Questions in the Chemical Enzymology of MAO
Abstract
1. Introduction
2. What Is the Chemical Mechanism of Amine Oxidation? Is It the Same for Both MAO A and MAO B?
3. The Oxidative Half-Reaction: How Does Oxygen Access the Reduced Flavin, and Is It Blocked or Facilitated by Ligands?
4. Kinetics: Can Simulation Explain the Alternative Pathways in Turnover; Why Substrate Accelerates Oxidation and the Influence of Two Forms for Binding Inhibitors?
4.1. Both the Oxidized and Reduced Forms of MAO Can Bind Ligands during Turnover
4.2. Substrate Stimulation of Reoxidation-Thermodynamic or Kinetic?
5. Is There Potential for Applied Use of MAO?
6. Importance of Flexibility
6.1. Is Flexibility Important in the Reaction?
6.2. The Influence of the Membrane on MAO Activity
6.3. Does the Oxidation of MAO Thiols Have Any Importance In Vivo?
7. Biological Questions
7.1. Does Variation in the Calcium Ion Concentration Change MAO Activity?
7.2. Does MAO A Moonliaght in the Cell?
7.3. Species Differences and Mutations?
7.4. The Pharma Debate-Reversible or Irreversible Inhibition?
8. Conclusions
Funding
Data Availability Statement
Conflicts of Interest
References
- Edmondson, D.E.; Binda, C.; Mattevi, A. The FAD binding sites of human monoamine oxidases A and B. NeuroToxicology 2004, 25, 63–72. [Google Scholar] [CrossRef]
- Hare, M.L.C. Tyramine oxidase: A new enzyme system in liver. Biochem. J. 1928, 62, 968–979. [Google Scholar] [CrossRef]
- Tipton, K.F. 90years of monoamine oxidase: Some progress and some confusion. J. Neural. Transm. 2018, 125, 1519–1551. [Google Scholar] [CrossRef]
- Kim, T.; Xu, C.; Amsterdam, J.D. Relative effectiveness of tricyclic antidepressant versus monoamine oxidase inhibitor monotherapy for treatment-resistant depression. J. Affect. Disord. 2019, 250, 199–203. [Google Scholar] [CrossRef]
- Mann, J.J.; Aarons, S.F.; Wilner, P.J.; Keilp, J.G.; Sweeney, J.A.; Pearlstein, T.; Frances, A.J.; Kocsis, J.H.; Brown, R.P. A controlled-study of the antidepressant efficacy and side-effects of (−)-deprenyl—A selective monoamine-oxidase inhibitor. Arch. Gen. Psychiatry 1989, 46, 45–50. [Google Scholar] [CrossRef]
- Quitkin, F.; Rifkin, A.; Klein, D.F. Mono-amine oxidase-inhibitors—Review of anti-depressant effectiveness. Arch. Gen. Psychiatry 1979, 36, 749–760. [Google Scholar] [CrossRef]
- Maycock, A.L.; Abeles, R.H.; Salach, J.I.; Singer, T.P. Structure of covalent adduct formed by interaction of 3-dimethylamino-1-propyne and flavin of mitochondrial amine oxidase. Biochemistry 1976, 15, 114–125. [Google Scholar] [CrossRef]
- Gartner, B.; Hemmerich, P.; Zeller, E.A. Structure of flavin adducts with acetylenic substrates—Chemistry of monoamine-oxidase and lactate oxidase Inhibition. Eur. J. Biochem. 1976, 63, 211–221. [Google Scholar] [CrossRef] [PubMed]
- Binda, C.; Newton-Vinson, P.; Hubalek, F.; Edmondson, D.E.; Mattevi, A. Structure of human monoamine oxidase B, a drug target for the treatment of neurological disorders. Nat. Struct. Biol. 2002, 9, 22–26. [Google Scholar] [CrossRef] [PubMed]
- Silverman, R.B. Radical ideas about monoamine-oxidase. Acc. Chem. Res. 1995, 28, 335–342. [Google Scholar] [CrossRef]
- Kalgutkar, A.S.; Castagnoli, N.; Testa, B. Selective inhibitors of monoamine-oxidase (MAO-A and MAO-B) as probes of its catalytic site and mechanism. Med. Res. Rev. 1995, 15, 325–388. [Google Scholar] [CrossRef] [PubMed]
- Finberg, J.P.M.; Gillman, K. Selective inhibitors of monoamine oxidase type B and the “cheese effect”. Int. Rev. Neurobiol. 2011, 100, 169–190. [Google Scholar] [CrossRef] [PubMed]
- Youdim, M.B.H.; Edmondson, D.; Tipton, K.F. The therapeutic potential of monoamine oxidase inhibitors. Nat. Rev. Neurosci. 2006, 7, 295–309. [Google Scholar] [CrossRef] [PubMed]
- Ramsay, R.R. Molecular aspects of the activity and inhibition of the FAD-containing monoamine oxidases. In Pharmaceutical Biocatalysis: Fundamentals, Enzyme Inhibitors, and Enzymes in Health and Diseases; Grunwald, P., Ed.; Pan Stanford Publishing Pte. Ltd.: Singapore, 2019; Volume 4. [Google Scholar]
- Ramsay, R.R.; Maniquet, A.; Hagenow, S.; Pappalardo, M.; Saija, M.C.; Bryant, S.D.; Albreht, A.; Guccione, S. Molecules Parameters for irreversible inactivation of monoamine oxidase. Molecules 2020, 25, 5908. [Google Scholar] [CrossRef] [PubMed]
- Binda, C.; Wang, J.; Li, M.; Hubalek, F.; Mattevi, A.; Edmondson, D.E. Structural and mechanistic studies of arylalkylhydrazine inhibition of human monoamine oxidases A and B. Biochemistry 2008, 47, 5616–5625. [Google Scholar] [CrossRef] [PubMed]
- Hubalek, F.; Binda, C.; Li, M.; Herzig, Y.; Sterling, J.; Youdim, M.B.H.; Mattevi, A.; Edmondson, D.E. Inactivation of purified human recombinant monoamine oxidases A and B by rasagiline and its analogues. J. Med. Chem. 2004, 47, 1760–1766. [Google Scholar] [CrossRef]
- Bautista-Aguilera, O.M.; Hagenow, S.; Palomino-Antolin, A.; Farre-Alins, V.; Ismaili, L.; Joffrin, P.L.; Jimeno, M.L.; Soukup, O.; Janockova, J.; Kalinowsky, L.; et al. Multitarget-Directed Ligands Combining Cholinesterase and Monoamine Oxidase Inhibition with Histamine H3R Antagonism for Neurodegenerative Diseases. Angew. Chem. Int. Ed. 2017, 56, 12765–12769. [Google Scholar] [CrossRef]
- Sablin, S.O.; Krueger, M.J.; Singer, T.P.; Bachurin, S.O.; Khare, A.B.; Efange, S.M.N.; Tkachenko, S.E. Interaction of Tetrahydrostilbazoles with Monoamine-Oxidase-a and Monoamine-Oxidase-B. J. Med. Chem. 1994, 37, 151–157. [Google Scholar] [CrossRef] [PubMed]
