Abstract
The Methionine Synthase process, in principle, can take an unlimited number of turnovers in the presence of the AdoMet substrate. In the absence of this substrate, the Methionine Synthase process lasts only about 2000 turnovers. During 2000 turnovers, the entire amount of methylcob(II)alamin cofactor is converted into inactive cob(II)alamin particles. Nevertheless, the mechanism of the Methionine Synthase process determined previously lacks the presence of the AdoMet substrate. On the other hand, the first step of this mechanism was only mentioned earlier without its analysis. The CASSCF geometry optimization of the inactive cob(II)alamin cofactor particle plus the AdoMet ion substrate and of the methylcob(II)alamin cofactor particle plus homocysteine ion and histidine molecule joint models have been performed. CASSCF calculations show that the AdoMet particle transfers the methyl radical to the biologically inactive cob(II)alamin particle during their interaction, transforming it into the biologically active particle of methylcob(II)alamin. CASSCF geometry optimization of the second model leads to the Co-N bond’s full cleavage. The two processes take place in the absence of the total energy barrier. The fully updated mechanism of the Methionine Synthase process has been drawn.
1. Introduction
The Methionine Synthase process’s general features have been determined theoretically and experimentally [1,2,3,4,5,6,7,8,9,10,11,12,13,14,15,16,17,18,19,20,21,22,23]. Nevertheless, the detailed in vivo mechanism of the Methionine Synthase process for a long time was unknown. One of the most debated stages of the Methionine Synthase process was the transfer of the methyl radical from methylcob(III)alamin cofactor to homocysteine (Figure 1). The mechanism of the Co-C bond cleavage was studied by Density Functional Theory (DFT) and Quantum Mechanics/Molecular Mechanics (QM/MM) DFT-based methods [11,12,13].
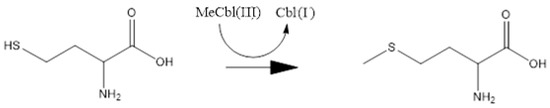
Figure 1.
The methyl radical transfer from methylcob(III)alamin cofactor to homocysteine in the methionine synthesis process. Cbl(I): cob(I)alamin, MeCbl(III): methylcob(III)alamin.
Although DFT and DFT-based QM/MM calculations obtained fairly high energy barriers in the SN1 Co-C bond cleavage reaction (24.4 kcal/mol.) [12], in the SN2 reaction of the methyl radical transfer from the methylcobalamin cofactor to homocysteine, the total energy barrier is lower [11,13,14] (10.5 kcal/mol., 8.5 kcal/mol. and 7.3 kcal/mol.). Still, it cannot explain the experimentally found practically unlimited number of Methionine Synthase turnovers in the presence of the S-adenosylmethionine (AdoMet) substrate [23]. On the other hand, DFT calculations show that the energy barrier of the Co-C bond cleavage reaction in the base-off species is much lower than in the base-on (Figure 2) species during the same reaction [12] in contradiction with the experimental data, which show that the relationship between the two barriers is the opposite [5,6,7,8,9,10,11,12,13,14,15,16,17,18,19]. Finally, according to these calculations [11,12,13,14], the base-off is the active particle in the Methionine Synthase process, excluding the base-on particle from the process in contradiction with the generally accepted mechanism [1,2,3,4,5,6,7,8,9,10]. The contradictions between the DFT-based methods results and experimental data have been explained by the limitations of the DFT method in the study of the processes where the pseudo-Jahn–Teller effect is effective [15,16,20]. Another part of the Methionine Synthase process mechanism, the transfer of the methyl radical from 5-methyltetrahydrofolate to cob(II)alamin cofactor (Figure 3), was studied by the DFT method [21]. Unfortunately, for the same reason, the total energy barrier of this obtained transfer was too high (38 kcal/mol.) [21] to be considered in the in vivo mechanism of this process.
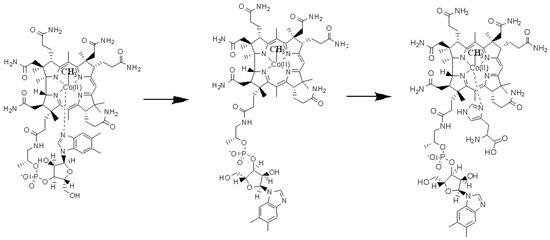
Figure 2.
