z.J. RNA Biomarkers: Frontier of Precision Medicine for Cancer. Noncoding RNA 2017, 3, 9. [Google Scholar]
- Vargas, A.J.; Harris, C.C. Biomarker development in the precision medicine era: Lung cancer as a case study. Nat. Rev. Cancer 2016, 16, 525–537. [Google Scholar] [PubMed]
- Bartel, D.P. MicroRNAs: Genomics, Biogenesis, Mechanism, and Function. Cell 2004, 116, 281–297. [Google Scholar] [PubMed]
- Croce, C.M. Causes and consequences of microRNA dysregulation in cancer. Nat. Rev. Genet. 2009, 10, 704–714. [Google Scholar] [PubMed]
- Ouyang, T.L.; Liu, Z.Y.; Han, Z.Y.; Ge, Q.Y. MicroRNA Detection Specificity: Recent Advances and Future Perspective. Anal. Chem. 2019, 91, 3179–3186. [Google Scholar] [PubMed]
- Gong, S.; Wang, X.; Zhou, P.; Pan, W.; Li, N.; Tang, B. AND Logic-Gate-Based CRISPR/Cas12a Biosensing Platform for the Sensitive Colorimetric Detection of Dual miRNAs. Anal. Chem. 2022, 94, 15839–15846. [Google Scholar]
- Kitte, S.A.; Bushira, F.A.; Xu, C.; Wang, Y.; Li, H.; Jin, Y. Plasmon-Enhanced Nitrogen Vacancy-Rich Carbon Nitride Electrochemiluminescence Aptasensor for Highly Sensitive Detection of miRNA. Anal. Chem. 2022, 94, 1406–1414. [Google Scholar] [CrossRef]
- Sun, Y.; Fang, L.; Han, Y.; Feng, A.; Liu, S.; Zhang, K.; Xu, J.J. Reversible Ratiometric Electrochemiluminescence Biosensor Based on DNAzyme Regulated Resonance Energy Transfer for Myocardial miRNA Detection. Anal. Chem. 2022, 94, 7035–7040. [Google Scholar]
- Liu, N.; Lu, H.; Liu, L.; Ni, W.; Yao, Q.; Zhang, G.J.; Yang, F. Ultrasensitive Exosomal MicroRNA Detection with a Supercharged DNA Framework Nanolabel. Anal. Chem. 2021, 93, 5917–5923. [Google Scholar] [CrossRef]
- Zhang, X.; Yang, Z.; Chang, Y.; Qing, M.; Yuan, R.; Chai, Y. Novel 2D-DNA-Nanoprobe-Mediated Enzyme-Free-Target-Recycling Amplification for the Ultrasensitive Electrochemical Detection of MicroRNA. Anal. Chem. 2018, 90, 9538–9544. [Google Scholar] [CrossRef]
- Xue, C.; Luo, M.; Wang, L.; Li, C.; Hu, S.; Yu, X.; Yuan, P.; Wu, Z.S. Stimuli-Responsive Autonomous-Motion Molecular Machine for Sensitive Simultaneous Fluorescence Imaging of Intracellular MicroRNAs. Anal. Chem. 2021, 93, 9869–9877. [Google Scholar]
- He, L.; Lu, D.Q.; Liang, H.; Xie, S.T.; Luo, C.; Hu, M.M.; Xu, L.J.; Zhang, X.B.; Tan, W.H. Fluorescence Resonance Energy Transfer-Based DNA Tetrahedron Nanotweezer for Highly Reliable Detection of Tumor-Related mRNA in Living Cells. Acs Nano 2017, 11, 4060–4066. [Google Scholar] [CrossRef]
- Tang, J.; Li, Q.; Yao, C.; Yang, D. DNA Nanomaterial-Based Optical Probes for Exosomal miRNA Detection. Chempluschem 2023, 88, e202200345. [Google Scholar] [CrossRef]
- Li, J.; Wang, W.; Zhang, H.; Lu, Z.; Wu, W.; Shu, M.; Han, H. Programmable DNA Tweezer-Actuated SERS Probe for the Sensitive Detection of AFB(1). Anal. Chem. 2020, 92, 4900–4907. [Google Scholar] [CrossRef]
- Ebrahimi, S.B.; Samanta, D.; Partridge, B.E.; Kusmierz, C.D.; Cheng, H.F.; Grigorescu, A.A.; Chavez, J.L.; Mirau, P.A.; Mirkin, C.A. Programming Fluorogenic DNA Probes for Rapid Detection of Steroids. Angew. Chem. Int. Ed. 2021, 60, 15260–15265. [Google Scholar] [CrossRef]
- Wei, Q.M.; Huang, J.; Li, J.; Wang, J.L.; Yang, X.H.; Liu, J.B.; Wang, K.M. A DNA nanowire based localized catalytic hairpin assembly reaction for microRNA imaging in live cells. Chem. Sci. 2018, 9, 7802–7808. [Google Scholar]
- Liu, L.; Rong, Q.M.; Ke, G.L.; Zhang, M.; Li, J.; Li, Y.Q.; Liu, Y.C.; Chen, M.; Zhang, X.B. Efficient and Reliable MicroRNA Imaging in Living Cells via a FRET-Based Localized Hairpin-DNA Cascade Amplifier. Anal. Chem. 2019, 91, 3675–3680. [Google Scholar] [CrossRef] [PubMed]
- Wu, Z.; Liu, G.Q.; Yang, X.L.; Jiang, J.H. Electrostatic nucleic acid nanoassembly enables hybridization chain reaction in living cells for ultrasensitive mRNA imaging. J. Am. Chem. Soc. 2015, 137, 6829–6836. [Google Scholar] [PubMed]