- Bocchinfuso, R.; Robinson, J.B. The stereoselectivity of inhibition of rat liver mitochondrial MAO-A and MAO-B by the enantiomers of 2-phenylpropylamine and their derivatives. Eur. J. Med. Chem. 1999, 34, 293–300. [Google Scholar] [CrossRef]
- Edmondson, D.E.; DeColibus, L.; Binda, C.; Li, M.; Mattevi, A. New insights into the structures and functions of human monoamine oxidases A and B. J. Neural. Transm. 2007, 114, 703–705. [Google Scholar] [CrossRef]
- Chajkowski-Scarry, S.; Rimoldi, J.M. Monoamine oxidase A and B substrates: Probing the pathway for drug development. Future Med. Chem. 2014, 6, 697–717. [Google Scholar] [CrossRef]
- Lu, X.; Rodriguez, M.; Ji, H.; Silverman, R.B.; Vintém, A.-P.B.; Ramsay, R.R. Irreversible inactivation of mitochondrial monoamine oxidases. In Flavins and Flavoproteins 2002; Chapman, S.K., Perham, R.N., Scrutton, N.S., Eds.; Rudolf Weber: Berlin, Germany, 2002; pp. 817–830. [Google Scholar]
- Erdem, S.S.; Buyukmenekse, B. Computational investigation on the structure-activity relationship of the biradical mechanism for monoamine oxidase. J. Neural. Transm. 2011, 118, 1021–1029. [Google Scholar] [CrossRef]
- Miller, J.R.; Edmondson, D.E. Structure-activity relationships in the oxidation of para- substituted benzylamine analogues by recombinant human liver monoamine oxidase A. Biochemistry 1999, 38, 13670–13683. [Google Scholar] [CrossRef]
- Orru, R.; Aldeco, M.; Edmondson, D.E. Do MAO A and MAO B utilize the same mechanism for the C–H bond cleavage step in catalysis? Evidence suggesting differing mechanisms. J. Neural. Transm. 2013, 120, 847–851. [Google Scholar] [CrossRef]
- MacMillar, S.; Edmondson, D.E.; Matsson, O. Nitrogen kinetic isotope effects for the Monoamine Oxidase B-catalyzed oxidation of benzylamine and (1,1-(2)H(2))benzylamine: Nitrogen rehybridization and CH bond cleavage are not concerted. J. Am. Chem. Soc. 2011, 133, 12319–12321. [Google Scholar] [CrossRef] [PubMed]
- Zenn, R.K.; Abad, E.; Kastner, J. Influence of the environment on the oxidative deamination of p-substituted benzylamines in monoamine oxidase. J. Phys. Chem. B 2015, 119, 3678–3686. [Google Scholar] [CrossRef] [PubMed]
- Umhau, S.; Pollegioni, L.; Molla, G.; Diederichs, K.; Welte, W.; Pilone, M.S.; Ghisla, S. The x-ray structure of D-amino acid oxidase at very high resolution identifies the chemical mechanism of flavin-dependent substrate dehydrogenation. Proc. Natl. Acad. Sci. USA 2000, 97, 12463–12468. [Google Scholar] [CrossRef]
- Edmondson, D.E.; Binda, C.; Mattevi, A. Structural insights into the mechanism of amine oxidation by monoamine oxidases A and B. Arch. Biochem. Biophys. 2007, 464, 269–276. [Google Scholar] [CrossRef]
- Akyuz, M.A.; Erdem, S.S. Computational modeling of the direct hydride transfer mechanism for the MAO catalyzed oxidation of phenethylamine and benzylamine: ONIOM (QM/QM) calculations. J. Neural. Transm. 2013, 120, 937–945. [Google Scholar] [CrossRef] [PubMed]
- Oanca, G.; Stare, J.; Vianello, R.; Mavri, J. Multiscale simulation of monoamine oxidase catalyzed decomposition of phenylethylamine analogs. Eur. J. Pharmacol. 2017, 817, 46–50. [Google Scholar] [CrossRef]
- Prah, A.; Purg, M.; Stare, J.; Vianello, R.; Mavri, J. How Monoamine Oxidase A Decomposes Serotonin: An Empirical Valence Bond Simulation of the Reactive Step. J. Phys. Chem. B 2020, 124, 8259–8265. [Google Scholar] [CrossRef]
- Vianello, R.; Repic, M.; Mavri, J. How are Biogenic Amines Metabolized by Monoamine Oxidases? Eur. J. Org. Chem. 2012, 36, 7057–7065. [Google Scholar] [CrossRef]
- Youngster, S.K.; McKeown, K.A.; Jin, Y.Z.; Ramsay, R.R.; Heikkila, R.E.; Singer, T.P. Oxidation of analogs of 1-methyl-4-phenyl-1,2,3,6-tetrahydropyridine by monoamine oxidase-A and oxidase-B and the inhibition of monoamine oxidases by the oxidation products. J. Neurochem. 1989, 53, 1837–1842. [Google Scholar] [CrossRef] [PubMed]
- Taft, R.W. Linear Steric Energy Relationships. J. Am. Chem. Soc. 1953, 75, 4538–4539. [Google Scholar] [CrossRef]
- Dunford, H.B.; Adeniran, A.J. Hammett ϱσ correlation for reactions of horseradish peroxidase compound II with phenols. Arch. Biochem. Biophys. 1986, 251, 536–542. [Google Scholar] [CrossRef]
- Zhang, H.; Dunford, H.B. Hammett pσ correlation for reactions of lactoperoxidase compound II with phenols. Can. J. Chem. 1993, 71, 1990–1994. [Google Scholar] [CrossRef]
- Santiago, C.B.; Milo, A.; Sigman, M.S. Developing a Modern Approach to Account for Steric Effects in Hammett-Type Correlations. J. Am. Chem. Soc. 2016, 138, 13424–13430. [Google Scholar] [CrossRef]
- Hart, H.; Sedor, E.A. Mechanism of cyclodehydration of 2-phenyltriarylcarbinols. J. Am. Chem. Soc. 1967, 89, 2342–2347. [Google Scholar] [CrossRef]
- Stein, A.R.; Tencer, M.; Moffatt, E.A.; Dawe, R.; Sweet, J. Nonlinearity of Hammett.sigma..rho. correlations for benzylic systems: Activation parameters and their mechanistic implications. J. Org. Chem. 1980, 45, 3539–3540. [Google Scholar] [CrossRef]
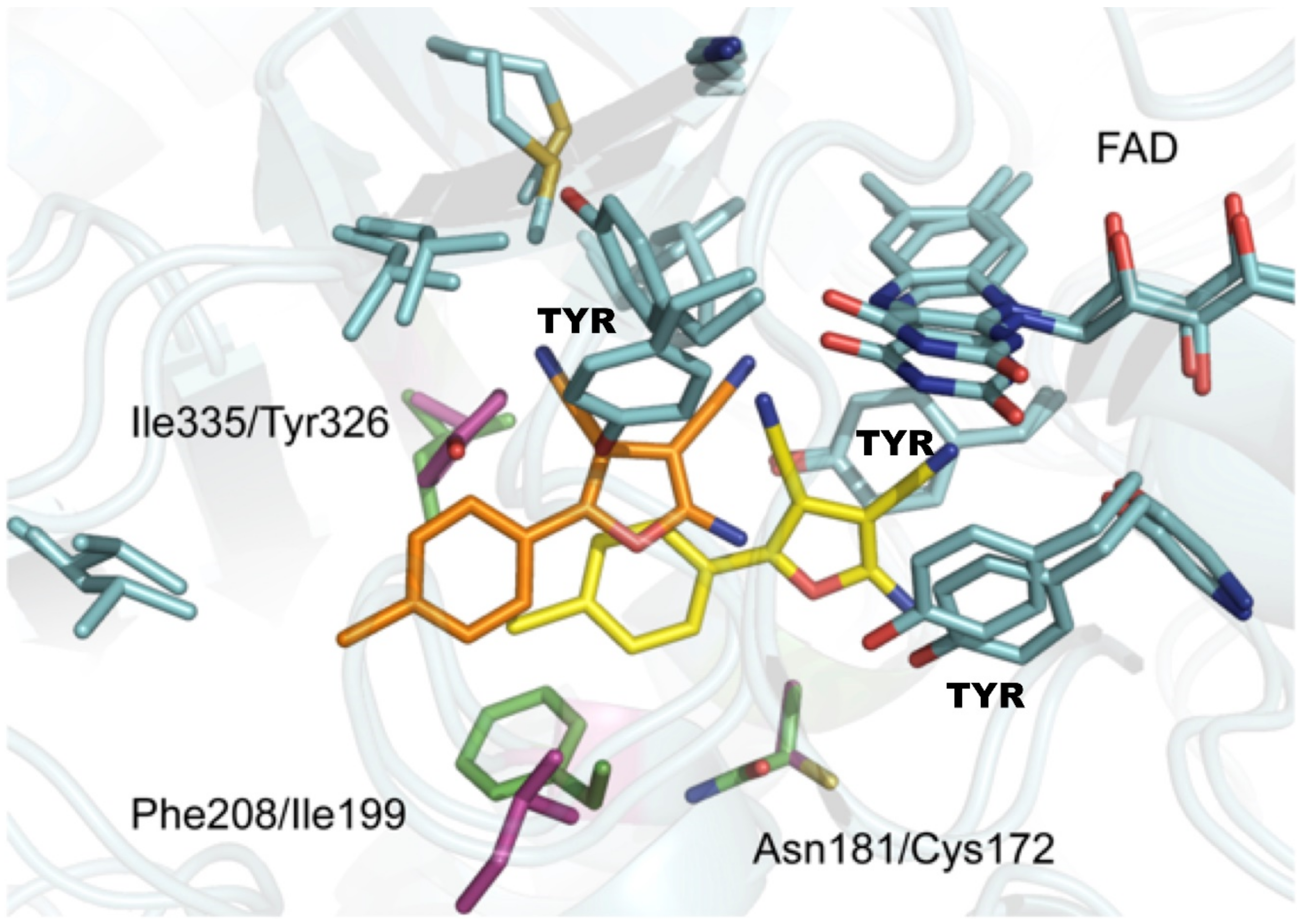
- Juarez-Jimenez, J.; Mendes, E.; Galdeano, C.; Martins, C.; Silva, D.B.; Marco-Contelles, J.; Carreiras, M.d.C.; Javier Luque, F.; Ramsay, R.R. Exploring the structural basis of the selective inhibition of Monoamine Oxidase A by dicarbonitrile aminoheterocycles: Role of Asn181 and Ile335 validated by spectroscopic and computational studies. Biochim. Biophys. Acta-Proteins Proteom. 2014, 1844, 389–397. [Google Scholar] [CrossRef]
- Ramsay, R.R.; Tipton, K.F. Assessment of Enzyme Inhibition: A Review with Examples from the Development of Monoamine Oxidase and Cholinesterase Inhibitory Drugs. Molecules 2017, 22, 1192. [Google Scholar] [CrossRef]