The schematic picture of the Methionine Synthase process’s first step. Left is a base-on structure, in the middle is a base-off structure, and on the right is a base-on structure of the methylcobalamin cofactor with a histidine ligand-bonded central cobalt atom.
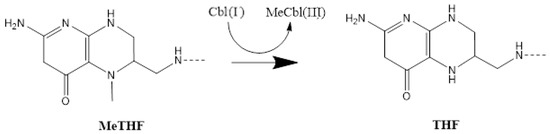
Figure 3.
The methyl radical transfer from 5-methyltetrahydrofolate to cob(III)alamin cofactor in the methionine synthesis process. MeTHF: 5-methyltetrahydrofolate, THF: tetrahydrofolate.
The mechanism of the Methionine Synthase process has recently been theoretically resolved [22]. The multi-configurational self-consistent field (MCSCF) was a key method for a detailed electronic structure and mechanism of the vitamin B12 cofactor-dependent process determinations since the pseudo-Jahn–Teller effect, which is effective in the in vivo active reactions of the vitamin B12 cofactors [15,16], is taken into account by the MCSCF methods. The MCSCF geometry optimization process shows that during the interaction of the 5-methyltetrahydrofolate protonated positive ion with the cob(I)alamin cofactor negative ion, the transfer of the methyl carbocation from the first compound to the second one takes place [22]. Another MCSCF geometry optimization process shows that the negative ion of cysteine transfers an electron to the methylcobalamin cofactor, under the influence of which the Co-C chemical bond is dramatically elongated and broken during the interaction with the negative ion of cysteine, reversely transferring the methyl radical to it [22]. Although the main reaction mechanisms of the Methionine Synthase process have been successfully determined, two more problems need to be solved for the complete description of its in vivo mechanism. It is known that the dimethylbenzimidazole ligand is substituted by the histidine molecule [6] in the methylcob(III)alamin cofactor (Figure 2), and the formed compound participates in the Methionine Synthase mechanism [22]. To substitute the dimethylbenzimidazole ligand with a histidine molecule, the Co-N bond must be cleaved first. Given that this reaction anticipates the entire Methionine Synthase process, we can consider the Co-N bond cleavage as the first-step reaction to this process. In our previous discussion, this reaction was only vaguely mentioned [22], so one of the goals of this article is to solve this first-step reaction mechanism of the Methionine Synthase process. Another problem, which is not yet determined, is the role of the AdoMet substrate in the Methionine Synthase process. Matthews and co-authors have studied the dependence of the Methionine Synthase process on its integrity, particularly on the presence of the AdoMet substrate. Surprisingly, the Methionine Synthase process, which naturally takes, in principle, an unlimited number of turnovers, has gradually slowed down to a total ceasing after about 2000 turnovers [23]. The cause of this cessation of the Methionine Synthase process is the gradual accumulation of cob(II)alamin particles, which are biologically inactive in this process so that at the 2000th turnover, their concentration is close to 100%. Initially, it was assumed that some of the cob(I)alamin cofactor particles are oxidized with flavodoxin in the in vivo Methionine Synthase process [8]. This is annoying because of some experimental evidence. First, explaining how the accumulation of inactive particles of the cob(II)alamin cofactor occurs at fixed rates is difficult. Second, other experimental evidence did not consider flavodoxin to participate in the mechanism of the Methionine Synthase process. Third, only the presence of the AdoMet substrate influences the number of turnovers running during the Methionine Synthase process [23]. The dependence of the Methionine Synthase turnover number on the presence of flavodoxin is unknown to our knowledge. The electronic structure of the methylcob(II)alamin cofactor has been calculated by the complete active space self-consistent field (CASSCF) method, and the total energy barrier of the Co-C bond cleavage reaction in the base-on methylcob(II)alamin cofactor has been found to be quite low (3.32 kcal/mol.) [22]. This energy barrier allows about 0.367% of the methylcob(II)alamin base-on cofactor particles to have the Boltzmann energy needed to break the Co-C bond at room temperature. Thus, 0.367% of methylcob(II)alamin cofactor during each Methionine Synthase process turnover is transformed into inactive cob(II)alamin particles, which are unable to participate in the SN2 methyl radical transfer reaction from methylcob(II)alamin cofactor to homocysteine. The simple calculation shows that after 2000 turnovers, only about 6.4 × 10–4% of methylcobalamin particles remain active, and the Methionine Synthase process is practically stopped. Although the cause of the effect of the lack of AdoMet in the Methionine Synthase process has been determined, the role of the AdoMet substrate is still unknown. This is the second scope of this article.
