Abstract
Training-anomaly-based, machine-learning-based, intrusion detection systems (AMiDS) for use in critical Internet of Things (CioT) systems and military Internet of Things (MioT) environments may involve synthetic data or publicly simulated data due to data restrictions, data scarcity, or both. However, synthetic data can be unrealistic and potentially biased, and simulated data are invariably static, unrealistic, and prone to obsolescence. Building an AMiDS logical model to predict the deviation from normal behavior in MioT and CioT devices operating at the sensing or perception layer due to adversarial attacks often requires the model to be trained using current and realistic data. Unfortunately, while real-time data are realistic and relevant, they are largely imbalanced. Imbalanced data have a skewed class distribution and low-similarity index, thus hindering the model’s ability to recognize important features in the dataset and make accurate predictions. Data-driven learning using data sampling, resampling, and generative methods can lessen the adverse impact of a data imbalance on the AMiDS model’s performance and prediction accuracy. Generative methods enable passive adversarial learning. This paper investigates several data sampling, resampling, and generative methods. It examines their impacts on the performance and prediction accuracy of AMiDS models trained using imbalanced data drawn from the UNSW_2018_IoT_Botnet dataset, a publicly available IoT dataset from the IEEEDataPort. Furthermore, it evaluates the performance and predictability of these models when trained using data transformation methods, such as normalization and one-hot encoding, to cover a skewed distribution, data sampling and resampling methods to address data imbalances, and generative methods to train the models to increase the model’s robustness to recognize new but similar attacks. In this initial study, we focus on CioT systems and train PCA-based and oSVM-based AMiDS models constructed using low-complexity PCA and one-class SVM (oSVM) ML algorithms to fit an imbalanced ground truth IoT dataset. Overall, we consider the rare event prediction case where the minority class distribution is disproportionately low compared to the majority class distribution. We plan to use transfer learning in future studies to generalize our initial findings to the MioT environment. We focus on CioT systems and MioT environments instead of traditional or non-critical IoT environments due to the stringent low energy, the minimal response time constraints, and the variety of low-power, situational-aware (or both) things operating at the sensing or perception layer in a highly complex and open environment.
1. Introduction
Training an AMiDS using an imbalanced dataset, where normal examples appear more frequently than malicious ones in a non-hostile environment, is normal and often desirable. The nature of the real-time network traffic in a non-critical or non-hostile IoT system or environment typically contains more normal events than malicious ones. However, the real-time network flow can exhibit a rise in malicious events in a CioT system that involves deploying and operating critical infrastructure. Consequently, in highly hostile environments such as MioT, it is possible to encounter a network flow where adversarial examples are notably higher than non-adversarial examples. Nonetheless, there is a tendency to train AMiDS models to protect nodes in CioT systems and MioT environments using imbalanced datasets where malicious examples appear less frequently than non-malicious examples.
Training AMiDS models using more normal examples than malicious ones is desirable when dealing with anomaly detection cases such as fraud detection and predictive maintenance. However, in mission-critical and highly hostile environments, there is a high possibility that real-time network traffic will contain more malicious examples than normal ones. The author of [1] described this case as a rare event prediction case and defined rare events, which constitute less than 5% of the dataset, as occurring infrequently. Moreover, the degree of imbalance based on the proportion of the minority class can be extreme, moderate, or mild [2]. When the proportion of the minority class is less than 1%, the degree of imbalance is extreme. However, proportions between 1 and 20% and 20 and 40% signal moderate and mild degrees of class imbalance, respectively.
Therefore, in this study, we consider the hostile case where the dataset contains more malicious events than normal ones with a moderate degree of imbalance. Similar to the authors of [1], we call this hostile case a rare event prediction case. Therefore, we use these two terms (hostile and rare) interchangeably in this paper. Likewise, a non-hostile case is where normal events occur more frequently than malicious events. In doing so, our main objective is to improve the performance of the PCA-based and oSVM-based AMiDS in hostile environments using imbalanced learning and passive adversarial learning. Our disposition towards considering the hostile case stems from the fact that behavior-based IDS typically detects intrusions based on a deviation from normal behavior instead of a malicious one.
Usually, datasets for intrusion detection systems and malware analyses consist of logs and network traffic captures, where the number of malicious and constant connections generated by an intruder is less than the number of normal connections generated by non-malicious applications [3]. In the hostile case, our underlying assumption is that a traffic flow instance may exhibit more malicious events than normal ones. Whether considering a hostile or a non-hostile case, insufficient data and a data imbalance invariably result in a skewed data distribution and, hence, the model’s inability to recognize the most significant features. Most notably, it can lead to biased predictive models, inaccurate predictions, and a performance degradation [4]. Therefore, training an AMiDS to protect nodes in CioT and MioT using insufficient or imbalanced data or both can adversely impact the model’s predictability and detection rate. In a hostile environment, an AMiDS that fails to predict and accurately detect malicious events due to a biased classification can have a catastrophic impact when used to secure, for example, smart transportation, and inevitably, it can have a fatal impact when used to secure Internet of Battle Things (IoBT) devices used by connected soldiers.
Unlike imbalanced learning, purpose-driven learning, where models are trainable using imbalanced real-time or simulated data, invariably leads to inefficient logical models that neither make accurate predictions nor generalize to highly hostile environments. In contrast, data-driven learning processes, such as imbalanced learning, attempt to solve the data imbalance problem by enabling intelligent systems to overcome their bias when trained using datasets containing disproportionate classes of data examples or points [5]. Additionally, imbalanced learning, i.e., the ability to learn the minority class, focuses on the performance of intelligent models trained using an unequal data distribution and significantly skewed class distributions [6,7].
In general, imbalanced learning uses data sampling or resampling methods to produce sufficiently balanced data from the imbalanced real-time data via augmentation; hence, it can lessen the impact of otherwise intractable issues resulting from a data imbalance. This paper investigates data sampling and resampling and generative methods to address the imbalanced data problem when training AMiDS to protect the sensing or perception (two terms we interchangeably use) nodes in CioT and MioT. We mainly use oversampling and generative methods to evaluate the performance of low-complexity AMiDS models designed and trained using imbalanced and passive adversarial learning. Furthermore, we compare their performance when trained using ordinary learning.
When applying imbalanced learning using selected data sampling and data generative methods, we aim to generate sufficient, realistic, and balanced data to train low-complexity ML-based models, despite the data rarity and restrictive data sharing, to predict existing and new attacks correctly. Data rarity occurs due to insufficient examples associated with one or more classes in the training dataset [7]. While data generative methods such as autoencoders do not necessarily balance the dataset, they can learn the patterns between the normal and anomalous classes. Our study is interested in learning whether the AMiDS models can correctly predict new attacks and whether the learned data pattern leads to a better performance when using imbalanced data. First, we apply the autoencoder to the imbalanced dataset without sampling the data and then implement a variational autoencoder that uses oversampling to sample the dataset. In doing so, we seek to determine the predictive ability in the presence of new attack examples and the performance gain from using a hybrid approach that combines the benefits of an autoencoder and data sampling. The main contributions of this paper are as follows:
- To improve the performance of low-complexity AMiDS models, i.e., PCA-based and oSVM-based models, trained on imbalanced data using regular learning to detect intrusions in the CioT and MioT sensing layer;
- To provide insights into the benefits of imbalanced learning using sampling, resampling, and generative methods to support learning in critical, hostile, and complex data environments, where small data samples, dirty data, and highly cluttered data are inevitable;
- To explore the impact of generative methods on the resiliency of the AMiDS models and their ability to realize new but similar hostile events, a type of learning we refer to as passive adversarial learning.
The remainder of this paper is organized as follows: Section 2 provides the background preliminaries, and Section 3 reports related work. Section 4 provides an overview of the data sampling and resampling methods. Section 5 describes the research methodology. Section 6 describes the experimental setup. Section 7 provides the results as well as the analysis of the results. Section 8 presents the evaluation analysis and discusses the AMiDS models’ performances using imbalanced learning versus ordinary learning. Finally, Section 9 provides the conclusion and highlights future work.
2. Background Preliminaries
This section defines the preliminaries related to imbalanced learning, the CioT and MioT environments, and low-complexity AMiDS.
2.1. Imbalanced Learning
Imbalanced learning is part of the machine learning pipeline in which the trained models learn from sampled or resampled data initially containing a high degree of imbalance. While many issues result from algorithm learning using imbalanced data, we focus on learning issues resulting from data rarity. While data rarity can be absolute or relative, in this paper, we are concerned with the relative data rarity, where the weight of imbalance in the minority class is relative to that in the majority class. A minority class contains less frequent examples than the majority class [8]. However, depending on the underlying theory behind a typical IDS, the minority class is either a positive or negative class of data examples. For example, if the dataset contains more normal examples than malicious ones, the normal example class is the majority class, and the malicious example class is the minority class.
Given that the underlying IDS theory is for the IDS to detect intrusions based on deviations from normal behavior, the majority class is the negative class, and the minority class is the positive one. Therefore, a detected malicious example that belongs to the minority class is positive. In this study, the malicious examples are more frequent than the normal examples in the dataset. Nevertheless, the malicious examples belong to the positive class, even though the malicious class is, indeed, the majority class.
In imbalanced learning, the learning process involves many learning forms as well as learning tasks. For example, imbalanced learning involves supervised and unsupervised learning and applies to classification and clustering tasks [9]. In addition to the performance degradation and high error rates, training learning algorithms using imbalanced data can lead to biased outcomes and measurements. For example, imbalanced classifiers can result in an imbalanced degree of predictive accuracy in which the majority class is closer to an accuracy of 100%, and the minority class is nearer to an accuracy of 0–10%. Overall, there are three approaches to imbalanced learning: the data-level, algorithm-level, and hybrid [5,10,11,12].
Algorithm-level methods modify the algorithm directly to reduce the bias towards the minority class. Data-level methods address these issues by modifying the data imbalance in the training dataset to yield a balanced distribution among the minority and majority classes, such that the majority class does not significantly outnumber the minority class. Examples of data-level methods are thesampling and resampling methods, such as random oversampling and the Synthetic Minority Oversampling Technique (SMOTE). Algorithm-level methods include ensemble-based methods, for example, EasyEnsemble and SMOTEBoost, to list a few. Section 4 gives an inexhaustible overview of several data-level sampling and resampling methods for imbalanced learning.
2.2. CioT Systems and MioT Environments
This study uses CioT as an umbrella term to define IoT environments that support mission-critical infrastructures or systems for mere civilian use and non-combative military use, such as smart transportation, the energy grid, and manufacturing plants. An industrial Internet of Things (IioT) is an example of a CioT. Notably, sixteen defined critical infrastructure sectors in the US span various domains, including industrial base defense [13]. In a CioT system, the various nodes are non-combative things operating in a non-combative mode to support combative and non-combative missions and operations, mainly in non-hostile environments.
On the contrary, nodes in an MioT environment are mainly deployed in a combative mode to directly support combative operations, both logistic and tactical, in hostile environments. MioT environments generally consist of a mission-critical battlefield infrastructure for combative military operations. The Internet of the Battle Things, or IoBT, is an example of such an environment. The IoBT is a constellation of intelligent and marginally intelligent devices and equipment used in the battlefield [14]. CioT and MioT offer many benefits to support civilian and military tasks and critical operations. For example, a CioT system enables predictive analytics in operation, pricing, distribution, management, and maintenance, to list a few. In contrast, an MioT environment enables predictive battlefield analytics [15].
CioT involves an integrated network of homogeneous and heterogeneous sensors and smart devices connected using edge and cloud infrastructures to support non-combative mission-critical tasks and operations. However, MioT involves a network of homogeneous and heterogeneous battlefield devices, intelligent and marginally intelligent, and wearable devices that are connected to support combative and battlefield operations. In an MioT environment, fog, edge, and cloud computing connect the ubiquitous sensors and IoT devices worn by military personnel and embedded in military equipment to create a cohesive fighting force to increase the situational awareness, risk assessment, and response time [14,15,16]. However, MioT uses mobile edge computing where IoT end devices transmit the data via a mobile edge server to a mobile base station [17].
While a CioT tends to be stable and predictable with controlled or semi-controlled actions and reactions, an MioT is highly dynamic and unpredictable with rapid and controlled actions and reactions. The dynamic and unpredictable nature of an MioT environment dictates the need to dynamically learn and update the characteristics of the environment in real time. Figure 1 shows the connection of various sensor-based IoT devices in an MioT environment of an IoBT in contrast to a CioT environment of an IioT, as shown in Figure 2.
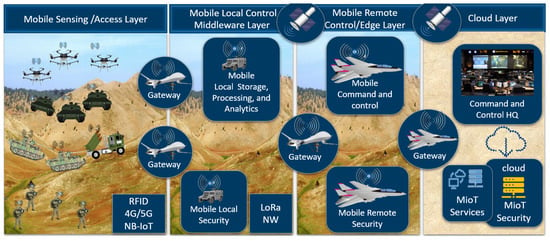
Figure 1.
Connected nodes in an MioT environment.
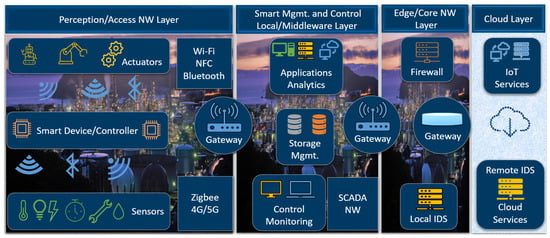
Figure 2.
Connected nodes in a CioT environment.
Due to their critical mission, IoT nodes in CioT and MioT are more susceptible to targeted cyber attacks at a high rate compared to IoT nodes deployed in a normal IoT environment. In addition, MioT environments marry things and humans, whereas the battlefield things and connected soldiers sense, communicate, negotiate, and collaborate to coordinate offensive and defensive tasks and operations. However, in both environments, a compromised sensor-based node, e.g., operating at the perception or sensing layer, can have a deadly or catastrophic impact.
2.3. Low-Complexity AMiDS
The building of scalable AMiDS to protect the sensing nodes in large, complex systems and environments requires robust yet simplistic and computationally inexpensive elements or constructs. Hence, the idea is to use low-complexity machine learning algorithms to build scalable and computationally inexpensive AMiDS models. The building of scalable and less costly AMiDS models often involves the use of low-complexity machine learning algorithms such as PCA and oSVM. Low-complexity security mechanisms are highly desirable when considering the protection of intelligent sensing devices with low power and computation cost requirements. This paper uses low-complexity machine learning algorithms, i.e., PCA and oSVM, to construct AMiDS models to protect IoT sensing nodes in CioT systems and MioT environments.
Given the stringent requirements of CioT and MioT in terms of their low latency, power, and computation overheads, the low-complexity ML algorithms have emerged as a viable option due to their low resource utilization and computation overhead compared to medium- and high-complexity models, such as the Random Forest and deep neural networks (DNNs). In particular, the PCA algorithm is inherently a dimensional reduction algorithm with low memory and CPU utilization. It uses statistical approaches to find data points with the highest variance, thus reducing the feature space by removing redundant and unnecessary features and organizing the remaining features in a new feature space known as the principal space [18]. oSVM, a variant of the SVM, uses kernel functions to reduce the calculations required without reducing the input dimensions. In addition, the SVM has a low bias and high variance and is suitable when the dataset is small.