- Husain, M.; Edmondson, D.E.; Singer, T.P. Kinetic-studies on the catalytic mechanism of liver Monoamine- Oxidase. Biochemistry 1982, 21, 595–600. [Google Scholar] [CrossRef] [PubMed]
- Tan, A.K.; Ramsay, R.R. Substrate-specific enhancement of the oxidative half-reaction of Monoamine-Oxidase. Biochemistry 1993, 32, 2137–2143. [Google Scholar] [CrossRef]
- Walker, M.C.; Edmondson, D.E. Structure-Activity-Relationships in the Oxidation of Benzylamine Analogs by Bovine Liver Mitochondrial Monoamine- Oxidase-B. Biochemistry 1994, 33, 7088–7098. [Google Scholar] [CrossRef] [PubMed]
- Nandigama, R.K.; Edmondson, D.E. Influence of FAD structure on its binding and activity with the C406A mutant of recombinant human liver monoamine oxidase A. J. Biol. Chem. 2000, 275, 20527–20532. [Google Scholar] [CrossRef] [PubMed]
- Dunn, R.V.; Marshall, K.R.; Munro, A.W.; Scrutton, N.S. The pH dependence of kinetic isotope effects in monoamine oxidase A indicates stabilization of the neutral amine in the enzyme-substrate complex. FEBS J. 2008, 275, 3850–3858. [Google Scholar] [CrossRef] [PubMed]
- Edmondson, D. Hydrogen peroxide produced by mitochondrial monoamine oxidase catalysis: Biological implications. Curr. Pharm. Des. 2014, 20, 155–160. [Google Scholar] [CrossRef]
- Romero, E.; Castellanos, J.R.G.; Gadda, G.; Fraaije, M.W.; Mattevi, A. Same Substrate, Many Reactions: Oxygen Activation in Flavoenzymes. Chem. Rev. 2018, 118, 1742–1769. [Google Scholar] [CrossRef]
- Baron, R.; Riley, C.; Chenprakhon, P.; Thotsaporn, K.; Winter, R.T.; Alfieri, A.; Forneris, F.; van Berkel, W.J.H.; Chaiyen, P.; Fraaije, M.W.; et al. Multiple pathways guide oxygen diffusion into flavoenzyme active sites. Proc. Natl. Acad. Sci. USA 2009, 106, 10603–10608. [Google Scholar] [CrossRef]
- Gadda, G. Oxygen Activation in Flavoprotein Oxidases: The Importance of Being Positive. Biochemistry 2012, 51, 2662–2669. [Google Scholar] [CrossRef]
- Chen, L.; Lyubimov, A.Y.; Brammer, L.; Vrielink, A.; Sampson, N.S. The binding and release of oxygen and hydrogen peroxide are directed by a hydrophobic tunnel in cholesterol oxidase. Biochemistry 2008, 47, 5368–5377. [Google Scholar] [CrossRef]
- Vrielink, A.; Ghisla, S. Cholesterol oxidase: Biochemistry and structural features. FEBS J. 2009, 276, 6826–6843. [Google Scholar] [CrossRef]
- Houslay, M.D.; Tipton, K.F. Reaction Pathway of Membrane-Bound Rat-Liver Mitochondrial Monoamine-Oxidase. Biochem. J. 1973, 135, 735–750. [Google Scholar] [CrossRef] [PubMed]
- Houslay, M.D.; Tipton, K.F. Rat-Liver Mitochondrial Monoamine-Oxidase—Change in Reaction-Mechanism on Solubilization. Biochem. J. 1975, 145, 311–321. [Google Scholar] [CrossRef][Green Version]
- Pearce, L.B.; Roth, J.A. Human-Brain Monoamine-Oxidase Type-B—Mechanism of Deamination as Probed by Steady-State Methods. Biochemistry 1985, 24, 1821–1826. [Google Scholar] [CrossRef] [PubMed]
- Ramsay, R.R. Kinetic mechanism of Monoamine Oxidase-A. Biochemistry 1991, 30, 4624–4629. [Google Scholar] [CrossRef] [PubMed]
- Edmondson, D.E.; Bhattacharyya, A.K.; Walker, M.C. Spectral and kinetic studies of imine product formation in the oxidation of p-(N,N-dimethylamino)benzylamine analogs by monoamine oxidase-B. Biochemistry 1993, 32, 5196–5202. [Google Scholar] [CrossRef] [PubMed]
- Woo, J.C.G.; Silverman, R.B. Monoamine oxidase B catalysis in low aqueous medium. Direct evidence for an imine product. J. Am. Chem. Soc. 1995, 117, 1663–1664. [Google Scholar] [CrossRef]
- Binda, C.; Mattevi, A.; Edmondson, D.E. Structural properties of human monoamine oxidases A and B. Int. Rev. Neurobiol. 2011, 100, 1–11. [Google Scholar]
- Dasgupta, S.; Mukherjee, S.; Mukhopadhyay, B.P.; Banerjee, A.; Mishra, D.K. Recognition dynamics of dopamine to human Monoamine oxidase B: Role of Leu171/Gln206 and conserved water molecules in the active site cavity. J. Biomol. Struct. Dyn. 2018, 36, 1439–1462. [Google Scholar] [CrossRef]
- Binda, C.; Hubalek, F.; Li, M.; Herzig, Y.; Sterling, J.; Edmondson, D.E.; Mattevi, A. Crystal structures of monoamine oxidase B in complex with four inhibitors of the N-propargylaminoindan class. J. Med. Chem. 2004, 47, 1767–1774. [Google Scholar] [CrossRef] [PubMed]
- Borštnar, R.; Repič, M.; Kamerlin, S.C.L.; Vianello, R.; Mavri, J. Computational Study of the pKa Values of Potential Catalytic Residues in the Active Site of Monoamine Oxidase B. J. Chem. Theory Comput. 2012, 8, 3864–3870. [Google Scholar] [CrossRef]
- Ramsay, R.; Olivieri, A.; Holt, A. An improved approach to steady-state analysis of monoamine oxidases. J. Neural. Transm. 2011, 118, 1003–1019. [Google Scholar] [CrossRef] [PubMed]
- Holt, A. On the practical aspects of characterising monoamine oxidase inhibition in vitro. J. Neural. Transm. 2018, 125, 1685–1705. [Google Scholar] [CrossRef] [PubMed]
- Carreau, A.; El Hafny-Rahbi, B.; Matejuk, A.; Grillon, C.; Kieda, C. Why is the partial oxygen pressure of human tissues a crucial parameter? Small molecules and hypoxia. J. Cell. Mol. Med. 2011, 15, 1239–1253. [Google Scholar] [CrossRef] [PubMed]
- Nandigama, R.K.; Edmondson, D.E. Structure-activity relations in the oxidation of phenethylamine analogues by recombinant human liver monoamine oxidase A. Biochemistry 2000, 39, 15258–15265. [Google Scholar] [CrossRef] [PubMed]
- Fitzpatrick, P.F. Insights into the mechanisms of flavoprotein oxidases from kinetic isotope effects. J. Label. Compd. Radiopharm. 2007, 50, 1016–1025. [Google Scholar] [CrossRef]