2. Computational Details
The geometry optimization of the Methionine Synthase bioprocess models, which includes either cob(II)alamin cofactor inactive particle and AdoMet substrate (Figure 4a) or methylcobalamin cofactor, histidine molecule, and homocysteine radical substrates (Figure 4b) have been performed by using the CASSCF method. Thirteen orbitals and thirteen electrons were taken into the CASSCF active space for a second model and twelve electrons and twelve orbitals for the first model of the studied methylcobalamin-dependent processes. The increase in the CASSCF active area to (13,14), (14,15) of the second model, and to (14,14) active areas of the first model does not lead to modifying electronic structure results for the calculated systems. Unfortunately, the further increase in the active area of the CASSCF calculations leads to the remarkable need for calculation time and memory thousands of times larger because of the number of determinants that must be solved at each iteration. For example, for (15,15) active areas, the number of determinants to be resolved at each iteration is three orders of magnitude higher than the same number of determinants to be solved for (13,13) and (12,12) active areas. To our knowledge, the CASSCF calculation of such a large system with (15,15) active areas with NwChem code is currently inaccessible to the existing computing resources in the university environment. On the other hand, we have demonstrated that increasing its activity leads to a greater pseudo-Jahn–Teller effect, responsible for the molecular transformations studied below [15]. A 6-31G** basis set for the cobalt and oxygen atoms and a 6-31G basis set for the remainder of the atoms have been used for all geometry optimization procedures. Such a basis set was used successfully for an electronic structure of the cofactor-dependent bio-process calculations [22,24]. NwChem computational software [25] was the main code used to optimize the geometry of the CASSCF calculated model. A quasi-newton optimization with line searches and approximate energy Hessian updates has been used as the optimization procedure. The side chains of the methylcobalamin cofactor have been replaced by hydrogen atoms (Figure 4). In principle, the calculations should include dynamic factors to have a more severe complete active space perturbation theory (CASPT2) approximation. However, the results of our calculations agree with the experimental evidence, which gives us confidence that our CASSCF calculations describe the studied systems well enough. We believe that our calculations allowed us to take into account all orbitals responsible for the Co-N bond cleavage.
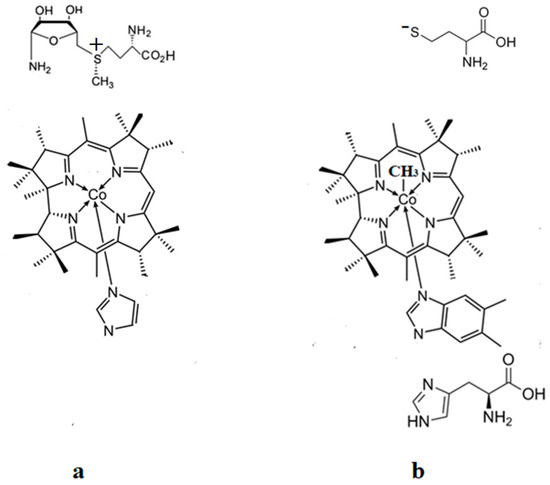
Figure 4.
The schematic model structures used in the CASSCF calculations: (a) model of the AdoMet substrate interaction with the inactive cob(II)alamin cofactor particle; (b) model of the Methionine Synthase process first step.