2.4. Overview of the Perception Layer Threats and Attacks
Frequently, CioT systems and MioT environments are subject to similar threats at the perception or sensing layer. However, attacks against the MioT environment sensing layer occur with ferocity, velocity, rapidity, and higher frequency. Hence, we focus our discussion on threats against the CioT perception layer while considering the IioT implementation of a CioT. Further, we consider some studies that have focused on the investigation of threats associated with the various layers of the IioT architecture to provide a quick overview.
Some modern attacks on IioT systems mainly target sensors, actuators, communication links, and control systems [19]. Therefore, identifying threats against the perception layer cannot occur in isolation; one must consider the abstraction layers that interface with and control the perception layer. While the study presented in [19] referred to the interface and control abstractions as the data-link layer, the study presented in [20] separated the interface and control abstractions into layers 2 and 3. Layer 2, which interfaces with the perception layer, or layer 1, included the distributed control system (DCS), programming logic control (PLCs), and gateways. Layer 3 included supervisory control and data acquisition (SCADA), data acquisition devices, and the human–machine interface (HMI). Other studies, such as the study presented in [21], described the IioT architecture using three layers: perception (or object), network, and applications. As a result, we consider threats against the perception layer in addition to the network layer.
Focusing on the IioT system, a CioT, the study in [20] investigated possible attacks at various levels of the IioT architecture. Attacks at the lower layers, including the perception layer, exploit the vulnerabilities in the communication protocols used by the various IioT/IoT nodes to communicate. At the perception layer, possible attacks include reverse engineering, malware, the injection of crafted packets, eavesdropping, and brute-force searches. Attacks at the layer interfacing with the perception layer include replay, Man-in-the-Middle (MitM), sniffing, wireless device attacks, and brute-force password guessing. The study identified IP spoofing, data sniffing, data manipulation, and malware as possible attacks at the control layer. However, possible attacks against the control layer include attacks against industrial control systems such as SCADA systems. In addition to MitM, these attacks include passive and active eavesdropping, masquerade, virus, trojan horse, worms, denial of service (DoS), fragmentation, Cinderella, and DoorKnob rattling.
While the study presented in [20], which limits the DoS attack to the interface layer or layer 2, the study presented in [19] considers DoS attacks to be among the most common threats in the perception and data-link (layers 2 and 3, respectively) layers. Further, the study classified the attacks on the perception and data-link layers based on the wireless communication protocol as jamming DoS attacks, collision DoS attacks, exhaustion DoS attacks, unfairness attacks, and data transit attacks. Collision DoS attacks result from continuous DoS attacks and unfairness attacks result from exhaustion DoS attacks that degrade the system’s ability in favor of the attacker. Usually, attacks against the data-link layer aim to speed up or slow down the ICS operations to damage the ICS devices permanently.
The study presented in [22] classified the attacks based on an IoT layer architecture consisting of three layers: the perception, network, and application layers. In addition to jamming and reply attacks, attacks against the perception layer include node capture, malicious code injection, and sleep deprivation. The network layer, which connects the perception layer to the upper layers, is vulnerable to selective forwarding (a form of DoS attack), the cloning of ID and Sybil, wormhole attacks, sinkhole attacks, blackhole attacks, spoofed, alter, and reply routing info attacks, as well as eavesdropping, DoS, and MitM.
While our study focuses on cyber attacks, the perception layer is subject to non-cyber attacks. The study presented in [23] focused on the vulnerabilities and threats associated with the various IioT layers and used the term edge to describe the perception layer. It argued that the greatest threat at the perception layer is not cyber but is rather electronic, such as jamming, or kinetic, such as physical attacks with the intent to damage, degrade, disrupt, or destroy the sensors, actuators, machines, controllers, and other devices at the perception layer. Attacks such as side-channel analysis, fault injection, and voltage glitch attacks are physical or hardware attacks [24]. However, this study focuses on cyber attacks on the perception layer.
Generally, in perception layer attacks, the attacker aims to gain unauthorized access to control the IioT devices to launch cyber attacks. While there are several types of attacks against IioT devices depending on the specific IioT layer, distributed denial of service (DDoS), data theft through keylogging, tracking through fingerprinting, and the scanning of open ports are some common attacks on IioT devices that are carried out by attackers through the use of botnets [21]. Other attacks against IioT devices include blackhole, Sybil, clone, and sinkhole attacks.
2.5. An Example of an IoT Botnet Attack
In general, CioT and MioT are subject to attacks that are common in traditional IoT environments, such as DDOS, IoT bot malware, flooding, jamming, hijacking, DDOS, and IoT device cloning, to list a few [25,26,27]. Figure 3 shows an IoT bot attack scenario against an IioT system where the hacker uses compromised nodes to scan for vulnerable IoT nodes to compromise, loads the malicious payload to compromise the nodes, and then uses the compromised or non-compromised nodes to launch a DoS, DDoS, or RDDoS attack. The illustrated IoT bot attack uses an attack pattern similar to the Mirai IoT bot malware attack, which compromised thousands of IoT devices and launched a large-scale DDoS attack [28,29,30,31]. A DoS attack against an IoT device invariably leads to system failure and can have a catastrophic impact [32,33].
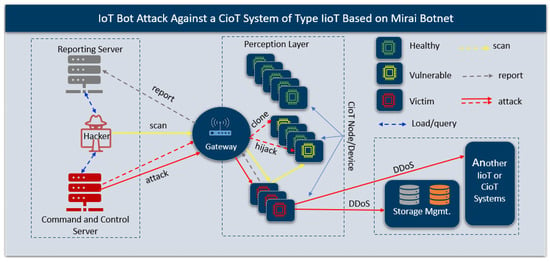
Figure 3.
An example of an IoT bot attack against a CioT system to launch a large-scale DDoS attack.
3. Related Work
Imbalanced datasets and improper data segmentation are the main contributors to a loss of the IDS detection accuracy. The authors of [34] proposed a machine learning framework in which they combined a variational autoencoder (VAE) and multilayer perceptron to simultaneously tackle the issues of imbalanced datasets (HDFS and TTP datasets) from multiple data sources and intrusions in complex, heterogeneous network environments. Using a hybrid learning approach, the authors used the variational autoencoder to address the imbalanced datasets issue in the training stage. However, they integrated the encoder from the unsupervised model into the multilayer perceptron to transfer imbalanced learning to the classifier model. They used supervised learning to accurately classify abnormal and normal examples. In addition, the authors had to balance the system’s performance and complexity due to using the multilayer perceptron for classification. In contrast, our non-hybrid approach uses supervised learning, simple data-level imbalanced learning methods, and an IoT imbalanced dataset of a relatively small size (2000 examples) to enable imbalanced learning while classifying normal and abnormal examples.
The proposed hybrid approach achieved better results than other solutions regarding the F1-score and recall rate, however, it exhibited substantial training and testing times. Generally, generative models, such as autoencoders, are more effective for solving the data imbalance problem than non-generative or traditional synthetic sampling methods when dealing with high-dimensional data. To that extent, the authors of [35] proposed a variational autoencoder method that generated new samples similar to, but not the same as, the original dataset by learning the data distribution. Given various evaluation metrics, the generative model outperformed the traditional sampling methods, such as SMOTE, ADASYN, and borderline SMOTE. Unlike the hybrid VAE method, which focuses on the sampling of network data, the non-hybrid VAE method considers an image dataset. This study uses non-hybrid generative methods to sample network data.
While the authors of [34] used a non-traditional approach to solve the data imbalance problem, the authors of [36] used traditional resampling methods (random undersampling, random oversampling, SMOTE, and ADA-SYN) to investigate the influences of these methods on the performance of artificial neural network (ANN) multi-class classifiers using benchmark cybersecurity datasets, including KDD99 and UNSW-NB18. Comparing the performance of the ANN multi-class classifiers using the resampling techniques, their study revealed that undersampling performed better than oversampling in terms of the training time, and oversampling performed better in terms of detecting minority data (abnormal examples). However, both methods significantly increased the recall when the dataset was highly imbalanced.
Furthermore, when comparing oversampling, undersampling, and SMOTE in terms of the resampling time, the authors found that oversampling and undersampling had the shortest time requirements, and the SMOTE resampling time was average. However, the ADASYN resampling time was the longest. In addition to the Random Oversampler and SMOTE methods, our study investigates the influence of other oversampling methods, such as KVSMOTE and KMeanSMOTE, hybrid methods such as SMOTEEN and SMOTETomek, ensemble methods such as EasyEnsemble and BalanceCascade, localized random affine shadow sampling known as LORAS, and generative methods such as autoencoders and variational autoencoders on the performance of two AMiDS models designed using PCA and one-class SVM (oSVM) and trained using a binary imbalanced IoT dataset exhibiting rare event cases, i.e., the number of abnormal examples (1874 or 93.7%) is proportionally higher than the number of normal examples (126 or 6.3%) in the dataset.
Similar to [36], the authors of [3] considered resampling methods with optimization techniques to address issues resulting from imbalanced or inadequate datasets, such as overfitting or poor classification rates. However, in addition to resampling methods, the authors of [3] used hyperparameter tuning (HyPaT) to increase the classifier’s sensitivity. In particular, using SMOTE, the authors proposed a combination of the oversampling, undersampling, and K-Nearest Neighbor methods with grid-search (GS) optimization and HyPaT to improve the classification results of different supervised learning algorithms, including SVM, among other supervised classifiers. Applying SMOTE with GS and HyPaT improved the results of the classifiers, while our study uses SMOTE, among other methods, to address the data imbalance by applying the random-search optimization method to reduce the computational overhead.
Using the KDD and NSL-KDD datasets with imbalanced examples, a deep learning intrusion detection system could not detect training samples in the minority classes. To handle this data imbalance issue, the authors of [37] proposed an IDS based on a Siamese neural network to detect intrusions without using traditional data sampling techniques, such as oversampling and undersampling. The proposed IDS achieved a higher recall rate for the minority classes of the attack than other neural-network-based IDSs trained using the imbalanced datasets. The author of [38] evaluated the effect of a class imbalance on the NSL_KDD intrusion dataset using four classification methods.
To address the data imbalance in network intrusion datasets, the authors of [39] used a combination of undersampling and oversampling techniques and clustering-based oversampling to evaluate the effect of imbalance on two intrusion datasets constructed from actual Notre Dame traffic. An analysis of the receiver operating characteristic (ROC) curves revealed that oversampling by synthetic generation outperformed oversampling by replication. To examine the influence of resampling methods on the performance of the multi-class neural network classifier, the authors of [36] used several data-level resampling methods on cybersecurity datasets. Using precision, recall, and the F1-score to evaluate the results, the authors found that oversampling increased the training time, while undersampling decreased the training time. In addition, resampling had a minor impact on the extreme data imbalance, and oversampling, in particular, detected more of the minority data.
The study presented in [40] considered the use of ensemble-based approaches to address the data imbalance problem for intrusion detection in industrial control networks using power grid datasets. The authors compared nine single and ensemble models specifically designed to handle imbalanced datasets, such as synthetic minority oversampling bagging and synthetic minority oversampling boosting. They evaluated their performance using balanced accuracy, Kappa statistics, and AUC metrics. Additionally, the authors listed the average training and testing times for each evaluated method or algorithm measured in seconds. The study revealed that undersampling and oversampling strategies were affected when using boosting-based ensembles. Our study uses ensemble-based methods not for classification purposes but rather for data sampling. For the classification task, we use PCA and oSVM.
4. Data-Level Sampling and Resampling Methods
Data-level sampling and resampling methods attempt to solve the data imbalance problem by adjusting the data distribution to obtain a balanced representation of the majority and minority examples in the training dataset. In general, data-level sampling and resampling often occur before learning takes place. Unlike algorithm-level methods, which focus on modifying the learning algorithm to reduce the bias towards the majority class, data-level methods do not solve the data imbalance directly, but solve it indirectly by modifying the training dataset to artificially balance the majority and minority examples in the dataset [5,7].
Data-level methods fall broadly into three main categories: undersampling, oversampling, and hybrid [41]. The hybrid method combines undersampling and oversampling methods [42]. It reduces the degree of imbalance in the dataset by synthetically creating a new class of minority examples. In contrast to the hybrid method, oversampling and undersampling methods use the same data to reduce the degree of data imbalance. Undersampling methods, however, balance the data distribution by increasing the weight of the minority examples, whereas oversampling methods reduce the weight of the majority examples to address the data imbalance [43].
Despite their promise, each type of method suffers from a particular issue; for example, oversampling causes overfitting due to the increased data size, and undersampling results in data loss due to a reduced data size [44]. Further, the combination of oversampling and undersampling causes overfitting and overlapping. However, overlapping, which occurs due to interpolation between relatively adjacent minority class examples, has a less significant impact on hybrid methods than overlapping. In general, oversampling methods are more accurate than undersampling methods when measuring their performance using ROC and AUC measurements.
In addition to the data sampling methods mentioned above, we consider a fourth category of methods known as generative methods to generate new synthetic data from the original data by reconstruction, which we then use to train the AMiDS models. We use the autoencoder (AE) and variational autoencoder (VAE), two unsupervised learning methods, to generate synthetic data that are closer to the original data but not quite the same. The main idea behind these two methods, each of which consists of two fully connected neural networks (an encoder and a decoder), is to first compress the input data using an encoder while ignoring the noise and then decompress it using a decoder to generate data as close as possible to the original [35,45]. Their main objective is to learn latent representations. Figure 4 shows the main components of an autoencoder network.
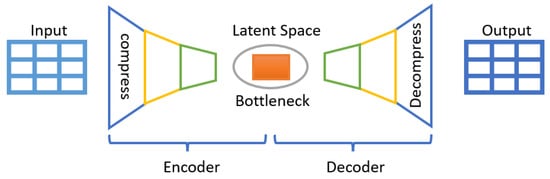
Figure 4.
A block diagram of an autoencoder model.
In general, autoencoders are trained to reconstruct the data at their output layer while minimizing the error between the input and reconstructed data. However, an autoencoder must first learn the data distribution to generate new data samples. While the two generative methods, AE ad VAE, are structurally similar, they are mathematically different [46,47,48]. Unlike AEs, VAEs use statistical means to interpolate by imposing a probability distribution on the latent space. Further, its inference model is a stochastic function of the input, i.e., the decoder is probabilistic.
On the other hand, an AE uses a separate variational distribution for each data point, which could be more efficient when using a large dataset. Additionally, an AE uses a computationally expensive inference approach, whereas a VAE uses a less expensive training approach. Perhaps the most important distinction between AEs and VAEs is that, unlike AEs, the hidden layer or latent space in the VAE network is smooth and continuous, making it easy to construct new data points using interpolation [45]. Although this study focuses on rare events where normal events are less frequent, we consider the generative methods to train the AMiDS model to examine the ability of the trained AMiDS to classify unseen or new anomalous events. Table 1 briefly describes each method, and Table 2 classifies various data sampling techniques based on their approach.