- Wang, J.; Edmondson, D.E. Topological Probes of Monoamine Oxidases A and B in Rat Liver Mitochondria: Inhibition by TEMPO-Substituted Pargyline Analogues and Inactivation by Proteolysis. Biochemistry 2011, 50, 2499–2505. [Google Scholar] [CrossRef]
- Prah, A.; Ogrin, P.; Mavri, J.; Stare, J. Nuclear quantum effects in enzymatic reactions: Simulation of the kinetic isotope effect of phenylethylamine oxidation catalyzed by monoamine oxidase A. Phys. Chem. Chem. Phys. 2020, 22, 6838–6847. [Google Scholar] [CrossRef]
- Sablin, S.O.; Ramsay, R.R. Substrates but not inhibitors alter the redox potentials of monoamine oxidases. Antioxid. Redox Signal. 2001, 3, 723–729. [Google Scholar] [CrossRef]
- Edmondson, D.E.; Newton-Vinson, P. The covalent FAD of monoamine oxidase: Structural and functional role and mechanism of the flavinylation reaction. Antioxid. Redox Signal. 2001, 3, 789–806. [Google Scholar] [CrossRef] [PubMed]
- Walsh, J.D.; Miller, A.F. Flavin reduction potential tuning by substitution and bending. J. Mol. Struct. 2003, 623, 185–195. [Google Scholar] [CrossRef]
- Hynson, R.; Kelly, S.; Price, N.; Ramsay, R. Conformational changes in monoamine oxidase A in response to ligand binding or reduction. Biochim. Biophys. Acta-Gen. Subj. 2004, 1672, 60–66. [Google Scholar] [CrossRef] [PubMed]
- Yu, P.H.; Bailey, B.A.; Durden, D.A.; Boulton, A.A. Stereospecific deuterium substitution at the alpha-carbon position of dopamine and its effect on oxidative deamination catalyzed by MAO-A and MAO-B from different tissues. Biochem. Pharm. 1986, 35, 1027–1036. [Google Scholar] [CrossRef]
- Yu, P.H.; Davis, B.A. Inversion of selectivity of N-substituted propargylamine monoamine oxidase inhibitors following structural modifications to quaternary salts. Int. J. Biochem. Cell Biol. 1999, 31, 1391–1397. [Google Scholar] [CrossRef]
- Edmondson, D.E.; Binda, C.; Wang, J.; Upadhyay, A.K.; Mattevi, A. Molecular and mechanistic properties of the membrane-bound mitochondrial monoamine oxidases. Biochemistry 2009, 48, 4220–4230. [Google Scholar] [CrossRef]
- Geha, R.M.; Rebrin, I.; Chen, K.; Shih, J.C. Substrate and inhibitor specificities for human monoamine oxidase A and B are influenced by a single amino acid. J. Biol. Chem. 2001, 276, 9877–9882. [Google Scholar] [CrossRef]
- Ma, J.C.; Yoshimura, M.; Yamashita, E.; Nakagawa, A.; Ito, A.; Tsukihara, T. Structure of rat monoamine oxidase A and its specific recognitions for substrates and inhibitors. J. Mol. Biol. 2004, 338, 103–114. [Google Scholar] [CrossRef]
- Milczek, E.M.; Binda, C.; Rovida, S.; Mattevi, A.; Edmondson, D.E. The ‘gating’ residues Ile199 and Tyr326 in human monoamine oxidase B function in substrate and inhibitor recognition. FEBS J. 2011, 278, 4860–4869. [Google Scholar] [CrossRef]
- Geha, R.M.; Chen, K.; Shih, J.C. Phe(208) and Ile(199) in human Monoamine Oxidase A and B do not determine substrate and inhibitor specificities as in rat. J. Neurochem. 2000, 75, 1304–1309. [Google Scholar] [CrossRef] [PubMed]
- Hong, R.; Li, X. Discovery of monoamine oxidase inhibitors by medicinal chemistry approaches. MedChemComm 2019, 10, 10–25. [Google Scholar] [CrossRef] [PubMed]
- Rehuman, N.A.; Mathew, B.; Jat, R.K.; Nicolotti, O.; Kim, H. A Comprehensive Review of Monoamine Oxidase-A Inhibitors in their Syntheses and Poteneies. Comb. Chem. High Throughput Screen. 2020, 23, 898–914. [Google Scholar] [CrossRef]
- Wu, S.-M.; Qiu, X.-Y.; Liu, S.-J.; Sun, J. Single Heterocyclic Compounds as Monoamine Oxidase Inhibitors: From Past to Present. Mini-Rev. Med. Chem. 2020, 20, 908–920. [Google Scholar] [CrossRef]
- Becerra-Hernandez, A.; Galindo-de-la-Rosa, J.; Martinez-Pimentel, Y.; Ledesma-Garcia, J.; Alvarez-Contreras, L.; Guerra-Balcazar, M.; Aguilar-Elguezabal, A.; Alvarez, A.; Chavez-Ramirez, A.U.; Vallejo-Becerra, V. Novel biomaterial based on monoamine oxidase-A and multi-walled carbon nanotubes for serotonin detection. Biochem. Eng. J. 2019, 149, 107240. [Google Scholar] [CrossRef]
- Kacar, C.; Erden, P.E.; Dalkiran, B.; Inal, E.K.; Kilic, E. Amperometric biogenic amine biosensors based on Prussian blue, indium tin oxide nanoparticles and diamine oxidase- or monoamine oxidase-modified electrodes. Anal. Bioanal. Chem. 2020, 412, 1933–1946. [Google Scholar] [CrossRef]
- Ramsay, R.R.; Dunford, C.; Gillman, P.K. Methylene blue and serotonin toxicity: Inhibition of monoamine oxidase A (MAO A) confirms a theoretical prediction. Br. J. Pharmacol. 2007, 152, 946–951. [Google Scholar] [CrossRef]
- Delport, A.; Harvey, B.H.; Petzer, A.; Petzer, J.P. The monoamine oxidase inhibition properties of selected structural analogues of methylene blue. Toxicol. Appl. Pharmacol. 2017, 325, 1–8. [Google Scholar] [CrossRef]
- Hiraka, K.; Tsugawa, W.; Sode, K. Alteration of Electron Acceptor Preferences in the Oxidative Half-Reaction of Flavin-Dependent Oxidases and Dehydrogenases. Int. J. Mol. Sci. 2020, 21, 3797. [Google Scholar] [CrossRef] [PubMed]
- Iacovino, L.G.; Manzella, N.; Resta, J.; Vanoni, M.A.; Rotilio, L.; Pisani, L.; Edmondson, D.E.; Parini, A.; Mattevi, A.; Mialet-Perez, J.; et al. Rational Redesign of Monoamine Oxidase A into a Dehydrogenase to Probe ROS in Cardiac Aging. ACS Chem. Biol. 2020, 15, 1795–1800. [Google Scholar] [CrossRef]
- Zajkoska, P.; Rosenberg, M.; Heath, R.; Malone, K.J.; Stloukal, R.; Turner, N.J.; Rebros, M. Immobilised whole-cell recombinant monoamine oxidase biocatalysis. Appl. Microbiol. Biotechnol. 2015, 99, 1229–1236. [Google Scholar] [CrossRef] [PubMed]
- Batista, V.F.; Galman, J.L.; Pinto, D.C.G.A.; Silva, A.M.S.; Turner, N.J. Monoamine Oxidase: Tunable Activity for Amine Resolution and Functionalization. ACS Catal. 2018, 8, 11889–11907. [Google Scholar] [CrossRef]