3. Results and Discussion
The preliminary step of the Methionine Synthase process. The most likely factor that would cause the cleavage of the Co-N axial bond, i.e., breaking the chemical bond between the dimethylbenzimidazole ligand and the central cobalt atom, is the influence of substrates on the vitamin B12 methylcobalamin cofactor. X-ray data [2,3,4,6,7,10] show a number of the substrates, which are near the vitamin B12 cofactors structures. One of the most present substrates near the methylcobalamin cofactor molecule in the Methionine Synthase process is that of negative homocysteine ion [22]. Second, according to X-ray data [2,3,4,6,7,10], there are several substrates in the proximity of the axial dimethylbenzimidazole ligand, such as histidine, glycine, tyrosine, arginine, threonine, serine, lysine, glutamine, leucine, asparagine, and tartrate, which can influence vitamin B12 cofactors. We have shown that the methylcobalamin-dependent enzyme process occurs in the absence of total energy barriers [22]. Therefore, we sought to find models in which the preliminary step of these processes also takes place in the absence of the total energy barrier. We have built all models in which, in addition to each structure of methylcobalamin cofactor, we added one, two, or three of the substrates presented above. The CASSCF geometry optimization of these joint models showed that the cleavage of the Co-N bond, in most cases, occurs with energy barriers, except for the model presented in Figure 4b. We have described in detail the whole mechanism of the Methionine Synthase bio-process [22]. According to this mechanism, the central cobalt atom of the methylcobalamin cofactor molecule is reduced by one electron modifying its oxidation state from +3 to +2 before the preliminary Methionine Synthase process step takes place. We have proven that the methylcobalamin base-on cofactor with histidine ligand bonded to the central cobalt atom is the active catalytic particle in the Methionine Synthase process [22]. Therefore, the Co-N(dimethylbenzimidazole) axial bond of the methylcobalamin cofactor must be cleaved in the preliminary step to allow the histidine ligand to bind to the central cobalt atom of the methylcobalamin cofactor before the Methionine Synthase process starts. It is well known that the negative homocysteine ion, which occurs in the cavity methylcobalamin cofactor after its de-coordination from the zinc coordinating compound, is part of the Methionine Synthase process [22]. We used the homocysteine ion, histidine molecule, and methylcobalamin cofactor structure as the first joint model for CASSCF geometry optimization (Figure 4b). We started the CASSCF geometry optimization of the Methionine Synthase preliminary step model with a Co-C axial bond distance equal to 2.08 Å and a Co-N axial bond distance equal to 2.35 Å, taking the Co-C bond distance from our previous CASSCF geometry optimization results [22], and a Co-N bond distance from the X-ray data [7] in biomaterials. We have shown that the behavior of the methylcobalamin cofactor, whose central atom is reduced by one electron, is governed by the pseudo-Jahn–Teller-effect by mixing the highest occupied molecular orbitals with the lowest unoccupied molecular orbitals [22]. At the beginning of the MCSCF geometry optimization of the Methionine Synthase preliminary step model (Figure 4b), the electron density population on the two molecular orbitals differs the most from integers 2,1 and 0 numbers among other molecular orbitals of the active space as a result of the orbital mixing process (Figure 5, see also Figure S1 in the supporting information). Mixing these two orbitals seems the largest during the CASSCF procedure process. The electronic density population on these two orbitals is equal to 1.72 e− and 0.28 e−. The CASSCF method has no formal HOMO, the highest occupied molecular orbital, or LUMO, the lowest unoccupied molecular orbital. However, without having the names of active orbitals in the reactions we are studying, it will be very difficult to give a reader a clear picture of the factors influencing them. For the convenience of further discussion, we will name the highest molecular orbital with an electronic population equal or greater than 1 e− the HOMO and then go lower with the next orbitals, HOMO1, HOMO2, etc., and the lower molecular orbital with a population less than 1 e− we will call the LUMO, going higher with the next orbitals, LUMO1, LUMO2, etc., through the entire paper. Therefore, we can call the molecular orbital with a population equal to 1.72 e− a HOMO2 and the molecular orbital with a population equal to 0.28 e− a LUMO for this particular model calculation. It should be noted that only the electronic density difference from 2 e− on HOMO2 and the electron density difference from 0 on the LUMO of the calculated systems is quite significant and can influence the calculated model’s CASSCF geometry optimization pathway. The electron