Table 1.
Description of data-level methods for imbalanced learning.

Table 2.
Classification of data-level methods for imbalanced learning.
5. Research Method
The employed research methodology is of the empirical type, where we used experiments to process and transform the imbalanced dataset, applied imbalanced methods to balance the data, trained the AMiDS models using imbalanced learning, scored (tested) and evaluated the models using standard ML metrics as well as time metrics, analyzed the evaluated models, and compared the models’ performances to the performances of the same AMiDS models trained and evaluated using regular learning. Figure 5 depicts the conceptual model based on the prescribed methodology.
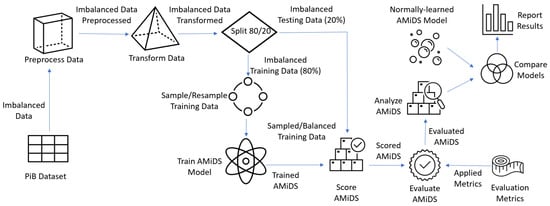
Figure 5.
The research methodology conceptual model.
Given the rare event prediction case, we used an empirical study to quantitatively assess the impact of imbalanced learning on low-complexity AMiDS models. As a result, we designed and ran a set of experiments using the data sampling and data generative methods listed in Table 2 to manipulate the portion (80%) of the imbalanced dataset used to train the AMiDS models. Then, we used the manipulated training dataset to train a PCA- and oSVM-based AMiDS on a supervised binary classification, which we later evaluated using the remaining portion (20%) of the imbalanced dataset, which was set aside for testing the trained models.
Furthermore, we used ML standard metrics as well as the training time and inference time as additional performance indicators. Further, we analyzed the performance of the AMiDS models trained using imbalanced learning with the AMiDS models’ performances when trained using regular learning (i.e., we trained the models using the training dataset (as-is) without applying data sampling to balance the class distribution).
The study presented in [69] provides detailed descriptions and performance analyses of the PCA- and oSVM-based AMiDS, referred to by the paper as AN-Intel-IDS models, using regular learning. To generate datasets for imbalanced learning using the sampling methods, we used the pre-processed IoT-Botnet (PiB) datasets used in [70]. The PiB dataset has two classes: the normal minority class and the anomalous majority class. The majority class contains DoS and DDoS intrusion data samples. According to the dataset statistics, there are 126.00 (6.3%) instances in the normal minority class and 1874.00 (93.7%) instances in the anomalous majority class.
While there are several proposed imbalanced learning methods in the literature to choose from, we selected methods from four different categories to obtain an early idea of which categories of methods are more suitable to apply when using imbalanced and passive adversarial learning, given the domain of applications (CioT and MioT) and the rare event prediction case.
5.1. ML and DL Methods
The study uses three ML methods and one deep learning (DL) method. The ML and DL methods used by this study are PCA-based anomaly detection, one-class SVM or oSVM-based anomaly detection, two-class SVM (tSVM), and two-class neural networks (tNN). The study uses PCA- and oSVM-based anomaly detection methods to build the PCA-based AMiDS and oSVM-based AMiDS models. We use the tSVM and tNN to create two baselines to compare the performance of the AMiDS models under the different data sampling and data generative methods.
5.2. Dataset for AMiDS Imbalanced Learning
To understand the impact of imbalanced learning on the performances of the AMiDS models in the rare event prediction case, we considered the UNSW_2018_IoT_Botnet dataset, which is available from the IEEE dataport [71]. However, we used the PiB datasets used in [70]. The dataset is a sample from one of the extracted dataset’s files but with fewer features. According to the creators of the dataset, the complete IoT-Botnet dataset, comprising benign and botnet-simulated IoT network traffic, contains about three million records and includes five types of attacks (DDoS, DoS, OS and service scan, keylogging, and data exfiltration) and forty-six features, including the attack label, attack category, and attack subcategory [72]. However, the PiB dataset consists of two thousand records and twenty-nine selected features and includes two attacks (DoS and DDoS) in addition to the normal examples. We used the PiB dataset rather than the extracted one because it has an imbalanced class distribution where the normal examples are less frequent than the attack examples, which is suitable for our study which focuses on imbalanced learning, given the rare prediction case. Figure 6 shows the distribution of the normal and attack classes in the dataset, and Table 3 shows the PiB dataset features.
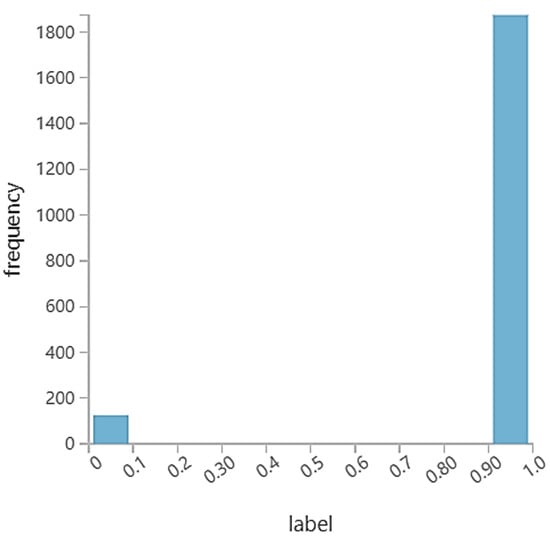
Figure 6.
PiB dataset with normal and anomalous class distributions.

Table 3.
Features of the PiB dataset.
5.3. Tools for Building the AMiDS Models
The Azure AI ML Studio from Microsoft was used to build, train, and deploy machine learning models and provided the platform to implement and run the experiments. Using Python, we implemented the code to process the PiB dataset, apply the data sampling and data generative methods to the PiB dataset, build the AMiDS models, and train and test the models using imbalanced learning. We used STANDARD_F4S_V2, a compute-optimized VMs, to code and run the experiments. The computer used an Intel(R) Xeon(R) Platinum 8168 CPU @ 2.70GHz and had 4 vCPU, 8 GiB RAM, 16 GiB SSD, 16 data disks, and 4 NICs [73].
5.4. Standard Performance Metrics
The study assessed the performance of the AMiDS models according to the applied data sampling and data generative methods using ML standard metrics, i.e., precision (P), accuracy (A), recall (R), F1-score (F1), ROC, and AUC, as well as using the training time and inference time as additional performance indicators.
6. Experimental Setup
This section describes the experimental framework used for assessing the impacts of imbalanced and passive adversarial learning on the PCA- and oSVM-based AMiDS models using four categories of data-level sampling methods. Consequently, we devised two experiments for each AMiDS, each consisting of four sub-experiments spanning the four categories of the data-level sampling methods. The four sub-experiments collectively covered eleven experimental runs, one for each selected data sampling and resampling method for imbalanced learning. While generative methods, such as autoencoders, usually train autoencoders on one class, i.e., the majority class, we trained two autoencoders for each class, similar to the study in [58].
We designed the experiments using the Microsoft Azure Machine Learning Studio (MS AMLS) platform. We built the models in Python using Azure Notebook, Azure ML curated environments and a Linux-based computer with four cores, 8 GB RAM, and 32 GB of storage. To build the PCA-based AMiDS model, we used the PCA anomaly detection module from the azureml-designer-classic-modules 0.0.176 package [74]. At the time of writing this paper, the current version of the azureml-designer-classic-modules is 0.0.177. However, we used the OneClassSVM from sklearn to build the oSVM-based AMiDS model due to the discontinuation of oSVM from Azure ML studio.
Additionally, we used the imbalanced-learn packages version 0.9.1, which contains the oversampling methods, and version 0.1.1, which contains the two ensemble-based methods used by this study. We used these ensemble-based methods even though the developer disapproves of them, according to to [75] either in favor of meta-estimators or algorithm-level methods that use boosting rules. This may be because these deprecated methods are mainly data samplers. For the LoRAS oversampling method, we used the pyloras package, version 0.1.0b5.
We adopted the autoencoder sampling method from the autoencoder developed by [76] to address the imbalance data problem for credit card fraud detection. Finally, we used the vae-oversampler package version 0.2 to implement the variational autoencoder generative method in [68]. Due to the number of methods used in the experiment, we do not present an analysis in this section but rather describe the experiments, categorizing them based on the applied methods and the sub-experiments to present the raw results. Then, we present a detailed analysis in the subsequent section.
6.1. Experimental Runs of the Oversampling Methods
The oversampling sub-experiment comprised five experimental runs (ERs) for each AMiDS type. We use the following notations to denote each experimental run: ER-KMS, ER-LRS, ER-ROS, ER-SMT, and ER-SVMSMT. ER stands for the experimental run and KMS, LRS, ROS, SMT, and SVMSMT stand for KMeanSMOTE, LoRAS, random oversampling, SMOTE, and SVM SMOTE, respectively. To distinguish between the PCA- and oSVM-based AMiDS, we use the subscripts and ; for example, ER-ROS and ER-ROS.
Before applying the oversampling methods, we normalized the PiB dataset’s numerical features. Then, we transformed the categorical features using one-hot encoding before splitting the transformed dataset into the training and testing datasets. As a result, before sampling, the training dataset had 90 features and 1600 examples, whereas the testing dataset set had a similar number of features but 400 examples. Additionally, the testing dataset was independent of oversampling; however, the training data depended on oversampling. Consequently, the number of examples in the training dataset changed after sampling.
In the training phase, the total number of examples after sampling using KMS, LRS, ROS, and SMOTE was 2992 with equally balanced normal and anomalous examples (i.e., 1496 each). The SVMSMT yielded 2161 examples but did not balance the normal and anomalous examples equally. As a result, the numbers of normal and anomalous examples were 665 and 1496, respectively. After sampling for the testing phase, the total number of samples was 2939 (1206 normal and 1187 anomalous) for all methods except the SVMSMT method, where the total number of samples was 1728 (529 normal and 1199 anomalous). The evaluation results after training and scoring (testing) the AMiDS models are shown in Table 4, Table 5, Table 6, Table 7, Table 8 and Table 9 for the PCA-based AMiDS and Table 10, Table 11, Table 12, Table 13, Table 14 and Table 15 for the oSVM-based AMiDS. Figure 7 shows the ROC curve and AUC values for the PCA-based AMiDS trained and scored models, and Figure 8 shows the ROC curve and AUC values for the oSVM-based AMiDS trained and scored models.