- Markosova, K.; Camattari, A.; Rosenberg, M.; Glieder, A.; Turner, N.J.; Rebros, M. Cloning and upscale production of monoamine oxidase N (MAO-N D5) by Pichia pastoris. Biotechnol. Lett. 2018, 40, 127–133. [Google Scholar] [CrossRef]
- Toscani, A.; Risi, C.; Black, G.W.; Brown, N.L.; Shaaban, A.; Turner, N.J.; Castagnolo, D. Monoamine Oxidase (MAO-N) Whole Cell Biocatalyzed Aromatization of 1,2,5,6-Tetrahydropyridines into Pyridines. ACS Catal. 2018, 8, 8781–8787. [Google Scholar] [CrossRef]
- Lauder, K.; Masci, D.; Toscani, A.; Al Mekdad, A.; Black, G.W.; Brown, N.L.; Turner, N.J.; Luisi, R.; Castagnolo, D. A facile and regioselective multicomponent synthesis of chiral aryl-1,2-mercaptoamines in water followed by monoamine oxidase (MAO-N) enzymatic resolution. Org. Biomol. Chem. 2019, 17, 8982–8986. [Google Scholar] [CrossRef] [PubMed]
- Knez, D.; Colettis, N.; Iacovino, L.G.; Sova, M.; Pišlar, A.; Konc, J.; Lešnik, S.; Higgs, J.; Kamecki, F.; Mangialavori, I.; et al. Stereoselective Activity of 1-Propargyl-4-styrylpiperidine-like Analogues That Can Discriminate between Monoamine Oxidase Isoforms A and B. J. Med. Chem. 2020, 63, 1361–1387. [Google Scholar] [CrossRef] [PubMed]
- Carradori, S.; Secci, D.; Petzer, J.P. MAO inhibitors and their wider applications: A patent review. Expert Opin. Ther. Pat. 2018, 28, 211–226. [Google Scholar] [CrossRef] [PubMed]
- Ramsay, R.R.; Bradley, R.J.E.; Giurato, L.; Guccione, S. The shape of flavin in monoamine oxidase. In Flavins and Flavoproteins 2008; Frago, S., Gomez-Moreno, C., Medina, M., Eds.; Prensas Universitarias de Zaragoza: Zaragoza, Spain, 2008; pp. 183–186. [Google Scholar]
- Eisenreich, W.; Kemter, K.; Bacher, A.; Mulrooney, S.B.; Williams, C.H., Jr.; Müller, F. 13C-, 15N- and 31P-NMR studies of oxidized and reduced low molecular mass thioredoxin reductase and some mutant proteins. Eur. J. Biochem. 2004, 271, 1437–1452. [Google Scholar] [CrossRef] [PubMed]
- Rojas, R.J.; Edmondson, D.E.; Almos, T.; Scott, R.; Massari, M.E. Reversible and irreversible small molecule inhibitors of monoamine oxidase B (MAO-B) investigated by biophysical techniques. Bioorganic Med. Chem. 2015, 23, 770–778. [Google Scholar] [CrossRef] [PubMed]
- Albreht, A.; Vovk, I.; Mavri, J.; Marco-Contelles, J.; Ramsay, R.R. Evidence for a Cyanine Link Between Propargylamine Drugs and Monoamine Oxidase Clarifies the Inactivation Mechanism. Front. Chem. 2018, 6, 169. [Google Scholar] [CrossRef]
- Upadhyay, A.K.; Borbat, P.P.; Wang, J.; Freed, J.H.; Edmondson, D.E. Determination of the oligomeric states of human and rat monoamine oxidases in the outer mitochondrial membrane and octyl beta-D-glucopyranoside micelles using pulsed dipolar electron spin resonance spectroscopy. Biochemistry 2008, 47, 1554–1566. [Google Scholar] [CrossRef] [PubMed]
- Esteban, G.; Allan, J.; Samadi, A.; Mattevi, A.; Unzeta, M.; Marco-Contelles, J.; Binda, C.; Ramsay, R.R. Kinetic and structural analysis of the irreversible inhibition of human monoamine oxidases by ASS234, a multi-target compound designed for use in Alzheimer’s disease. Biochim. Biophys. Acta-Proteins Proteom. 2014, 1844, 1104–1110. [Google Scholar] [CrossRef] [PubMed]
- Upadhyay, A.K.; Wang, J.; Edmondson, D.E. Comparison of the structural properties of the active site cavities of human and rat monoamine oxidase A and B in their soluble and membrane-bound forms. Biochemistry 2008, 47, 526–536. [Google Scholar] [CrossRef] [PubMed]
- Son, S.Y.; Ma, A.; Kondou, Y.; Yoshimura, M.; Yamashita, E.; Tsukihara, T. Structure of human Monoamine Oxidase A at 2.2-angstrom resolution: The control of opening the entry for substrates/inhibitors. Proc. Natl. Acad. Sci. USA 2008, 105, 5739–5744. [Google Scholar] [CrossRef] [PubMed]
- Apostolov, R.; Yonezawa, Y.; Standley, D.M.; Kikugawa, G.; Takano, Y.; Nakamura, H. Membrane Attachment Facilitates Ligand Access to the Active Site in Monoamine Oxidase A. Biochemistry 2009, 48, 5864–5873. [Google Scholar] [CrossRef]
- Allen, W.J.; Bevan, D.R. Steered Molecular Dynamics Simulations Reveal Important Mechanisms in Reversible Monoamine Oxidase B Inhibition. Biochemistry 2011, 50, 6441–6454. [Google Scholar] [CrossRef]
- Jones, H.B.L.; Crean, R.M.; Mullen, A.; Kendrick, E.G.; Bull, S.D.; Wells, S.A.; Carbery, D.R.; MacMillan, F.; van der Kamp, M.W.; Pudney, C.R. Exposing the Interplay Between Enzyme Turnover, Protein Dynamics, and the Membrane Environment in Monoamine Oxidase B. Biochemistry 2019, 58, 2362–2372. [Google Scholar] [CrossRef]
- Burlakova, E.B.; Kayrane, C.B.; Molochkina, E.M.; Khokhlova, A.P. Modifications of external mitochondrial-membrane lipids in mice liver and kinetic patterns of membrane-bound monoamine-oxidase in vivo and in vitro. Vopr. Meditsinskoi Khimii 1984, 30, 66–72. [Google Scholar]
- Ekstedt, B.; Oreland, L. Effect of lipid-depletion on different forms of monoamine-oxidase in rat-liver mitochondria. Biochem. Pharmacol. 1976, 25, 119–124. [Google Scholar] [CrossRef]
- Houslay, M.D. Lipid substitution of mitochondrial monoamine-oxidase can lead to the abolition of clorgyline selective-inhibition without alteration in the A-B ratio assessed by substrate utilization. Biochem. Pharmacol. 1980, 29, 3211–3213. [Google Scholar] [CrossRef]
- Medvedev, A.; Kirkel, A.; Kamyshanskaya, N.; Gorkin, V. Lipid-peroxidation affects catalytic properties of rat-liver mitochondrial Monoamine Oxidases and their sensitivity to proteolysis. Int. J. Biochem. 1993, 25, 1791–1799. [Google Scholar] [CrossRef]