density difference from 2, 1, or 0 on other occupied molecular orbitals of the active CASSCF space is lower compared to that of HOMO2 and LUMO orbitals and, therefore, less influences the CASSCF geometry optimization procedure of the calculated system. Here, we admit from the fact that in the absence of orbital mixing, or in other words, in the absence of the pseudo-Jahn–Teller effect, the separate constituent structural parts of the model are stable, and their structure does not change. Interestingly, both the electronic density population of HOMO2, which is equal to approximately 1.72 e−, and the electronic density population of LUMO, which is equal to approximately 0.28 e−, remain unchanged throughout the whole CASSCF geometry optimization procedure of the calculated model. It can be seen from Figure 5 (see also Figure S1) that HOMO2 is a substrate molecular orbital, and LUMO is an antibonding orbital, which includes the atomic orbitals of the corrin ring and the dimethylbenzimidazole ligand. It turns out that the HOMO–LUMO mixing process creates a repulsive force of the dimethylbenzimidazole ligand from the cobalt central atom and from the corrin ring of the methylcobalamin cofactor molecule. On the other hand, the Co-N axial bond is the weakest coordination bond in vitamin B12 cofactors, its length being placed between 2.30 Å and 2.50 Å in biological materials [6,7,10]. The repulsive force between the dimethylbenzimidazole ligand and the central cobalt atom (and the corrin ring) increases the distance of the Co-N chemical bond during the CASSCF Methionine Synthase model preliminary step geometry optimization. The CASSCF geometry optimization leads to the permanent Co-N bond distance increasing up to more than 4.00 Å distance, where there is no cobalt–axial nitrogen or corrin ring–axial nitrogen interaction. Taking into account the above-mentioned nature and electronic density on the most mixing molecular orbitals, we can conclude that the summary charge on the substrate and on the methylcobalamin cofactor are fractional; that is, the transfer of electron density from the substrate to the methylcobalamin cofactor takes place, and this stimulates the breaking of the Co-N chemical bond. Thus, two factors in the electronic structure of the calculated CASSCF model lead to the complete rupture of the Co-N bond in the CASSCF geometry optimization process and the removal of the dimethylbenzimidazole ligand from the central atom and the corrin ring (Figure 6), the active pseudo-Jahn–Teller effect, and the weakness of the Co-N bond.
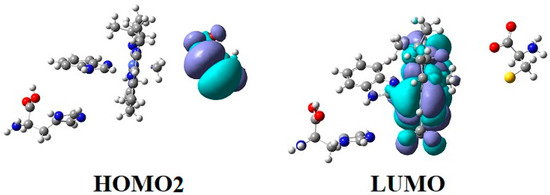
Figure 5.
The HOMO2 and LUMO surfaces of the Methionine Synthase process preliminary step model (Figure 1a) at the beginning of the CASSCF geometry optimization process (R (Co-C) = 2.08 Å; R (Co-N) = 2.35 Å).
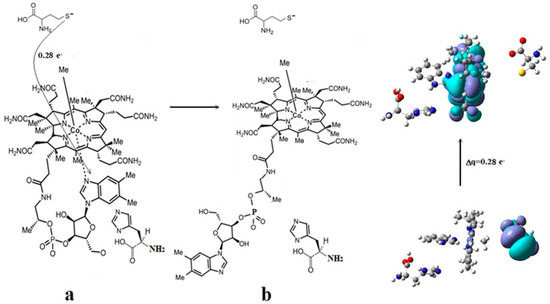
Figure 6.
The mechanism of the Co-N bond cleavage in the methylcobalamin cofactor under the influence of the homocysteine and histidine substrates: (a) substrate–adenosylcobalamin charge transfer; (b) the cleavage of the Co-N bond.
The AdoMet substrate’s role in the Methionine Synthase process. It is known that the Methionine Synthase process in the presence of the AdoMet substrate can run, in principle, an unlimited number of turnovers. However, from the above, it appears that in the absence of the AdoMet substrate, the process runs only about 2000 turnovers. Therefore, the AdoMet substrate plays an important role in the proper conduct of the Methionine Synthase process. The AdoMet substrate can exist in the basic cation, zwitterion, and dissociated hydrogen bond specie. To find the role of AdoMet in the Methionine Synthase process, each of these AdoMet states, together with the cob(II)alamin cofactor inactive particle, were used as the common models of the CASSCF geometry optimization. The biologically inactive cob(II)alamine particle actively interacts with the first two forms of the AdoMet substrate through the coulombic interaction (each of the two AdoMet sulfonic cations and the inactive cob(II)alamine particle anion are attracted to each other), and by their occupied and unoccupied orbitals mixing. Meanwhile, in the case of the interaction of the cob(II)alamine particle with that of AdoMet, which is lost the proton, the systems attract each other only due to their common orbital mixing. Here, we will consider only the interaction of two opposite-charged ions, the inactive cob(II)alamine particle anion, and the basic state of the AdoMet sulfonic cation (the behavior of the zwitterion is similar) because, in this case, the proximity of the two structures of the calculated model is granted by two factors, their opposite charges, and their orbital mixing process. At Co-S and Co-C distances equal or greater than 4.35 Å and 2.50 Å, the ionic attraction ensuring the proximity of the two structures remains most important. At smaller distances, their interaction is ensured mostly by orbital mixing.