Table 4.
Trained PCA-based AMiDS evaluation results for the oversampling methods based on the weighted average.

Table 5.
Trained PCA-based AMiDS evaluation results for the oversampling methods based on the normal case.

Table 6.
Trained PCA-based AMiDS evaluation results for the oversampling methods based on the anomalous case.

Table 7.
Scored PCA-based AMiDS evaluation results for the oversampling methods based on the weighted average.

Table 8.
Scored PCA-based AMiDS evaluation results for the oversampling methods based on the normal case.

Table 9.
Scored PCA-based AMiDS evaluation results for the oversampling methods based on the anomalous case.

Table 10.
Trained oSVM-based AMiDS evaluation results for the oversampling methods based on the weighted average.

Table 11.
Trained oSVM-based AMiDS evaluation results for the oversampling methods based on the normal case.

Table 12.
Trained oSVM-based AMiDS evaluation results for the oversampling methods based on the anomalous case.

Table 13.
Scored oSVM-based AMiDS evaluation results for the oversampling methods based on the weighted average.

Table 14.
Scored oSVM-based AMiDS evaluation results for the oversampling methods based on the normal case.

Table 15.
Scored oSVM-based AMiDS evaluation results for the oversampling methods based on the anomalous case.
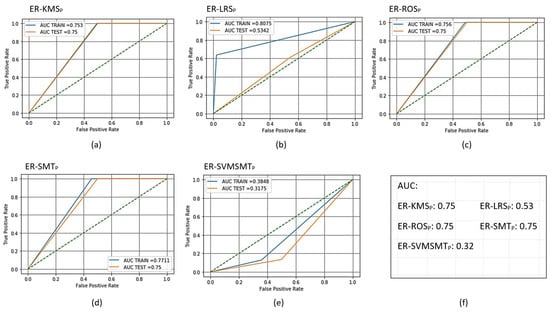
Figure 7.
Receiver operating characteristic (ROC) curves for the trained and scored PCA-based AMiDS using oversampling methods based on the five experimental runs, respectively: (a) ER-KMS, (b) ER-LRS, (c) ER-ROS, (d) ER-SMT, and (e) ER-SVMSMT. Summary of the AUC values based on the experimental runs according to the applied methods is shown in (f).
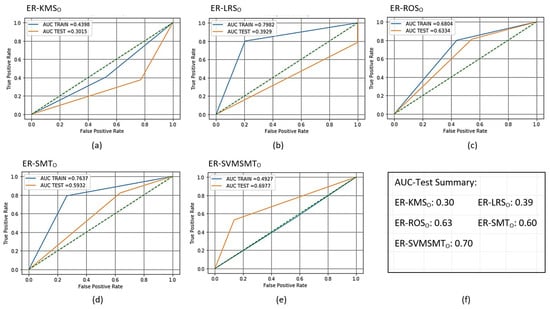
Figure 8.
Receiver operating characteristic (ROC) curves for the trained and scored oSVM-based AMiDS using oversampling methods based on the five experimental runs, respectively: (a) ER-KMS, (b) ER-LRS, (c) ER-ROS, (d) ER-SMT, and (e) ER-SVMSMT. Summary of the AUC values based on the experimental runs according to the applied methods is shown in (f).
6.2. Experimental Runs of the Hybrid Methods
We applied two hybrid methods SMOTEEN and SMOTETomek, to train and evaluate the two AMiDS models. As a result, the hybrid method sub-experiment consisted of two experimental runs for each model. In the first experimental run, we applied the SMOTEEN method to the PCA- and oSVM-based AMiDS. We denote the experiments as ER-SMTEN and ER-SMTEN, respectively. Similarly, we applied the SMOTETomek method to design two experimental runs. We named the runs using the ER-SMTEK and ER-SMTEK notations to distinguish the PCA-based AMiDS experiment from the oSVM-based AMiDS experiment. We applied the methods after normalizing and encoding the PiB dataset, as was also performed for the oversampling categories of the methods.
After applying the sampling methods, the resulting PiB data included 2992 samples for the PCA-based methods, with normal and anomalous examples being equally distributed (1496 each). However, for the oSVM-based methods, the resulting PiB data included 2393 samples with normal and anomalous examples being almost equally distributed (1193 normal and 1200 anomalous for ER-SMTEN, and 1206 normal and 1187 anomalous for ER-SMTEK). Table 16, Table 17, Table 18, Table 19, Table 20 and Table 21 show the evaluation results for the two AMiDS models for the training and testing phases. Figure 9 shows the ROC curves and AUC values.

Table 16.
Trained PCA-based and oSVM-based AMiDS evaluation results for the hybrid methods based on the weighted average.

Table 17.
Trained PCA-based and oSVM-based AMiDS evaluation results for the hybrid methods based on the normal case.

Table 18.
Trained PCA-based and oSVM-based AMiDS evaluation results for the hybrid methods based on the anomalous case.

Table 19.
Scored PCA-based and oSVM-based AMiDS evaluation results for the hybrid methods based on the weighted average.

Table 20.
Scored PCA-based and oSVM-based AMiDS evaluation results for the hybrid methods based on the normal case.

Table 21.
Scored PCA-based and oSVM-based AMiDS evaluation results for the hybrid methods based on the anomalous case.
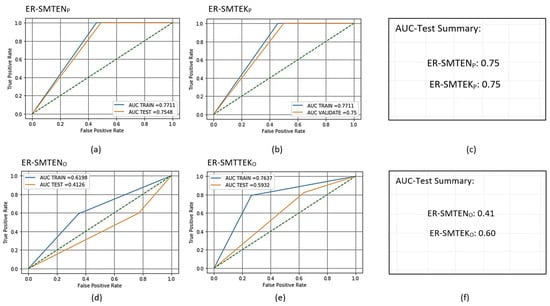
Figure 9.
Receiver operating characteristic (ROC) curves for the trained and scored PCA- and oSVM-based AMiDS using hybrid methods based on the four experimental runs, respectively: (a) ER-SMTEN for PCA, (b) ER-SMTEK for PCA, (d) ER-SMTEN for oSVM, and (e) ER-SMTEK for oSVM. Summary of the AUC values based on the experimental runs according to the applied methods is shown in (c) and (f) for the PCA- and oSVM-based AMiDS, respectively.
6.3. Experimental Runs of the Ensemble-Based Methods
Like the hybrid-based methods, we conducted two experiments with four experimental runs to train and evaluate the PCA- and oSVM-based AMiDS models using two data sampler ensemble-based methods, i.e., EasyEnsemble and BalanceCascade. We applied the EasyEnsemble method first, followed by the BalanceCascade. Then, using notations consistent with those used in the previous runs, we named the experimental runs using the following notations ER-EENSMB, ER-BALCAS, ER-EENSMB, and ER-BALCAS, where EENSMB and BALCAS stand for the EasyEnsemble and BalanceCascade, respectively.
Similar to the hybrid methods, we applied the ensemble-based methods after transforming the PiB data. The size of the resulting balanced data for the PCA-based AMiDS model, after applying the EENSMB and BALCAS methods, was 208 examples for both methods. However, for the oSVM-based model, the size of the resulting PiB data was 166 examples for both methods. Given the training and scoring phases, Table 22, Table 23, Table 24, Table 25, Table 26 and Table 27 show the evaluation results for the two AMiDS models. Figure 10 shows the ROC curves and AUC values.

Table 22.
Trained PCA- and oSVM-based AMiDS evaluation results for the ensemble-based methods based on the weighted average.

Table 23.
Trained PCA- and oSVM-based AMiDS evaluation results for the ensemble-based methods based on the normal case.

Table 24.
Trained PCA- and oSVM-based AMiDS evaluation results for the ensemble-based methods based on the anomalous case.

Table 25.
Scored PCA- and oSVM-based AMiDS evaluation results for the ensemble-based methods based on the weighted average.

Table 26.
Scored PCA- and oSVM-based AMiDS evaluation results for the ensemble-based methods based on the normal case.

Table 27.
Scored PCA- and oSVM-based AMiDS evaluation results for the ensemble-based methods based on the anomalous case.
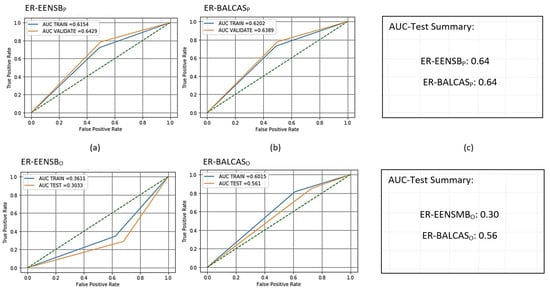
Figure 10.
Receiver operating characteristic (ROC) curves for trained and scored PCA- and oSVM-based AMiDS using ensemble-based methods based on the four experimental runs, respectively: (a) ER-EENSB for PCA, (b) ER-BALCAS for PCA, (d) ER-EENSB for oSVM, and (e) ER-BALCAS for oSVM. Summary of the AUC values based on the experimental runs according to the applied methods is shown in (c) and (f) for the PCA- and oSVM-based AMiDS, respectively.
6.4. Experimental Runs of the Generative Methods
We conducted two sub-experiments using AE with L1 regularization and VAE methods with oversampling to account for imbalanced and passive adversarial learning. Given the two AMiDS models (PCA- and oSVM-based), the generative model sub-experiments consisted of four experimental runs. We describe these four runs using similar notations to the ones stated previously. As a result, we use the following notations: ER-AE, ER-AE, ER-VAEOS, and ER-VAEOS, respectively. For the two AE experimental runs, we trained the AE using the normalized and encoded imbalanced PiB dataset after splitting the PiB into training and testing sets with 1600 and 400 examples, respectively.
To build the AE, we used a shallow neural network consisting of an input layer, one hidden layer, and an output layer. The size of the input and output layers was 90. To build the encoder, latent space, and decoder, we used the Dense() function from Keras. To encode the input layer, we used the ReLU activation function, L1, or Lasso Regularization (L1-type) activity_regularization with 64 units to specify the dimensionality of the latent space. Similar to the encoding space, we used the ReLU activation function to define the latent and encoding spaces. Further, we optimized the autoencoder using adadelta optimization and used the mean square error to define the loss function. Figure 11 shows the AE model’s training history, layer type, output shape, and number of parameters, including the total and trainable parameters.
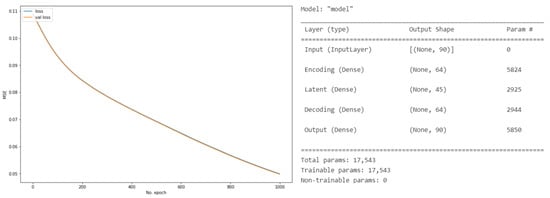
Figure 11.
The Autoencoder (AE) model history and parameters for the PCA- and OSVM-based AMiDS models.
As for the VAE’s two experimental runs, we used the vae_oversampler API introduced in [68]. The API provides methods to generate synthetic data using a VAE and an oversampler. First, it builds the VAE using Keras to learn the probability distribution of the data and then samples the learned distribution to generate the minority examples. To instantiate the vae_oversampler, we set the number of epochs to 100, the input dimension to 90, the latent dimension to 45, the batch size to 64, the dimension to 45, the batch size to 64, and the minority class identity to 0 (i.e., normal class). We then sampled the data using the fit_resample() method, which we used to build the training dataset to train the AMiDS models. After learning the distribution and resampling, the training dataset, which consisted of 2992 samples, showed balanced normal and anomalous examples, each with 1496 samples.
According to [68], the VAEOversampler initializes the encoder network using a ReLU activation function and defines the hidden layer’s size using the mean and log of the latent variable. For the decoder, the VAEOversampler uses a sigmoid activation function and the mean and log of the latent variable to sample new data similar to the original data. Figure 12 shows the VAEOversampler encoder and decoder parameters, including the total and trainable parameters.