- Huang, R.H. Lipid-protein interactions in the multiple forms of monoamine-oxidase—Enzymatic and electron-spin-resonance studies with purified intact rat-brain mitochondria. Mol. Pharmacol. 1980, 17, 192–198. [Google Scholar]
- Kearney, E.B.; Salach, J.I.; Walker, W.H.; Seng, R.; Singer, T.P. Structure of the covalently bound flavin of monoamine oxidase. Biochem. Biophys. Res. Commun. 1971, 42, 490–496. [Google Scholar] [CrossRef]
- Hubalek, F.; Pohl, J.; Edmondson, D.E. Structural comparison of human monoamine oxidases A and B—Mass spectrometry monitoring of cysteine reactivities. J. Biol. Chem. 2003, 278, 28612–28618. [Google Scholar] [CrossRef]
- Paulsen, C.E.; Carroll, K.S. Cysteine-Mediated Redox Signaling: Chemistry, Biology, and Tools for Discovery. Chem. Rev. 2013, 113, 4633–4679. [Google Scholar] [CrossRef] [PubMed]
- Nietzel, T.; Mostertz, J.; Hochgraefe, F.; Schwarzlaender, M. Redox regulation of mitochondrial proteins and proteomes by cysteine thiol switches. Mitochondrion 2017, 33, 72–83. [Google Scholar] [CrossRef] [PubMed]
- Go, Y.-M.; Chandler, J.D.; Jones, D.P. The cysteine proteome. Free Radic. Biol. Med. 2015, 84, 227–245. [Google Scholar] [CrossRef] [PubMed]
- Weyler, W.; Hsu, Y.P.P.; Breakefield, X.O. Biochemistry and genetics of monoamine-oxidase. Pharmacol. Ther. 1990, 47, 391–417. [Google Scholar] [CrossRef]
- Hubalek, F.; Edmondson, D.E. Thiol reactivities as probes of MAO A and MAO B structure and function. In Flavins and Flavoproteins 2002; Chapman, S.K., Perham, R.N., Scrutton, N.S., Eds.; Rudolf Weber: Berlin, Germany, 2002; pp. 217–222. [Google Scholar]
- Vintem, A.; Price, N.; Silverman, R.; Ramsay, R. Mutation of surface cysteine 374 to alanine in monoamine oxidase A alters substrate turnover and inactivation by cyclopropylamines. Bioorganic Med. Chem. 2005, 13, 3487–3495. [Google Scholar] [CrossRef] [PubMed]
- Wu, H.F.; Chen, K.; Shih, J.C. Site-directed mutagenesis of Monoamine Oxidase-A and Oxidase-B—Role of cysteines. Mol. Pharmacol. 1993, 43, 888–893. [Google Scholar]
- Zhong, B.Y.; Silverman, R.B. Identification of the active site cysteine in bovine liver monoamine oxidase B. J. Am. Chem. Soc. 1997, 119, 6690–6691. [Google Scholar] [CrossRef]
- Murphy, M.P. Mitochondrial Thiols in Antioxidant Protection and Redox Signaling: Distinct Roles for Glutathionylation and Other Thiol Modifications. Antioxid. Redox Signal. 2012, 16, 476–495. [Google Scholar] [CrossRef] [PubMed]
- Alves-Figueiredo, H.; Silva-Platas, C.; Lozano, O.; Vazquez-Garza, E.; Guerrero-Beltran, C.E.; Zarain-Herzberg, A.; Garcia-Rivas, G. A systematic review of post-translational modifications in the mitochondrial permeability transition pore complex associated with cardiac diseases. Biochim. Biophys. Acta-Mol. Basis Dis. 2021, 1867, 165992. [Google Scholar] [CrossRef] [PubMed]
- Cao, X.; Li, X.-M.; Mousseau, D.D. Calcium alters monoamine oxidase-A parameters in human cerebellar and rat glial C6 cell extracts: Possible influence by distinct signalling pathways. Life Sci. 2009, 85, 262–268. [Google Scholar] [CrossRef] [PubMed]
- Egashira, T.; Sakai, K.; Sakurai, M.; Takayama, F. Calcium disodium edetate enhances type A monoamine oxidase activity in monkey brain. Biol. Trace Elem. Res. 2003, 94, 203–211. [Google Scholar] [CrossRef]
- Kosenko, E.A.; Venediktova, N.I.; Kaminsky, Y.G. Calcium and ammonia stimulate Monoamine Oxidase A activity in brain mitochondria. Biol. Bull. 2003, 30, 449–452. [Google Scholar] [CrossRef]
- Samantaray, S.; Chandra, G.; Mohanakumar, K.P. Calcium channel agonist, (+/−)-Bay K8644, causes a transient increase in striatal monoamine oxidase activity in Balb/c mice. Neurosci. Lett. 2003, 342, 73–76. [Google Scholar] [CrossRef]
- Ramsay, R.R.; Armstrong, S. Calcium ions alter monoamine oxidase A activity. In Flavins and Flavoproteins 2011; Miller, S.H.R., Ed.; Lulu Press: Raleigh, NC, USA, 2011. [Google Scholar]
- Cao, X.; Wei, Z.; Gabriel, G.G.; Li, X.; Mousseau, D.D. Calcium-sensitive regulation of monoamine oxidase-A contributes to the production of peroxyradicals in hippocampal cultures: Implications for Alzheimer disease-related pathology. BMC Neurosci. 2007, 8, 73. [Google Scholar] [CrossRef]
- Pennington, P.R.; Wei, Z.; Rui, L.; Doig, J.A.; Graham, B.; Kuski, K.; Gabriel, G.G.; Mousseau, D.D. Alzheimer disease-related presenilin-1 variants exert distinct effects on monoamine oxidase-A activity in vitro. J. Neural. Transm. 2011, 118, 987–995. [Google Scholar] [CrossRef]
- Wei, Z.; Gabriel, G.G.; Rui, L.; Cao, X.; Pennington, P.R.; Chlan-Fourney, J.; Nazarali, A.J.; Baker, G.B.; Mousseau, D.D. Monoamine Oxidase-A Physically Interacts with Presenilin-1(M146V) in the Mouse Cortex. J. Alzheimers Dis. 2012, 28, 403–422. [Google Scholar] [CrossRef]
- Shih, J.C.; Chen, K. MAO-A and -B gene knock-out mice exhibit distinctly different behavior. Neurobiology 1999, 7, 235–246. [Google Scholar] [PubMed]
- Godar, S.C.; Fite, P.J.; McFarlin, K.M.; Bortolato, M. The role of monoamine oxidase A in aggression: Current translational developments and future challenges. Prog. Neuro-Psychopharmacol. Biol. Psychiatry 2016, 69, 90–100. [Google Scholar] [CrossRef] [PubMed]
- Naoi, M.; Maruyama, W.; Shamoto-Nagai, M. Type A and B monoamine oxidases distinctly modulate signal transduction pathway and gene expression to regulate brain function and survival of neurons. J. Neural Transm. 2018, 125, 1635–1650. [Google Scholar] [CrossRef]