The geometry optimization procedures were performed at different distances, starting with the distance between the closest reagent atoms slightly greater than 4.00 Å. Several mixing common intermolecular molecular orbitals have already been formed at this distance, with a significant observable electron density population different from 2 and 0 numbers (see Figure S2). The HOMOs and LUMOs orbitals include both the S-C bond atoms, which must break to transfer the methyl radical from the AdoMet ion to the inactive cob(II)alamin particle, and the Co-C bond atoms, which should form to revitalize the cob(II)alamin cofactor inactive particle (Figure 7 and Figure S2).
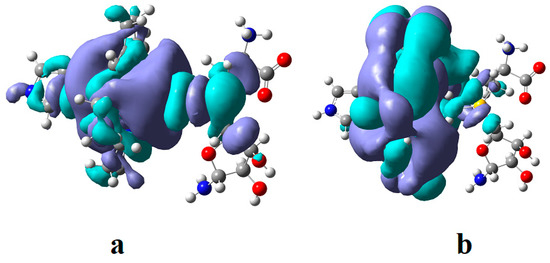
Figure 7.
The HOMO (a) and LUMO (b) surfaces of the cob(II)alamin inactive particle and AdoMet substrate common model at the beginning of the CASSCF geometry optimization process.
The HOMOs are more S-C bonding orbitals, while LUMOs are more S-C antibonding orbitals in the AdoMet substrate. The largest orbital mixing occurs between the HOMO and LUMO orbitals, with the largest population change in their electronic density (see Figure 7 and Figure S2). At long distances, it equals 0.12 e−, reaching the value of 0.30 e− in the intermediate compound. This shows these two orbitals’ influence on the methyl radical’s transfer process. It should be mentioned that during geometry optimizations, the charge of the transferred methyl radical remained equal to zero, proving that the cobalt ion in the methylcobalamin product remains in the +2 oxidation state after transferring the methyl radical. Mixing several intermolecular orbitals is nothing but the pseudo-Jahn–Teller effect, which influences the length of the S-C bond and the length of the Co-C bond distance. The two systems are moving toward each other until the Co-C distance reaches a value of approximately 2.50 Å. Then, the sharp increase in the length of the S-C bond occurs, seconded by a simultaneous decrease in the Co-C bond length, reaching about 2.00 Å of the Co-C bond distance in the newly formed methylcob(II)alamin compound. Simultaneously, the S-C bond distance increases up to more than 4.00 Å, where the S-C bond is fully cleaved. The rupture of the S-C bond and the formation of the Co-C bond occur under the influence of the pseudo-Jahn–Teller effect mentioned above. Considering that during the entire geometry optimization procedure, the amount of electronic density on the HOMO decreases by the same value as the increase in the electronic density on the LUMO and that the difference in their electron population differs the most from the 2 e− and 0 e−, it is obvious that during orbital mixing, the greater mixing takes place between these two molecular orbitals, and in chemical terms, we can assume that during their mixing, their electronic density population changes as a result of HOMO and LUMO orbital mixing. Both HOMO and LUMO, from the beginning, resemble the σ* and σ orbitals of the isolated methylcob(II)alamin cofactor. This shows additionally that the S-C bond is cleaved homolytically. This explains why the AdoMet substrate does not transfer the methyl radical to the cob(I)alamin cofactor particle, which is present at this stage of the Methionine Synthase process turnover. Such an eventual homolytic methyl transfer from AdoMet to cob(I)alamin particles is impossible since cob(I)alamin particles do not bond methyl radicals without the oxidation of the central Co atom. In conclusion, the pseudo-Jahn–Teller effect, e.g., the mixing of HOMO bonding and LUMO antibonding intermolecular orbitals during the interaction of the AdoMet ion with the inactive particle of the cob(II)alamin cofactor, leads to the S-C bond cleavage in the AdoMet ion and to creating the Co-C bond in the methylcob(II)alamin cofactor. This is transforming the inactive particles of the cob(II)alamin cofactor into active particles of the methylcob(II)alamin cofactor (Figure 8), able to participate in the reaction with homocysteine, revitalizing the Methionine Synthase process.