- Xue, C.; Zhang, S.X.; Ouyang, C.H.; Chang, D.; Salena, B.J.; Li, Y.; Wu, Z.S. Target-Induced Catalytic Assembly of Y-Shaped DNA and Its Application for In Situ Imaging of MicroRNAs. Angew. Chem. Int. Ed. 2018, 57, 9739–9743. [Google Scholar] [CrossRef] [PubMed]
- Wang, K.M.; Tang, Z.W.; Yang CY, J.; Kim, Y.M.; Fang, X.H.; Li, W.; Wu, Y.R.; Medley, C.D.; Cao, Z.H.; Li, J.; et al. Molecular Engineering of DNA: Molecular Beacons. Angew. Chem. Int. Ed. 2009, 48, 856–870. [Google Scholar]
- He, L.; Lu, D.Q.; Liang, H.; Xie, S.T.; Zhang, X.B.; Liu, O.L.; Yuan, Q.; Tan, W.H. mRNA-Initiated, Three-Dimensional DNA Amplifier Able to Function inside Living Cells. J. Am. Chem. Soc. 2018, 140, 258–263. [Google Scholar] [CrossRef]
- Duan, Z.; Tan, L.; Duan, R.; Chen, M.; Xia, F.; Huang, F. Photoactivated Biosensing Process for Dictated ATP Detection in Single Living Cells. Anal. Chem. 2021, 93, 11547–11556. [Google Scholar] [CrossRef]
- Shao, Y.; Zhao, J.; Yuan, J.; Zhao, Y.; Li, L. Organelle-Specific Photoactivation of DNA Nanosensors for Precise Profiling of Subcellular Enzymatic Activity. Angew. Chem. Int. Ed. 2021, 60, 8923–8931. [Google Scholar]
- Zhao, X.X.; Zhang, L.L.; Gao, W.Y.; Yu, X.L.; Gu, W.; Fu, W.L.; Luo, Y. Spatiotemporally Controllable MicroRNA Imaging in Living Cells via a Near-Infrared Light-Activated Nanoprobe. ACS Appl. Mater. Interfaces 2020, 12, 35958–35966. [Google Scholar] [CrossRef]
- Meng, X.; Wang, H.; Yang, M.; Li, J.; Yang, F.; Zhang, K.; Dong, H.; Zhang, X. Target-Cell-Specific Bioorthogonal and Endogenous ATP Control of Signal Amplification for Intracellular MicroRNA Imaging. Anal. Chem. 2021, 93, 1693–1701. [Google Scholar]
- Mo, R.; Jiang, T.; Gu, Z. Enhanced anticancer efficacy by ATP-mediated liposomal drug delivery. Angew. Chem. Int. Ed. 2014, 53, 5815–5820. [Google Scholar]
- Meng, X.; Dai, W.; Zhang, K.; Dong, H.; Zhang, X. Imaging multiple microRNAs in living cells using ATP self-powered strand-displacement cascade amplification. Chem. Sci. 2018, 9, 1184–1190. [Google Scholar]
- Wu, H.; Chen, T.T.; Wang, X.N.; Ke, Y.; Jiang, J.H. RNA imaging in living mice enabled by an in vivo hybridization chain reaction circuit with a tripartite DNA probe. Chem. Sci. 2020, 11, 62–69. [Google Scholar]
- Ren, K.; Xu, Y.; Liu, Y.; Yang, M.; Ju, H. A Responsive “Nano String Light” for Highly Efficient mRNA Imaging in Living Cells via Accelerated DNA Cascade Reaction. ACS Nano 2018, 12, 263–271. [Google Scholar]
- Wan, Y.; Li, G.; Zou, L.; Wang, H.; Wang, Q.; Tan, K.; Liu, X.; Wang, F. A Deoxyribozyme-Initiated Self-Catalytic DNA Machine for Amplified Live-Cell Imaging of MicroRNA. Anal. Chem. 2021, 93, 11052–11059. [Google Scholar] [CrossRef]



Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Zhong, W.; Zheng, Y.; Huang, L.; Xing, C.; Lu, C. Construction of an ATP-Activated Y-Shape DNA Probe for Smart miRNA Imaging in Living Cells. Chemistry 2023, 5, 1634-1644. https://doi.org/10.3390/chemistry5030112
Zhong W, Zheng Y, Huang L, Xing C, Lu C. Construction of an ATP-Activated Y-Shape DNA Probe for Smart miRNA Imaging in Living Cells. Chemistry. 2023; 5(3):1634-1644. https://doi.org/10.3390/chemistry5030112
Chicago/Turabian StyleZhong, Wukun, Yanlin Zheng, Lei Huang, Chao Xing, and Chunhua Lu. 2023. "Construction of an ATP-Activated Y-Shape DNA Probe for Smart miRNA Imaging in Living Cells" Chemistry 5, no. 3: 1634-1644. https://doi.org/10.3390/chemistry5030112
APA StyleZhong, W., Zheng, Y., Huang, L., Xing, C., & Lu, C. (2023). Construction of an ATP-Activated Y-Shape DNA Probe for Smart miRNA Imaging in Living Cells. Chemistry, 5(3), 1634-1644. https://doi.org/10.3390/chemistry5030112