- Cheng, Y.; Buchan, M.; Vitanova, K.; Aitken, L.; Gunn-Moore, F.J.; Ramsay, R.R.; Doherty, G. Neuroprotective actions of leptin facilitated through balancing mitochondrial morphology and improving mitochondrial function. J. Neurochem. 2020, 155, 191–206. [Google Scholar] [CrossRef] [PubMed]
- Quartey, M.O.; Nyarko, J.N.K.; Pennington, P.R.; Heistad, R.M.; Klassen, P.C.; Baker, G.B.; Mousseau, D.D. Alzheimer Disease and Selected Risk Factors Disrupt a Co-regulation of Monoamine Oxidase-A/B in the Hippocampus, but Not in the Cortex. Front. Neurosci. 2018, 12, 419. [Google Scholar] [CrossRef]
- Fitzgerald, J.C.; Ufer, C.; Billett, E.E. A link between monoamine oxidase-A and apoptosis in serum deprived human SH-SY5Y neuroblastoma cells. J. Neural Transm. 2007, 114, 807–810. [Google Scholar] [CrossRef] [PubMed]
- Rybaczyk, L.A.; Bashaw, M.J.; Pathak, D.R.; Huang, K. An indicator of cancer: Downregulation of Monoamine Oxidase-A in multiple organs and species. BMC Genom. 2008, 9, 134. [Google Scholar] [CrossRef]
- Li, P.C.; Siddiqi, I.N.; Mottok, A.; Loo, E.Y.; Wu, C.H.; Cozen, W.; Steidl, C.; Shih, J.C. Monoamine oxidase A is highly expressed in classical Hodgkin lymphoma. J. Pathol. 2017, 243, 220–229. [Google Scholar] [CrossRef]
- Wu, J.B.; Shao, C.; Li, X.; Li, Q.; Hu, P.; Shi, C.; Li, Y.; Chen, Y.T.; Yin, F.; Liao, C.P.; et al. Monoamine oxidase A mediates prostate tumorigenesis and cancer metastasis. J. Clin. Investig. 2014, 124, 2891–2908. [Google Scholar] [CrossRef]
- Gross, M.E.; Agus, D.B.; Dorff, T.B.; Pinski, J.K.; Quinn, D.I.; Castellanos, O.; Gilmore, P.; Shih, J.C. Phase 2 trial of monoamine oxidase inhibitor phenelzine in biochemical recurrent prostate cancer. Prostate Cancer Prostatic Dis. 2021, 24, 61–68. [Google Scholar] [CrossRef]
- Krueger, M.J.; Mazouz, F.; Ramsay, R.R.; Milcent, R.; Singer, T.P. Dramatic species differences in the susceptibility of Monoamine-Oxidase-B to a group of powerful inhibitors. Biochem. Biophys. Res. Commun. 1995, 206, 556–562. [Google Scholar] [CrossRef]
- Egashira, T.; Takayama, F.; Yamanaka, Y. The inhibition of monoamine oxidase activity by various antidepressants: Differences found in various mammalian species. Jpn. J. Pharmacol. 1999, 81, 115–121. [Google Scholar] [CrossRef]
- Inoue, H.; Castagnoli, K.; van der Schyf, C.; Mabic, S.; Igarashi, K.; Castagnoli, N. Species-dependent differences in monoamine oxidase A and B-catalyzed oxidation of various C4 substituted 1-methyl-4-phenyl-1,2,3,6-tetrahydropyridinyl derivatives. J. Pharmacol. Exp. Ther. 1999, 291, 856–864. [Google Scholar] [PubMed]
- Ben Ramadan, Z.; Wrang, M.L.; Tipton, K.F. Species differences in the selective inhibition of monoamine oxidase (1-methyl-2-phenylethyl)hydrazine and its potentiation by cyanide. Neurochem. Res. 2007, 32, 1783–1790. [Google Scholar] [CrossRef] [PubMed]
- Yagodina, O.V.; Basova, I.N. Comparative enzymological study of catalytic properties of liver monoamine oxidases in frogs. J. Evol. Biochem. Physiol. 2010, 46, 350–356. [Google Scholar] [CrossRef]
- Senatori, O.; Pierucci, F.; Parvez, S.H.; Scopelliti, R.; Nicotra, A. Monoamine oxidase in Teleosts. Biog. Amines 2003, 17, 199–213. [Google Scholar] [CrossRef]
- Wang, J.; Edmondson, D.E. H-2 Kinetic Isotope Effects and pH Dependence of Catalysis as Mechanistic Probes of Rat Monoamine Oxidase A: Comparisons with the Human Enzyme. Biochemistry 2011, 50, 7710–7717. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Santillo, M.F.; Liu, Y.T.; Ferguson, M.; Vohra, S.N.; Wiesenfeld, P.L. Inhibition of monoamine oxidase (MAO) by beta-carbolines and their interactions in live neuronal (PC12) and liver (HuH-7 and MH1C1) cells. Toxicol. In Vitro 2014, 28, 403–410. [Google Scholar] [CrossRef]
- Brummett, B.H.; Krystal, A.D.; Siegler, I.C.; Kuhn, C.; Surwit, R.S.; Zuechner, S.; Ashley-Koch, A.; Barefoot, J.C.; Williams, R.B. Associations of a regulatory polymorphism of monoamine oxidase-A gene promoter (MAOA-uVNTR) with symptoms of depression and sleep quality. Psychosom. Med. 2007, 69, 396–401. [Google Scholar] [CrossRef]
- Kunugi, H.; Ishida, S.; Kato, T.; Tatsumi, M.; Sakai, T.; Hattori, M.; Hirose, T.; Nanko, S. A functional polymorphism in the promoter region of monoamine oxidase-A gene and mood disorders. Mol. Psychiatry 1999, 4, 393–395. [Google Scholar] [CrossRef] [PubMed]
- Manca, M.; Pessoa, V.; Lopez, A.I.; Harrison, P.T.; Miyajima, F.; Sharp, H.; Pickles, A.; Hill, J.; Murgatroyd, C.; Bubb, V.J.; et al. The Regulation of Monoamine Oxidase A Gene Expression by Distinct Variable Number Tandem Repeats. J. Mol. Neurosci. 2018, 64, 459–470. [Google Scholar] [CrossRef]
- Roohi, J.; DeVincent, C.; Hatchwell, E.; Gadow, K. Association of a Monoamine Oxidase-A Gene Promoter Polymorphism with ADHD and Anxiety in Boys with Autism Spectrum Disorder. J. Autism Dev. Disord. 2009, 39, 67–74. [Google Scholar] [CrossRef]
- Bortolato, M.; Floris, G.; Shih, J.C. From aggression to autism: New perspectives on the behavioral sequelae of monoamine oxidase deficiency. J. Neural Transm. 2018, 125, 1589–1599. [Google Scholar] [CrossRef]
- Pregeljc, D.; Jug, U.; Mavri, J.; Stare, J. Why does the Y326I mutant of monoamine oxidase B decompose an endogenous amphetamine at a slower rate than the wild type enzyme? Reaction step elucidated by multiscale molecular simulations. Phys. Chem. Chem. Phys. 2018, 20, 4181–4188. [Google Scholar] [CrossRef] [PubMed]
- Fowler, J.S.; Logan, J.; Shumay, E.; Alia-Klein, N.; Wang, G.-J.; Volkow, N.D. Monoamine oxidase: Radiotracer chemistry and human studies. J. Label. Compd. Radiopharm. 2015, 58, 51–64. [Google Scholar] [CrossRef] [PubMed]