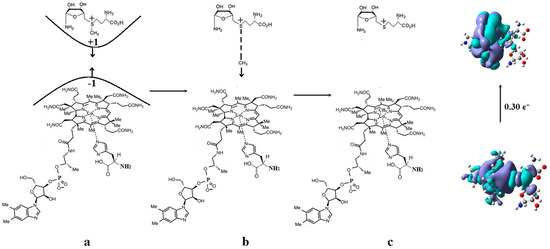
Figure 8.
The mechanism of the –CH3 group transfer from the AdoMet substrate to the inactive cob(II)alamin cofactor particle: (a) substrate–cob(II)alamin starting model; (b) intermediate structures; (c) the products.
The complete mechanism of the Methionine Synthase process. As we have shown before [22], the active particle in the process is the base-on species of the methylcob(II)alamin cofactor with a histidine ligand bonded to a cobalt atom [22]. Above, we have presented the mechanism of the Methionine Synthase process’s first step. On the other hand, the DFT method with the BP86 functional and 6-311** basis set and the CASSCF method geometry optimizations with the above-presented parameters show that the Co-N bond reaction formation with histidine in the methylcobalamin cofactor occurs in the absence of the energy barrier only in the case of the +3 oxidation state of the central cobalt atom. Besides that, the Methionine Synthase process needs the AdoMet methylating substrate to have an unbreakable evolution, as shown above. So, the mechanism of the Methionine Synthase process presented before [22] suffered from certain assumptions in the absence of the here-presented data [22]. In particular, the AdoMet substrate was absent from the full mechanism of the Methionine Synthase process [22]. Here, we present, in the absence of total energy barriers, a fully updated mechanism of the Methionine Synthase process determined by the MCSCF method (Figure 9).
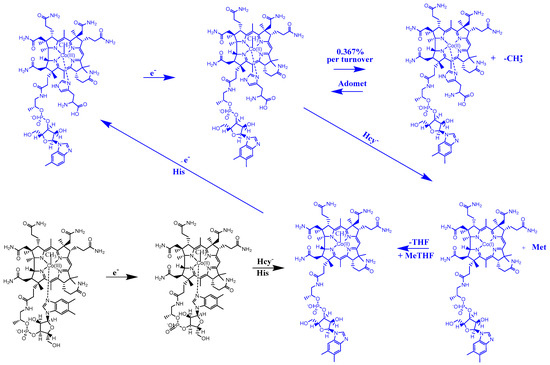
Figure 9.
The proposed mechanism for the in vivo Methionine Synthase process. Hcy—homocysteine; His—histidine; MeTHF—5-methyltetrahydrofolate; THF—tetrahydrofolate; Met—methionine; Adomet-S—adenosylmethionine. The preliminary step is black, while the turnover process is blue.