Figure 12.
Variational Autoencoder Oversampler (VAEOversampler) encoder and decoder parameters for the PCA- and OSVM-based AMiDS models.
To train the two AMiDS models, we applied the new data generated by the AE and VAEOversampler and scored the two trained models to obtain the training and testing metrics. Figure 10 shows the ROC curves and AUC values. The rest of the results are shown in Table 28, Table 29, Table 30, Table 31, Table 32 and Table 33.

Table 28.
Trained PCA- and oSVM-based AMiDS evaluation results for the generative-based methods based on the weighted average.

Table 29.
Trained PCA- and oSVM-based AMiDS evaluation results for the generative-based methods based on the normal case.

Table 30.
Trained PCA- and oSVM-based AMiDS evaluation results for the generative-based methods based on the anomalous case.

Table 31.
Scored PCA- and oSVM-based AMiDS evaluation results for the generative-based methods based on the weighted average.

Table 32.
Scored PCA- and oSVM-based AMiDS evaluation results for the generative-based methods based on the normal case.

Table 33.
Scored PCA- and oSVM-based AMiDS evaluation results for the generative-based methods based on the anomalous case.
7. Imbalanced Learning Models Result Analysis
This section presents the results of the imbalanced learning experiments and analyzes the outcomes for each experiment. First, we analyze the performances of the two AMiDS models based on the different categories of methods within the categories, subsequently comparing their performances across the categories. Finally, we compare the performances of the two models when trained using imbalance learning versus regular learning.
7.1. Oversampling-Based Imbalanced Learning Models Results Analysis
To analyze the classification performance of the PCA- and oSVM-based AMiDS using oversampling methods, we first considered the degree of separability (AUC) and the probability curve (ROC). Then, we examined the other metrics, including the training time and inference time. According to Figure 7, oversampling the unbalanced PiB dataset using the KMS, ROS, and SMT methods improved the performance of the PCA-based AMiDS compared to its performance when using the LRS and SVMSMT methods. When applying the KMS, ROS, and SMT methods, the PCA-based AMiDS could distinguish between the normal and anomalous examples with a probability of 75%.
Furthermore, the difference in the AUC values between the training and testing data was substantially small, thus suggesting that the model was able to learn a generalizable pattern from the training data. On the other hand, in contrast to the three methods mentioned above, the model exhibited a poor performance when applying the SVMSMT and LRS methods. The model had probabilities of 32% and 53%, respectively, thus indicating a model with a low degree of separability in the case of SVMSMT or no capacity to distinguish between examples in the case of LRS.
An observation to note is that when applying imbalanced learning using the LRS method, the model exhibited a high AUC for the training data (0.86) but a relatively low AUC for the testing data (0.53). The big gap between the two values suggests that the PCA-based AMiDS model either exhibited overfitting where the model fitted the training data but not the testing data, failing to learn a generalizable pattern from the training data that could be applied when using the testing data, or the distributions of the training and testing data differed. On the other hand, the model performance when using the SVMSMT method suggests underfitting, since both the training and testing data AUC values are less than 0.5.
Based on Figure 8, the oSVM-based AMiDS distinguished between the normal and anomalous examples with 63% and 60% probabilities when using the ROS and SMT methods, respectively. Although it performed slightly better when using the SVMSMT method with a 70% chance of distinguishing between the different examples, the model had a low AUC value (0.50) for the training data compared to the AUC value (0.70) for the testing data. When the testing data’s AUC value was larger than the training data’s AUC values, random variation or sampling variability was often the underlying reason.
Generally, a trained model with a 50% chance of separating normal from anomalous examples randomly classifies the examples; hence, there is no capacity to distinguish between the two types of examples. Applying the LRS method, on the other hand, improved the trained model’s degree of separability. The trained model could distinguish between examples with an 80% probability; however, it showed a poor performance when tested.
The tested model was able to distinguish between the examples with a 40% probability. In the LRS method case, the big gap between the AUC values suggests model overfitting or the inability to learn a generalizable pattern from the training data.
Invariably, determining the AMiDS models’ probabilities of distinguishing between normal and anomalous examples under the different oversampling methods for both the training and testing data helps to determine how well the models could distinguish between the classes and whether the models could learn a generalizable pattern from the data to classify new data correctly or not. However, quantitatively determining the models’ classification performances using metrics such as A, P, R, and F1 is essential to understand the usefulness and completeness of the obtained results. Additionally, determining the inference time is vital to assess the model’s suitability to the application and type of problem.
According to the weighted averages presented in Table 4 and Table 7, the PCA-based AMiDS showed a superior performance when applying the KMS, ROS, and SMT with 97% A, P, R, and F1 for the testing data, but it had different values for the training data with SMT performing slightly better with 77% A, 84% P, 77% R, and 76% F1 compared to the KMS and ROS. A detailed interpretation of the results revealed that the percentage of correct predictions given the total number of predictions was 97% for the testing data compared to 77% for the training data. Furthermore, the ratio of true positives (anomalous) and total positives predicted was 97% for the testing data, compared with 84% for the true data. Further, the ratio of true positives to the number of total positives in the dataset was 97% for the testing data, compared to 77% for the training data, indicating that the model had a high level of sensitivity for the testing data compared to the training data. Among the examples that the model could identify, given the testing data, 97% of them were correct compared to 76% for the training data.
When applying the LRS method, the PCA-based AMiDS performed more poorly than the KMS, ROS, and SMT methods. The percentage of correct predictions, given the total number of predictions, was 81% for the training data and 60% for the testing data. The ratio of true positives to total positives predicted was 85% and 90% for the training and testing data, respectively. However, the ratio of true positives and total positives in the dataset was 81% for the training data, compared to 60% for the test data. Given the harmonic mean, 80% of the examples that the model could identify were correct for the training data, compared to 71% for the testing data. While the performance of the PCA-based AMiDS was the worst when applying LRS compared to the other methods, it had an inference time of 0.21 s for the testing data, thus ranking it second after the ROS method, which recorded the lowest inference time of 0.18 s.
According to the weighted averages presented in Table 10 and Table 13, the oSVM-based AMiDS performance was dissimilar to the PCA-based AMiDS model. For example, it exhibited a poor performance when applying the KMS method compared to the PCA-based AMiDS model. However, it performed relatively better when applying the LRS, ROS, and SMT. The model could correctly predict 74% of the examples given the total number of predictions for the training data and 80% for the testing data using the LRS method. Additionally, the ratio of true positives to total positives predicted was 88% for the training data and 80% for the testing data, and the ratio of true positives to total positives in the data was 80% for the training data and 74% for the testing data. Furthermore, among all examples that the model could identify, it correctly predicted 80% of them for the training data and 81% for the testing data. The SMT method slightly improved the performance of the OSVM-based AMiDS. The percentages of correct predictions given the total number of predictions were 76% and 80% for the training and testing data, respectively. The ratio of true positives to the total number of positives predicted was 76% for the training data and 91% for the testing data.
In addition, the ratios of true positives to the total number of all positives in the data were 76% and 80% for the training and testing data, respectively. Finally, among all examples that the model could identify, it correctly predicted 76% of the examples for the training data and 85% for the testing data. Finally, when applying the ROS method, the oSVM-based model could correctly predict 68% of the examples of the total number of predictions for the training data and 79% for the testing data. The ratios of true positives and the total number of positives predicted were 69% and 92% for the training and testing data, respectively. However, the ratios of true positives to the total number of all the positives in the data were 68% and 79%, respectively. Finally, among all the examples that the model could identify, it correctly predicted 67% and 84% of the examples for the training and testing data, respectively.
The AUC values were 0.40, 0.63, and 0.60 for the LRS, ROS, and SMT on the testing data compared to 0.79, 0.68, and 0.76 on the training data, respectively. The degree of separability suggests a random variation or sampling variability when the degree of separability for the testing data is higher than that of the training data. Generally, the AUC for the testing data is smaller than that of the training data when the gap between the two is relatively small. A large gap may signal a sample bias, whereby the testing data sample is easier to classify than the training data.
7.2. Hybrid-Based Imbalanced Learning Models Results Analysis
Given the two hybrid methods, SMTEN and SMTTEK, the PCA-based AMiDS generally performed better than the oSVM-based AMiDS when fitting the testing data. Based on Figure 10, the AUC value for the training and testing data when considering the PCA-based model was 0.75 for both methods, and the gap between the AUC for the testing and training data was slight. Thus, the model could learn a generalizable pattern from the training data. On the other hand, the oSVM-based AMiDS showed a poor performance when applying the SMTEN method. The AUC value for the testing data was 0.41 compared to the AUC value of 0.62 for the training data. The trained model had a 62% chance of distinguishing between normal and anomalous examples. However, the tested model could not learn a generalized pattern or, perhaps, exhibited overfitting. When applying the SMTTEK method, the oSVM-based AMiDS exhibited a slightly better performance with a 60% probability of distinguishing between examples using the testing data. Given the training data, the model could distinguish between examples with a probability of 76%.
To analyze the classification performance of the AMiDS models, we considered the values of the other standard ML metrics and inference times recorded in Table 16 and Table 19. The percentages of correct predictions given the total number of predictions for the PCA- and oSVM-based AMiDS models when applying the SMTEN method on the training data were 77% and 62%, respectively. When we applied the testing data, the percentages were 97% and 58%, respectively.
The ratios of true positives to the total number of positives predicted for the PCA-based AMiDS for the training and testing data were 84% and 97%, respectively. For the oSVM-based AMiDS, the ratios were 62% and 88% for the training and testing data, respectively. When applying the training data, the ratios of true positives to the number of true positives were 77% and 62% for the PCA- and oSVM-based AMiDS, respectively. When we applied the testing data, the ratios were 97% and 58%, respectively. Finally, among all the examples that both models could identify, they correctly predicted 76% (PCA-based) and 62% (oSVM-based) when applying the training data and 97% (PCA-based) and 69% (oSVM-based) when applying the testing data.
When applying the SMTEK method using the training data, the percentages of correct predictions given the total number of predictions were 77% and 76% for the PCA- and oSVM-based AMiDS, respectively. For the testing data, the percentages were 97% and 80%, respectively. The ratios of true positives to the total true positives predicted were 84% and 76% for the PCA- and oSVM-based AMiDS, respectively. In addition, the ratios of true positives to the total number of true positives in the data were 77% and 76%, respectively. However, among all examples that the models could identify, both models correctly predicted 76% of the examples.
When applying the testing data, the PCA- and oSVM-based AMiDS could predict 97% and 80% of all predicted examples correctly. The ratios of the true positives given the total number of true positives predicted and all true positives in the dataset were 91% and 80% for the oSVM-based AMiDS, respectively. The PCA-based AMiDS reported a value of 97% for both cases. Additionally, among all examples that the models could identify, the PCA-based AMiDS correctly predicted 97% of the examples compared to the oSVM-based model, which was able to predict 85% of them correctly.
While the ML metrics suggest that the PCA-based AMiDS had a better classification performance than the oSVM-based AMiDS, it incurred a slightly higher inference time than the oSVM-based AMiDS, if we consider the difference to be significant in the context of the application domain. Considering the inference time for the testing phases, the oSVM-based AMiDS inference times were 0.04 and 0.06 s for the SMTN and MSTEK, respectively. For both methods, The PCA-based AMiDS inference times were 0.24 and 0.21 s, respectively.
7.3. Ensemble-Based Imbalanced Learning Models Results Analysis
Like the hybrid methods, the PCA-based AMiDS performed better than the oSVM-based AMiDS when applying imbalanced learning using the ensemble-based methods EENSMB and BALCAS. For example, given Figure 10, the model had a 64% chance of distinguishing between the normal and anomalous examples using the testing data for both ensemble-based methods. Using the training data, the model could distinguish between examples with a probability of 62%. The oSVM-based AMiDS had probabilities of 30% and 56% and 36% and 58% when using the EENSMB and BALCAS, respectively, for the testing and testing data. The oSVM-based AMiDS model’s performance when using the BALCAS was much better than that when using the EENSMB; however, it performed relatively poorly compared to the PCA-based AMiDS for the same method.
While it is common for the AUC value for the training data to be higher than the AUC value for the testing data, the gaps between the two values are relatively small. The gap between the AUC values for the testing and training data is generally attributable to random variation or sampling variability due to the statistical nature of the function used to calculate AUC when the testing AUC value is slightly higher than the training AUC value. Therefore, we attribute the small value to the models making an arbitrary distinction between a few examples among the total number of examples in the testing dataset.