- Fowler, J.S.; Logan, J.; Wang, G.-J.; Volkow, N.D.; Telang, F.; Ding, Y.-S.; Shea, C.; Garza, V.; Xu, Y.; Li, Z.; et al. Comparison of the binding of the irreversible monoamine oxidase tracers, [11C]clorgyline and [11C]l-deprenyl in brain and peripheral organs in humans. Nucl. Med. Biol. 2004, 31, 313–319. [Google Scholar] [CrossRef] [PubMed]
- Muller, T.; Mohr, J.D. Pharmacokinetics of monoamine oxidase B inhibitors in Parkinson’s disease: Current status. Expert Opin. Drug Metab. Toxicol. 2019, 15, 429–435. [Google Scholar] [CrossRef] [PubMed]
- Arnett, C.D.; Fowler, J.S.; MacGregor, R.R.; Schlyer, D.J.; Wolf, A.P.; Långström, B.; Halldin, C. Turnover of Brain Monoamine Oxidase Measured In Vivo by Positron Emission Tomography Using l-[11C]Deprenyl. J. Neurochem. 1987, 49, 527. [Google Scholar] [CrossRef] [PubMed]
- Naoi, M.; Maruyama, W.; Shamoto-Nagai, M. Rasagiline and selegiline modulate mitochondrial homeostasis, intervene apoptosis system and mitigate alpha-synuclein cytotoxicity in disease-modifying therapy for Parkinson’s disease. J. Neural Transm. 2020, 127, 131–147. [Google Scholar] [CrossRef]
- Müller, T. Pharmacokinetic drug evaluation of safinamide mesylate for the treatment of mid-to-late stage Parkinson’s disease. Expert Opin. Drug Metab. Toxicol. 2017, 13, 693–699. [Google Scholar] [CrossRef]
- Orayj, K.; Lane, E. Patterns and Determinants of Prescribing for Parkinson’s Disease: A Systematic Literature Review. Parkinsons Dis. 2019, 2019, 9237181. [Google Scholar] [CrossRef]
- Binde, C.D.; Tvete, I.F.; Gasemyr, J.; Natvig, B.; Klemp, M. A multiple treatment comparison meta-analysis of monoamine oxidase type B inhibitors for Parkinson’s disease. Br. J. Clin. Pharmacol. 2018, 84, 1917–1927. [Google Scholar] [CrossRef]
- Gillman, P.K. A reassessment of the safety profile of monoamine oxidase inhibitors: Elucidating tired old tyramine myths. J. Neural. Transm. 2018, 125, 1707–1717. [Google Scholar] [CrossRef]
- Gillman, P.K.; Feinberg, S.S.; Fochtmann, L.J. Revitalizing monoamine oxidase inhibitors: A call for action. CNS Spectr. 2020, 25, 452–454. [Google Scholar] [CrossRef] [PubMed]
- Mathew, B.; Carradori, S.; Guglielmi, P.; Uddin, M.S.; Kim, H. New Aspects of Monoamine Oxidase B Inhibitors: The Key Role of Halogens to Open the Golden Door. Curr. Med. Chem. 2021, 28, 266–283. [Google Scholar] [CrossRef] [PubMed]
- Mellado, M.; Mella, J.; Gonzalez, C.; Vina, D.; Uriarte, E.; Matos, M.J. 3-Arylcoumarins as highly potent and selective monoamine oxidase B inhibitors: Which chemical features matter? Bioorganic Chem. 2020, 101, 103964. [Google Scholar] [CrossRef] [PubMed]
- Stefanachi, A.; Leonetti, F.; Pisani, L.; Catto, M.; Carotti, A. Coumarin: A Natural, Privileged and Versatile Scaffold for Bioactive Compounds. Molecules 2018, 23, 250. [Google Scholar] [CrossRef] [PubMed]
- Mladenović, M.; Patsilinakos, A.; Pirolli, A.; Sabatino, M.; Ragno, R. Understanding the Molecular Determinant of Reversible Human Monoamine Oxidase B Inhibitors Containing 2H-Chromen-2-One Core: Structure-Based and Ligand-Based Derived Three-Dimensional Quantitative Structure–Activity Relationships Predictive Models. J. Chem. Inf. Modeling 2017, 57, 787–814. [Google Scholar]
- Lutsenko, K.; Hagenow, S.; Affini, A.; Reiner, D.; Stark, H. Rasagiline derivatives combined with histamine H-3 receptor properties. Bioorganic Med. Chem. Lett. 2019, 29, 126612. [Google Scholar] [CrossRef] [PubMed]


| MAO A | Compound | MAO B | |||
|---|---|---|---|---|---|
| KM (μM) | Vmax (pmol.min−1.mg protein−1) | Substrates A | KM (μM) | Vmax (pmol.min−1.mg protein−1) | |
| 137 | 228 | Serotonin | 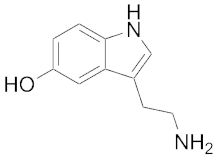 | 1093 | 6.6 |
| 212 | 680 | Dopamine | 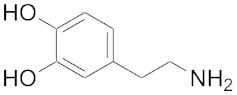 | 229 | 702 |
| 140 | 20 | 2-Phenylethylamine | 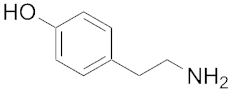 | 4 | 309 |
| 127 | 182 | Tyramine | 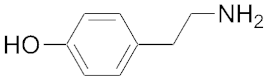 | 107 | 343 |
| Ki (μM) | Reversible Inhibitors B | Ki (μM) | |||
| 18 | D-Amphetamine | 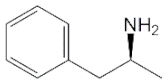 | 250 | ||
| 3 | MPP+ c | 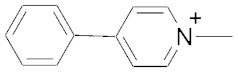 | 230 | ||
| 365 | Safinamide |  | 0.45 | ||
| 11.5 | Moclobemide | 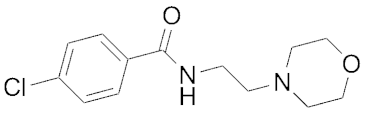 | >100 | ||
| 0.005 | Harmine | 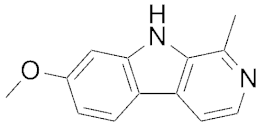 | 121 | ||
| 0.028 | Methylene Blue | 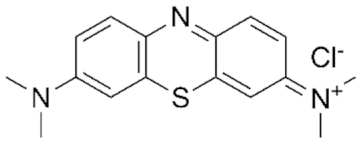 | 5.5 | ||
| KI (μM) | kinact (min−1) | Irreversible Inhibitors C | KI (μM) | kinact (min−1) | |
| 3.1 | 0.12 | Phenelzine | 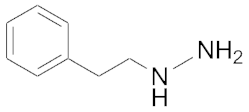 | 50 a | 0.9 a |
| 7.7 | 0.78 | Tranylcypromine | 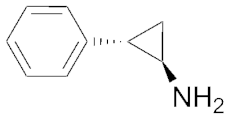 | 0.7 | 0.16 |
| 193 | 0.25 | Selegiline d | 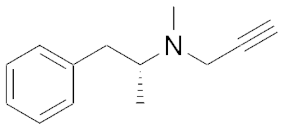 | 0.1 | 0.53 |
| 9.7 | 0.007 | Rasagiline b |  | 0.7 | 0.05 |
| 0.03 | 1.8 | Clorgyline | 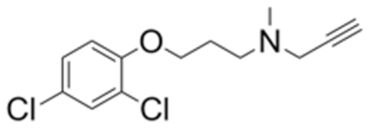 | 1.8 | 0.02 |
| 3.0 | 0.39 | Contilisant e | 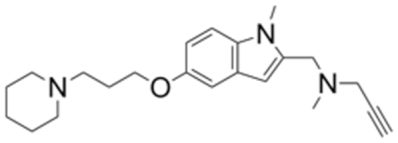 | 4.6 | 0.19 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Ramsay, R.R.; Albreht, A. Questions in the Chemical Enzymology of MAO. Chemistry 2021, 3, 959-978. https://doi.org/10.3390/chemistry3030069
Ramsay RR, Albreht A. Questions in the Chemical Enzymology of MAO. Chemistry. 2021; 3(3):959-978. https://doi.org/10.3390/chemistry3030069
Chicago/Turabian StyleRamsay, Rona R., and Alen Albreht. 2021. "Questions in the Chemical Enzymology of MAO" Chemistry 3, no. 3: 959-978. https://doi.org/10.3390/chemistry3030069
APA StyleRamsay, R. R., & Albreht, A. (2021). Questions in the Chemical Enzymology of MAO. Chemistry, 3(3), 959-978. https://doi.org/10.3390/chemistry3030069