Given the training and testing data, the oSVM-based model’s capacity to distinguish between the two types of examples when applying imbalanced learning using the EENSB method is poor. According to Figure 10, the model’s probabilities of distinguishing between normal and anomalous examples for the training and testing data were 36% and 30%, respectively. On the other hand, the model exhibited a better performance when using the BALCAS method. The AUC values for the training and testing data were 0.60 and 0.56, respectively. Nonetheless, the model still suffered from the inability to learn a generalizable pattern.
Consistent with the AUC and ROC curve results, the oSVM-based AMiDS model had a poor classification performance when applying the EENSMB method compared to the BALCAS method, which enabled the model to perform relatively better. The same was true for the other metrics. For example, the percentages of correct predictions, given the total number of predictions when applying the EENSMB method, were 36% and 29% for the training and testing data, respectively, compared to the BALCAS method, where the percentages were 60% and 82% for the training and testing data, respectively (see Table 22 and Table 25). The rest of the metrics show similar values for the training data when applying the EENSMB method.
The ratio of true positives predicted to the total number of true positives predicted and the total number of true positives in the dataset was 36%. Additionally, among all examples that the model could identify, the model predicted 36% of those examples correctly. However, for the testing data, the values fluctuated between small and relatively high. For example, the ratio of true positives predicted to the total number of true positives predicted was 83%. However, the ratio of true positives predicted to the total number of true positives in the dataset was only 29%.
Further, among all examples that the model could identify, the model correctly predicted 41% of the examples. When applying the BALCAS method, the ratios of true positives correctly predicted to the total number of true positives predicted and the total number of true positives in the dataset were 62% and 60% for the training data and 91% and 82% for the testing data, respectively. The percentages of correctly predicted examples compared to the total number of examples that the model would have identified were 58% and 86% for the training and testing data, respectively.
In general, the classification performance of the PCA-based AMiDS model was relatively better than the oSVM-based AMiDS model’s performance when applying both ensemble methods. In addition, the PCA-based AMiDS model showed similar performances across the two ensemble methods for the training and testing data (see Table 22 and Table 25). The percentage of correct predictions given the total number of predictions when applying the training data was 62% for both methods. The ratios of true positives correctly predicted to the total number of true positives predicted were 62% and 63% when applying the EENSMB and BALCAS methods, respectively. The ratio of true positives correctly predicted to the total number of true positives in the dataset was 62% when applying both methods. The percentages of correctly predicted examples among all examples that the PCA-based model could identify were 61% and 62% when applying the EENSMB and BALCAS methods, respectively.
For the testing data, the results were close to those of the training data when applying both ensemble-based methods. The percentage of correct predictions given the total number of predictions was 77% for the EENSB and 76% for the BALCAS. The ratio of correctly predicted true positives to the total number of positives predicted was 92% for both methods. However, the ratios of correctly predicted true positives to the total number of positives in the dataset were higher: 77% and 76% for EENSB and BALCAS, respectively. Finally, the percentage of correctly predicted examples among all examples that the model could identify was 83% when applying the EENSB method and 82% when applying the BALCAS methods.
Given the PCA-based AMiDS model, the inference time when applying imbalanced learning using the EENSMB method (0.22 s) was less than when applying the BALCAS method (0.42 s). Despite the oSVM-based AMiDS model’s low performance compared to that of the PCA-based model, the inference times when using the oSVM-based AMiDS were relatively low. The inference times of the model when using the EENSMB methods were 0.008 s and 0.004 s when applying the BALCAS method.
7.4. Generative-Based Imbalanced Learning Models Results Analysis
Invariably, the underlying reason behind selecting the generative methods, AE and VAEOS, for imbalanced learning is to train the model to predict unseen examples, i.e., new examples that are similar to the original but are not the original; hence, this is termed passive adversarial learning. We consider it passive because we do not generate examples in a perturbational manner to trick the model. Instead, we use the existing data to generate similar data points that are not the same. On the other hand, we view active adversarial learning as trying to trick the model into making incorrect predictions using perturbed examples.
Given the AE method, both AMiDS models recognized the anomalous examples in the training and testing data but did not recognize the normal examples. In contrast, the two AMiDS models could recognize the normal examples when applying imbalanced learning using the VAEOS method. When considering the AE method and the AUC and ROC curve metrics shown in Figure 13, the PCA- and oSVM-based AMiDS models could not distinguish between the normal and anomalous examples for the training and testing data. The probability of the model was 50%. We attribute the results to the inability of the models to recognize normal examples.
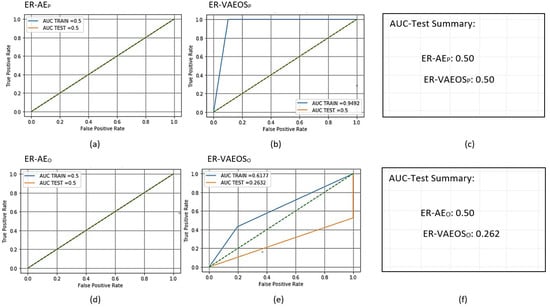
Figure 13.
Receiver operating characteristic (ROC) curves for trained and scored PCA- and oSVM-based AMiDS using generative-based methods based on the four experimental runs, respectively: (a) ER-AE for PCA, (b) ER-VAEO for PCA, (d) ER-AE for oSVM, and (e) ER-VAEO for oSVM. Summary of the AUC values based on the experimental runs according to the applied methods is shown in (c) and (f) for the PCA- and oSVM-based AMiDS, respectively.
While both AMiDS models performed the same in the case of the AE method, the two models exhibited different performances when applying imbalanced and passive adversarial learning using the VAEOS method. The PCA-based model had a probability of 94% for distinguishing between anomalous examples for the training data compared to a 50% probability for the testing data. However, the oSVM-based model had a probability of 61% for the training data and only a 26% chance of distinguishing between examples for the testing data.
Analyzing the classification performance of the two generative methods based on the degree of separability and the probability of distinguishing between the two classes of examples is an essential step in assessing the classification capability of the models. However, looking at the other metrics provides a more granular look into the overall performance of the AMiDS models. To that effect, the values of the A, P, R, and F1-score metrics, when considering the weighted average, indicate that the PCA- and oSVM-based AMiDS exhibited an excellent classification performance for the training and testing data when applying the AE method.
However, looking at the values of these metrics based on the normal case reveals that the AMiDS models can identify only the anomalous examples (see Table 29 and Table 32). Given the inability of the AMiDS models to identify normal examples in the training and testing data, one could conclude that this impacted the model’s ability to separate between examples and its probability of distinguishing between the normal and anomalous examples when looking at the AUC and ROC curve metrics.
Considering the anomalous case for the training and testing data, the PCA- and oSVM-based models performed well. The ratio of correctly predicted true positives to the total number of positives predicted in the training and testing data was 94% for both models. However, the ratio of correctly predicted true positives to the total number of positives in the training and testing data was 100% for both models. Given all the examples for the training data, the percentage of correctly predicted examples was 91% for both models. For the testing data, the percentage was 97% for both models. Looking at the weighted average of the metrics (see Table 28 and Table 31, the values of the metrics, given both AMiDS models, consistently exhibited a good classification performance for the training and testing data.
When applying the VAEOS method, we observed similar model behaviors. The PCA- and oSVM-based AMiDS models could not recognize normal examples in the training and testing data. However, both models were able to identify anomalous examples. During the training and testing phases, the PCA-based AMiDS performed better than the oSVM-based AMiDS. The accuracy of the PCA-based model was 95% compared to 73% for the oSVM. The precision, recall, and F1-score of the PCA-based AMiDS model had a value of 95% compared to 73% for the accuracy and recall for the oSVM model. The F1-score for the oSVM-based model was 72%.
The PCA-based model consistently performed better during the training and testing phases than the oSVM-based model. During testing, the PCA-based model achieved an accuracy, recall, and F1-score of 94% and a precision of 89%. On the other hand, the oSVM-based model had an accuracy and recall of only 50% and a precision and F1-score of 85% and 63%, respectively. When comparing the performances of the two models during the training and testing phases, the PCA-based model superseded the oSVM-model, thus suggesting that the PCA-based model was able to learn the data pattern and hence generalize better.
While the PCA-based AMiDS consistently performed better than the oSVM-based AMiDS model during the training and testing phases for both methods, the oSVM-based model reported shorter inference times than the PCA-based model. For example, the oSVM-based model inference times for the testing phase were 0.03 s and 0.02 s when applying the AE and VAEROS methods, respectively, compared to 0.20 s for the PCA-based model. In addition, the oSVM-based training time when applying the AE methods was 0.26 s compared to 1.61 s for the PCA-based model. However, the training time of the oSVM-based model was slightly higher when applying the VAEROS. The oSVM-based model training time was 0.73 s compared to 0.67 s for the PCA-based model.
7.5. Analysis Results across the Categories of the Methods
The PCA-based AMiDS model generally had a better classification performance when applying the KMS, ROS, and SMT oversampling methods, SMTEN and SMTEK hybrid methods, and EENSMB and BALCAS ensemble methods. However, when comparing the performance of the PCA-based AMiDS model based on the categories of the methods, the performance of the AMiDS when applying the ensemble-based models ranked second compared with the oversampling and hybrid methods, which showed parallel performance levels, hence ranking first.
The minimum inference time within the categories was observed during the testing phase when applying the ROS (0.18 s), SMTEN (0.25 s), and EENSB (0.21 s). On the other hand, the oSVM AMiDS model had a relatively better classification performance when applying the ROS and SMT oversampling methods, the SMTEK hybrid method, and the BALCAS ensemble method. The minimum inference time within the oversampling category was observed during testing when applying the SMT method (0.039 s). When comparing the oSVM-based AMiDS model classification performance to the PCA-based AMiDS performance, given the ROS, SMT, and SMTEK, the PCA-based AMiDS performed better than the oSVM-based AMiDS.
The classification performance of the PCA-AMiDS model when applying the generative methods was generally better than that of the oSVM-based AMiDS model. The performance of the PCA-AMiDS model under the generative category of methods showed a similar trend to that observed when considering the other three categories and the oSVM-based AMiDS model’s performance. In addition, the PCA-based AMiDS model performed better than the oSVM-based AMiDS model when applying the VAEOS method. However, the two models performed similarly when applying the AE method with the oSVM-based AMiDS model, recording the minimum inference time (0.033 s) during the testing phase.
8. Performance Evaluation and Discussion
In this section, we focus on evaluating the performances of the AMiDS models when applying imbalanced learning to address the rare event case using two baseline models and a worst-case AMiDS model. Given the rare event case, the worst-case AMiDS model refers to an AMiDS model that did not perform well under regular learning. In doing so, we consider the performance of the AMiDS models when applying regular learning, as presented by the comparative study in [70], to quantify the improvement gained from applying imbalanced learning. Further, we used the two-class neural network IDS (tNN IDS) model used by the authors of [70] as a baseline to evaluate the performance of the AMiDS models under regular learning conditions.
In this study, we used the tNN IDS model as a baseline to evaluate the performance of the AMiDS models under imbalanced learning conditions. We called this class of baseline models ADiDS, i.e., anomaly-based deep learning IDS. In addition, we designed a two-class SVM (tSVM) AMiDS model to create an AMiDS baseline model to be used in addition to the ADiDS baseline model to evaluate the performances of the models presented in this paper. We refer to these baseline models as the best-case AMiDS and ADiDS models. The classification results of the worst-case AMiDS models and the best-case baseline models are shown in Table 34, and the ROC curve of the tSVM AMiDS model is shown in Figure 14.

Table 34.
Performance of the worst-case AMiDS and the ADiDS and tSVM AMiDS baseline models under regular learning conditions.
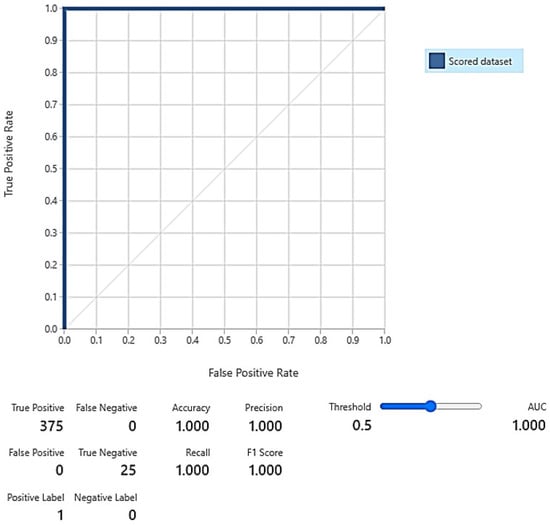
Figure 14.
tSVM AMiDS baseline model performance.
Based on the analysis results across the categories of the imbalanced learning methods presented in Section 6, we considered the performance of the PCA-based AMiDS model when applying KMS, ROS, SMT, SMTEN, SMTEK, EENSMB, and BALCAS, as well as the performance of the oSVM-based AMiDS when applying the ROS, SMTEK, and BALCAS when assessing the improvement gain. We selected these AMiDS models because they performed the best out of all of the trained AMiDS. This means that, among all the trained PCA- and oSVM-based AMiDS, the listed AMiDS performed better than the rest. According to Table 34, the two AMiDS models’ performances as a result of regular learning were poorer compared to their performances when applying imbalanced learning. In contrast, the better-case AMiDS models exhibited better performances, with the PCA-based AMiDS model exhibiting a performance near to that of the best-case model.
Invariably, we view the generative methods in light of two training objectives: imbalanced learning and passive adversarial learning. Consequently, we considered the ADiDS baseline model when evaluating the impact of generative methods on the performance of the low-complexity AMiDS models. In addition, we considered the better-case performance, i.e., the PCA-based AMiDS model’s performance when using the AE and VAEOS methods and the oSVM-based AMiDS model’s performance when using the AE method. Although the best-case models’ performances were superlative, both AMiDS models exhibited comparative performances to the best-case models and superlative performances when compared to their performances using regular learning (see Table 34 and Table 35). Figure 15 shows the performance of the better-case scored AMiDS model based on the weighted average.

Table 35.
Performance of the best-case scored AMiDS model based on the weighted average under imbalanced learning conditions.
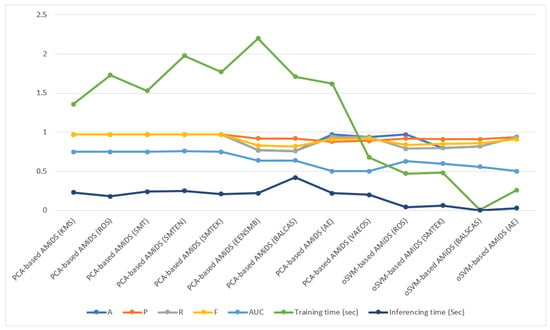
Figure 15.
Better-case scored AMiDS model performance based on the weighted average.
9. Conclusions and Future Work
Building efficient and effective AMiDS systems to detect intrusions in MioT and CioT environments requires the AMiDS model to be trained using low-complexity models and a sufficient dataset representing real-world events. However, such ground truth data may not be available due to data scarcity, data restrictions, or both. As a result, the use of synthesized and simulated data is common in such situations. However, synthetic data can be unrealistic and potentially biased, and simulated data can become irrelevant and obsolete.
While using ground truth data to train models is highly desirable, the existing real-time data are usually imbalanced, thus leading to undesired results, such as prediction inaccuracies. Data-driven learning using imbalanced learning methods to sample, resample, and generate ground truth data can lessen the adverse impact of imbalanced data on the performance and prediction accuracy of low-complexity AMiDS models. This study examined the impact of imbalanced learning on the performance of two low-complexity AMiDS models.
Given the distribution of the normal and anomalous events, these ground truth data can be highly imbalanced. Imbalanced learning using the ROS and SMTEK methods significantly improved the performance of the PCA-based AMiDS model. Additionally, it boosted the performance of the oSVM-based AMiDS model compared to regular learning. On the other hand, the oSVM-based model performed better than the PCA-based model when using imbalanced learning when the BALCAS ensemble-based method was applied. When considering regular learning, both models significantly improved under imbalanced learning conditions. In addition, the PCA-based AMiDS model performed significantly better than the oSVM-based AMiDS model when using imbalanced learning when applying the KMS and SMTEN methods. However, using the EENSMB methods only boosted the performance of the PCA-based AMiDS model.
Invariably, imbalanced learning improved the performance of the AMiDS models compared to regular learning, where the models performed poorly. Nonetheless, the methods used in this study did not achieve equally significant performance gains. Therefore, choosing the appropriate sampling or resampling method for imbalanced learning depends primarily on the AMiDS model. Furthermore, while low-complexity AMiDS models can benefit from imbalanced learning, the sampling of imbalanced data using oversampling methods can result in slow model training compared to that produced with hybrid- and ensemble-based methods due to the large amount of data generated to increase the distribution of the minority class. However, the models did not incur a high training time in this study.
MioT and CioT devices generally have low energy and storage requirements. Moreover, they operate in hostile or semi-hostile environments where anomalous events are usually more frequent than in regular IoT environments. Therefore, it is essential to consider imbalanced learning methods that are not memory or computationally intense. Further, characterizing the MioT and CioT as environments and systems operating under hostile events dictates a critical need to train low-complexity AMiDS models using passive and active adversarial learning.
In the future, we plan to expand the pool of generative methods to apply methods such as the Wasserstein GAN (WGAN) and examine their impacts on the performance of the AMiDS models. Furthermore, we will explore the use of online or stream ML learning versus batch or static ML learning to improve the AMiDS models’ predictability in the case of new or unseen anomalous examples. In doing so, we aim to select the most suitable imbalanced learning method to build highly predictive AMiDS models to address the rare prediction case. Then, we will compare the resulting models with state-of-the-art AMiDS models. In addition, we will consider the mapping of MioT and CioT data, which, unlike image data, are non-continuous, containing both categorical and numeric data for adversarial learning.
Finally, this study considered the data imbalance problem in light of the rare event case to examine the impact of imbalanced learning on the performance of low-complexity AMiDS models to detect intrusions in CioT systems and MioT environments.
Author Contributions
Conceptualization, G.A. and D.B.R.; methodology, G.A. and D.B.R.; investigation, G.A., D.B.R. and A.R.; resources, D.B.R. and A.R.; writing—original draft preparation, G.A.; writing—review and editing, G.A., D.B.R. and A.R.; supervision, D.B.R.; project administration, D.B.R. and A.R.; funding acquisition, D.B.R. All authors contributed equally to this work. All authors have read and agreed to the published version of the manuscript.
Funding
This research was funded in part by the DoD Center of Excellence in AI and Machine Learning (CoE-AIML) at Howard University under Contract Number W911NF-20-2-0277 with the U.S. Army Research Laboratory, the Microsoft Research and the U.S. National Science Foundation under grant number 1828811 and CNS/SaTC 2039583 and Microsoft Research Gift Funds. Any opinions, findings, conclusions, or recommendations expressed in this document are those of the authors and should not be interpreted as necessarily representing the official policies, either expressed or implied, of the funding agencies.
Institutional Review Board Statement
Not applicable.
Informed Consent Statement
Not applicable.
Data Availability Statement
Not applicable.
Acknowledgments
Special thanks is given to my colleague Uchenna Ezeboi for sharing their programming expertise in fixing bugs and resolving packages and programming conflicts.
Conflicts of Interest
The authors declare no conflict of interest.
References
- Ranjan, C. Understanding Deep Learning: Application in Rare Event Prediction. In Imbalanced Learning; Connaissance Publishing: Atlanta, GA, USA, 2020; pp. 19–27. [Google Scholar]
- Kumar, V.; Lalotra, G.S.; Sasikala, P.; Rajput, D.S.; Kaluri, R.; Lakshmanna, K.; Shorfuzzaman, M.; Alsufyani, A.; Uddin, M. Addressing Binary Classification over Class Imbalanced Clinical Datasets Using Computationally Intelligent Techniques. Healthcare 2022, 10, 1293. [Google Scholar] [CrossRef] [PubMed]
- Gonzalez-Cuautle, D.; Hernandez-Suarez, A.; Sanchez-Perez, G.; Toscano-Medina, L.K.; Portillo-Portillo, J.; Olivares-Mercado, J.; Perez-Meana, H.M.; Sandoval-Orozco, A.L. Synthetic Minority Oversampling Technique for Optimizing Classification Tasks in Botnet and Intrusion-Detection-System Datasets. Appl. Sci. 2020, 10, 794. [Google Scholar] [CrossRef]
- Navidan, H.; Moshiri, P.F.; Nabati, M.; Shahbazian, R.; Ghorashi, S.A.; Shah-Mansouri, V.; Windridge, D. Generative Adversarial Networks (GANs) in networking: A comprehensive survey & evaluation. Comput. Netw. 2021, 194, 108149. [Google Scholar] [CrossRef]
- Krawczyk, B. Learning from imbalanced data: Open challenges and future directions. Prog. Artif. Intell. 2016, 5, 221–232. [Google Scholar] [CrossRef]
- He, H.; Garcia, E.A. Learning from Imbalanced Data. IEEE Trans. Knowl. Data Eng. 2009, 21, 1263–1284. [Google Scholar] [CrossRef]
- Weiss, G.M. Foundations of Imbalanced Learning. In Imbalanced Learning; John Wiley & Sons, Ltd.: Hoboken, NJ, USA, 2013; pp. 13–41. [Google Scholar]
- Ling, C.X.; Sheng, V.S. Class Imbalance Problem. In Encyclopedia of Machine Learning; Sammut, C., Webb, G.I., Eds.; Springer: Boston, MA, USA, 2010; p. 171. [Google Scholar]
- He, H. Introduction. In Imbalanced Learning; John Wiley & Sons, Ltd.: Hoboken, NJ, USA, 2013; Chapter 1; pp. 1–12. [Google Scholar]
- Ahmed, S.; Mahbub, A.; Rayhan, F.; Jani, R.; Shatabda, S.; Farid, D.M. Hybrid Methods for Class Imbalance Learning Employing Bagging with Sampling Techniques. In Proceedings of the 2017 2nd International Conference on Computational Systems and Information Technology for Sustainable Solution (CSITSS), Bengaluru, India, 21–23 December 2017; pp. 1–5. [Google Scholar] [CrossRef]
- Johnson, J.M.; Khoshgoftaar, T.M. Survey on deep learning with class imbalance. J. Big Data 2019, 6, 27. [Google Scholar] [CrossRef]
- Desuky, A.S.; Hussain, S. An Improved Hybrid Approach for Handling Class Imbalance Problem. Arab. J. Sci. Eng. 2021, 46, 3853–3864. [Google Scholar] [CrossRef]
- Laplante, P.; Amaba, B. Artificial Intelligence in Critical Infrastructure Systems. Computer 2021, 54, 14–24. [Google Scholar] [CrossRef]
- Kott, A.; Swami, A.; West, B.J. The Internet of Battle Things. Computer 2016, 49, 70–75. [Google Scholar] [CrossRef]
- Cameron, L. IoT Meets the Military and Battlefield: Connecting Gear and Biometric Wearables for an IOMT and IOBT. Available online: https://www.computer.org/publications/tech-news/research/internet-of-military-battlefield-things-iomt-iobt (accessed on 11 October 2021).
- Castiglione, A.; Choo, K.R.; Nappi, M.; Ricciardi, S. Context Aware Ubiquitous Biometrics in Edge of Military Things. IEEE Cloud Comput. 2017, 4, 16–20. [Google Scholar] [CrossRef]
- Lin, D.; Wu, W. Heuristic Algorithm for Resource Allocation in an Internet of Battle Things. IEEE Syst. J. 2022, 17, 1–12. [Google Scholar] [CrossRef]
- Almaiah, M.A.; Almomani, O.; Alsaaidah, A.; Al-Otaibi, S.; Bani-Hani, N.; Hwaitat, A.K.A.; Al-Zahrani, A.; Lutfi, A.; Awad, A.B.; Aldhyani, T.H.H. Performance Investigation of Principal Component Analysis for Intrusion Detection System Using Different Support Vector Machine Kernels. Electronics 2022, 11, 3571. [Google Scholar] [CrossRef]
- Tsiknas, K.; Taketzis, D.; Demertzis, K.; Skianis, C. Cyber Threats to Industrial IoT: A Survey on Attacks and Countermeasures. IoT 2021, 2, 163–186. [Google Scholar] [CrossRef]
- Panchal, A.C.; Khadse, V.M.; Mahalle, P.N. Security Issues in IIoT: A Comprehensive Survey of Attacks on IIoT and Its Countermeasures. In Proceedings of the 2018 IEEE Global Conference on Wireless Computing and Networking (GCWCN), Lonavala, India, 23–24 November 2018; pp. 124–130. [Google Scholar] [CrossRef]
- Mudassir, M.; Unal, D.; Hammoudeh, M.; Azzedin, F. Detection of Botnet Attacks against Industrial IoT Systems by Multilayer Deep Learning Approaches. Wirel. Commun. Mob. Comput. 2022, 2022, e2845446. [Google Scholar] [CrossRef]
- Abosata, N.; Al-Rubaye, S.; Inalhan, G.; Emmanouilidis, C. Internet of Things for System Integrity: A Comprehensive Survey on Security, Attacks and Countermeasures for Industrial Applications. Sensors 2021, 21, 3654. [Google Scholar] [CrossRef] [PubMed]
- Hoffman, F. Industrial internet of things vulnerabilities and threats: What stakeholders need to consider. Issues Inf. Syst. 2019, 20, 119–133. [Google Scholar] [CrossRef]
- Jiang, W. Machine Learning Methods to Detect Voltage Glitch Attacks on IoT/IIoT Infrastructures. Comput. Intell. Neurosci. 2022, 2022, 6044071. [Google Scholar] [CrossRef]
- Wu, C.K. IoT Perception Layer Security. In Internet of Things Security: Architectures and Security Measures; Wu, C.K., Ed.; Advances in Computer Science and Technology; Springer: Singapore, 2021; pp. 95–106. [Google Scholar]
- Nasralla, M.M.; Garcia-Magarino, I.; Lloret, J. Defenses Against Perception-Layer Attacks on IoT Smart Furniture for Impaired People. IEEE Access 2020, 8, 119795–119805. [Google Scholar] [CrossRef]
- Singh, D.; Mishra, M.K.; Lamba, A.; Swagatika, S. Security Issues In Different Layers Of IoT And Their Possible Mitigation. Int. J. Sci. Technol. Res. 2020, 9, 10. [Google Scholar]
- Osborne, C. Mirai Splinter Botnets Dominate IoT Attack Scene. 2022. Available online: https://www.zdnet.com/article/mirai-splinter-botnets-dominate-iot-attack-scene/ (accessed on 3 April 2022).
- Zhang, X.; Upton, O.; Beebe, N.L.; Choo, K.K.R. IoT Botnet Forensics: A Comprehensive Digital Forensic Case Study on Mirai Botnet Servers. Forensic Sci. Int. Digit. Investig. 2020, 32, 300926. [Google Scholar] [CrossRef]
- Bursztein, E. Inside the Infamous Mirai IoT Botnet: A Retrospective Analysis. 2017. Available online: http://blog.cloudflare.com/inside-mirai-the-infamous-iot-botnet-a-retrospective-analysis/ (accessed on 30 March 2022).
- De Donno, M.; Dragoni, N.; Giaretta, A.; Spognardi, A. DDoS-Capable IoT Malwares: Comparative Analysis and Mirai Investigation. Secur. Commun. Netw. 2018, 2018, e7178164. [Google Scholar] [CrossRef]
- Horak, T.; Strelec, P.; Huraj, L.; Tanuska, P.; Vaclavova, A.; Kebisek, M. The Vulnerability of the Production Line Using Industrial IoT Systems under DDoS Attack. Electronics 2021, 10, 381. [Google Scholar] [CrossRef]
- Verma, A.; Ranga, V. Machine Learning Based Intrusion Detection Systems for IoT Applications. Wirel. Pers Commun. 2020, 111, 2287–2310. [Google Scholar] [CrossRef]
- Lin, Y.D.; Liu, Z.Q.; Hwang, R.H.; Nguyen, V.L.; Lin, P.C.; Lai, Y.C. Machine Learning With Variational AutoEncoder for Imbalanced Datasets in Intrusion Detection. IEEE Access 2022, 10, 15247–15260. [Google Scholar] [CrossRef]
- Wan, Z.; Zhang, Y.; He, H. Variational autoencoder based synthetic data generation for imbalanced learning. In Proceedings of the 2017 IEEE Symposium Series on Computational Intelligence (SSCI), Honolulu, HI, USA, 27 November–1 December 2017; IEEE: Piscataway, NJ, USA, 2017; pp. 1–7. [Google Scholar]
- Sikha, B.; Kunqi, L. Resampling imbalanced data for network intrusion detection datasets. J. Big Data 2021, 8, 6. [Google Scholar] [CrossRef]
- Bedi, P.; Gupta, N.; Jindal, V. Siam-IDS: Handling class imbalance problem in Intrusion Detection Systems using Siamese Neural Network. Procedia Comput. Sci. 2020, 171, 780–789. [Google Scholar] [CrossRef]
- Rodda, S.; Erothi, U.S.R. Class imbalance problem in the Network Intrusion Detection Systems. In Proceedings of the 2016 International Conference on Electrical, Electronics, and Optimization Techniques (ICEEOT), Chennai, India, 3–5 March 2016; pp. 2685–2688. [Google Scholar]
- Cieslak, D.; Chawla, N.; Striegel, A. Combating imbalance in network intrusion datasets. In Proceedings of the 2006 IEEE International Conference on Granular Computing, Atlanta, GA, USA, 10–12 May 2006; pp. 732–737. [Google Scholar] [CrossRef]
- Louk, M.H.L.; Tama, B.A. Exploring Ensemble-Based Class Imbalance Learners for Intrusion Detection in Industrial Control Networks. Big Data Cogn. Comput. 2021, 5, 72. [Google Scholar] [CrossRef]
- Agustianto, K.; Destarianto, P. Imbalance Data Handling using Neighborhood Cleaning Rule (NCL) Sampling Method for Precision Student Modeling. In Proceedings of the 2019 International Conference on Computer Science, Information Technology, and Electrical Engineering (ICOMITEE), Jember, Indonesia, 16–17 October 2019; pp. 86–89. [Google Scholar] [CrossRef]
- Leevy, J.L.; Khoshgoftaar, T.M.; Bauder, R.A.; Seliya, N. A survey on addressing high-class imbalance in big data. J. Big Data 2018, 5, 42. [Google Scholar] [CrossRef]
- Lee, J.; Park, K. GAN-based imbalanced data intrusion detection system. Pers. Ubiquit. Comput. 2021, 25, 121–128. [Google Scholar] [CrossRef]
- Li, B.; Xiaofeng, G.; Wang, C.; Ruijuan, W.; Tong, B.; Li, Y.; Wang, Z.; Ruisen, L. MMD-encouraging convolutional autoencoder: A novel classification algorithm for imbalanced data. Appl. Intell. 2021, 51, 7384–7401. [Google Scholar] [CrossRef]
- Fuertes, T. Variational Autoencoder as a Method of Data Augmentation Quantdare. 2020. Available online: https://quantdare.com/variational-autoencoder-as-a-method-of-data-augmentation/ (accessed on 11 December 2022).
- Wan, Z.; He, H.; Tang, B. A Generative Model for Sparse Hyperparameter Determination. IEEE Trans. Big Data 2018, 4, 2–10. [Google Scholar] [CrossRef]
- Kingma, D.P.; Welling, M. An Introduction to Variational Autoencoders. FNT Mach. Learn. 2019, 12, 307–392. [Google Scholar] [CrossRef]
- MathWorks, T. Train Variational Autoencoder (VAE) to Generate Images. 2023. Available online: https://www.mathworks.com/help/deeplearning/ug/train-a-variational-autoencoder-vae-to-generate-images.html (accessed on 21 January 2023).
- Baldi, P. Autoencoders, Unsupervised Learning, and Deep Architectures. In ICML Workshop on Unsupervised and Transfer Learning. JMLR Workshop and Conference Proceedings; JMLR: Washington, DC, USA, 2012; pp. 37–49. ISSN 1938-7228. [Google Scholar]
- Feng, W.; Huang, W.; Ren, J. Class Imbalance Ensemble Learning Based on the Margin Theory. Appl. Sci. 2018, 8, 815. [Google Scholar] [CrossRef]
- Liu, X.Y.; Wu, J.; Zhou, Z.H. Exploratory Undersampling for Class-Imbalance Learning. IEEE Trans. Syst. Man Cybern. Part B 2009, 39, 539–550. [Google Scholar] [CrossRef]
- Last, F.; Douzas, G.; Bacao, F. Oversampling for Imbalanced Learning Based on K-Means and SMOTE. Inf. Sci. 2018, 465, 1–20. [Google Scholar] [CrossRef]
- Bej, S.; Davtyan, N.; Wolfien, M.; Nassar, M.; Wolkenhauer, O. LoRAS: An oversampling approach for imbalanced datasets. Mach. Learn. 2021, 110, 279–301. [Google Scholar] [CrossRef]
- Menardi, G.; Torelli, N. Training and assessing classification rules with imbalanced data. Data Min. Knowl. Discov. 2014, 28, 92–122. [Google Scholar] [CrossRef]
- Chawla, N.V.; Bowyer, K.W.; Hall, L.O.; Kegelmeyer, W.P. SMOTE: Synthetic minority over-sampling technique. J. Artif. Int. Res. 2002, 16, 321–357. [Google Scholar] [CrossRef]
- Batista, G.E.A.P.A.; Prati, R.C.; Monard, M.C. A study of the behavior of several methods for balancing machine learning training data. SIGKDD Explor. Newsl. 2004, 6, 20–29. [Google Scholar] [CrossRef]
- Kingma, D.P.; Welling, M. Auto-Encoding Variational Bayes. arXiv 2022, arXiv:1312.6114. [Google Scholar]
- Tomescu, V.I.; Czibula, G.; Nitica, S. A study on using deep autoencoders for imbalanced binary classification. Procedia Comput. Sci. 2021, 192, 119–128. [Google Scholar] [CrossRef]
- Raghuwanshi, B.S.; Shukla, S. Classifying imbalanced data using BalanceCascade-based kernelized extreme learning machine. Pattern Anal. Appl. 2020, 23, 1157–1182. [Google Scholar] [CrossRef]
- Lemaitre, G.; Nogueira, F.; Aridas, C.K. Imbalanced-Learn API—Imbalanced-Learn 0.3.0.dev0 Documentation. 2016. Available online: http://glemaitre.github.io/imbalanced-learn/api.html (accessed on 26 November 2022).
- Lemaître, G.; Nogueira, F.; Aridas, C.K. Imbalanced-learn: A Python Toolbox to Tackle the Curse of Imbalanced Datasets in Machine Learning. J. Mach. Learn. Res. 2017, 18, 1–5. [Google Scholar]
- zoj613. LoRAS. 2022. Available online: https://github.com/zoj613/pyloras (accessed on 26 November 2022).
- Pradipta, G.A.; Wardoyo, R.; Musdholifah, A.; Sanjaya, I.N.H.; Ismail, M. SMOTE for Handling Imbalanced Data Problem: A Review. In Proceedings of the 2021 Sixth International Conference on Informatics and Computing (ICIC), Jakarta, Indonesia, 3–4 November 2021; pp. 1–8. [Google Scholar] [CrossRef]
- Fernandez, A.; Garcia, S.; Herrera, F.; Chawla, N.V. SMOTE for Learning from Imbalanced Data: Progress and Challenges, Marking the 15-year Anniversary. J. Artif. Intell. Res. 2018, 61, 863–905. [Google Scholar] [CrossRef]
- Kovacs, G. An empirical comparison and evaluation of minority oversampling techniques on a large number of imbalanced datasets. Appl. Soft Comput. 2019, 83, 105662. [Google Scholar] [CrossRef]
- Swana, E.F.; Doorsamy, W.; Bokoro, P. Tomek Link and SMOTE Approaches for Machine Fault Classification with an Imbalanced Dataset. Sensors 2022, 22, 3246. [Google Scholar] [CrossRef]
- Wang, Z.; Wu, C.; Zheng, K.; Niu, X.; Wang, X. SMOTETomek-based resampling for personality recognition. IEEE Access 2019, 7, 129678–129689. [Google Scholar] [CrossRef]
- Yanni, D. vae-oversampler: Oversample a Minority Class Using a Variational Autoencoder. 2019. Available online: https://github.com/dyanni3/vae_oversampler (accessed on 25 November 2022).
- Abdelmoumin, G.; Rawat, D.B.; Rahman, A. On the Performance of Machine Learning Models for Anomaly-Based Intelligent Intrusion Detection Systems for the Internet of Things. IEEE Internet Things J. 2021, 9, 6. [Google Scholar] [CrossRef]
- Abdelmoumin, G.; Rawat, D.B. SmartIDS: A Comparative Study of Intelligent Intrusion Detection Systems for Internet of Things; Lecture Notes in Networks and Systems; Springer Nature: London, UK, 2021. [Google Scholar]
- Moustafa, N. The Bot-IoT Dataset. 2019. Available online: https://ieee-dataport.org/documents/bot-iot-dataset (accessed on 31 October 2020).
- Koroniotis, N.; Moustafa, N.; Sitnikova, E.; Turnbull, B. Towards the development of realistic botnet dataset in the Internet of Things for network forensic analytics: Bot-IoT dataset. Future Gener. Comput. Syst. 2019, 100, 779–796. [Google Scholar] [CrossRef]
- Shan, P.; Downer, R.; McKittrick, M.; Haas, S.; Coulter, D.; Verma, R.; Borsecnik, J.; Hughes, L. Fsv2-Series—Azure Virtual Machines. Microsoft: Redmond, WA, USA, 2022. Available online: https://learn.microsoft.com/en-us/azure/virtual-machines/fsv2-series (accessed on 16 August 2023).
- Crop, M. Azureml-Designer-Classic-Modules: A Variety of Modules for Data Processing, Model Training, Inferencing and Evaluation. 2022. Available online: https://pypi.org/project/azureml-designer-classic-modules/ (accessed on 2 December 2022).
- Lemaitre, G. Release History—Version 0.9.1. Available online: https://imbalanced-learn.org/stable/whats_new.html (accessed on 2 December 2022).
- Engel, J.N. Dealing with Imbalance: Autoencoder & Sampling; Kaggle: San Francico, CA, USA, 2020. [Google Scholar]
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).