4. Conclusions
Two main conclusions can be drawn from the above-presented calculations: 1. The AdoMet substrate cation forms several common HOMOs and LUMOs intermolecular orbitals with the inactive cob(II)alamine anion particles cofactor starting with distances slightly greater than 4.00 Å. Although, the interaction and proximity of the two oppositely charged systems is caused by two factors, the coulombic interaction between them and the mixing of their HOMO-LUMO orbitals. The structure of the HOMO and LUMO is such that the process of the CASSCF geometry optimization of the inactive particle cob(II)alamin–AdoMet substrate anion common model leads to the breaking of the S-C bond in the AdoMet anion substrate and to the formation of the Co-C chemical bond in the newly formed methylcob(II)alamin cofactor particle. During the mixing of the HOMO–LUMO orbitals, their overlapping led to an impressive electron density difference from 2 and 0 population of up to 0.30 e− at average distances in the transition model. At the same time as the -CH3 radical approaches the cobalt atom, the remaining part of the AdoMet anion moves away from the same -CH3 radical. When the Co-C bond is formed, the remaining part of the AdoMet ion is at a greater distance than 4.00 Å from the -CH3 radical. Thus, the formation of the Co-C bond in the methylcob(II)alamin cofactor particle and the breaking of the S-C bond in the AdoMet substrate anion are concerted reactions. 2. The methylcobalamin cofactor loses its dimethylbenzimidazole axial ligand in the preliminary phase of the enzymatically active act. The rupture of the Co-N axial bond and the removal of the axial ligand dimethylbenzimidazole takes place under the joint influence of the histidine molecule, on the one hand, and of the negative ion of the homocysteine, on the other hand. The process of the Co-N axial bond breaking occurs under the influence of HOMO2–LUMO mixing, leading to the electronic density transfer from the substrate negative ion atoms to the methylcobalamin cofactor in chemical interpretation. A major role is playing the fact that LUMO represents antibonding molecular π-orbitals, composed of the atomic orbitals of the corrin ring and cobalt atom. The rupture of the Co-N axial bond in the methylcobalamin occurs in the absence of an energy barrier. The DFT and CASSCF geometry optimization show that the Co-N bond formation in the base-on methylcobalamin cofactor with histidine in the absence of the energy barrier occurs only in the species with a +3 oxidation state of the central cobalt atom. As a result of the above-mentioned reactions, the fully updated final mechanism of the Methionine Synthase process was drawn. The whole mechanism is based on theoretical data, being in full agreement with the experimental ones.
Supplementary Materials
The following supporting information can be downloaded at: https://www.mdpi.com/article/10.3390/reactions4020016/s1, Figure S1: The active space orbitals surfaces, energy, and their electronic density population at the beginning of the CASSCF geometry optimization of the homocysteine negative ion methylcob(II)alamin cofactor and histidine common model: (a) The Co-C bond distance is equal to 2.08 Å; (b) The Co-N bond distance is equal to 2.35 Å; Figure S2: The active space orbitals surfaces, energy, and their electronic density population at the beginning of the CASSCF geometry optimization of the inactive particle of the cob(II)alamin cofactor and Adomet substrate common model. The Co-N bond distance is equal to 4.00 Å; Table S1: The methylcob(II)alamin, homocysteine ion, and histidine substrates common model CASSCF geometry optimization starting coordinates; Table S2: The cob(II)alamin bio inactive particle plus AdoMet substrate common model CASSCF geometry optimization starting coordinates.
Funding
This research was supported, in part, under the National Science Foundation Grants CNS-0958379 and CNS0855217 and the City University of New York High-Performance Computing Center at the College of Staten Island and by the National Science Foundation through TeraGrid resources provided by the TeraGrid Science Gateways program under grants CHE090082 and CHE140071.
Data Availability Statement
Additional data is stored in the form of initial files and files resulting from calculations in the personal directories of the Texas Advanced Computing Center.
Conflicts of Interest
The authors declare no conflict of interest.
References
- Pratt, J.M. The roles of Co, corrin, and protein. I. Co-ligand bonding and the trans effect. In Chemistry and Biochemistry of B12; Banerjee, R., Ed.; John Wiley & Sons: New York, NY, USA, 1999; pp. 73–112. [Google Scholar]
- Matthews, R.G. Cobalamin-dependent methionine synthase. In Chemistry and Biochemistry of B12; Banerjee, R., Ed.; John Wiley & Sons: New York, NY, USA, 1999; pp. 681–706. [Google Scholar]
- Banerjee, R.V.; Matthews, R.G. Cobalamin-dependent methionine synthase. FASEB J. 1990, 4, 1450–1459. [Google Scholar] [CrossRef]
- Matthews, R.G. Cobalamin-Dependent Methyltransferases. Acc. Chem. Res. 2001, 34, 681–689. [Google Scholar] [CrossRef]
- Matthews, R.G.; Koutmos, M.; Datta, S. Cobalamin-dependent and cobamidedependent methyltransferases. Curr. Opin. Struct. Biol. 2008, 18, 658–666. [Google Scholar] [CrossRef]
- Drennan, C.L.; Huang, S.; Drummond, J.T.; Matthews, R.G.; Ludwig, M.L. How a protein binds B12: A 3.0 Å x-ray structure of B12-binding domains of methionine synthase. Science 1994, 266, 1669–1674. [Google Scholar] [CrossRef]
- Mancia, F.; Keep, N.M.; Nakagawa, A.; Leadlay, P.F.; McSweeney, S.; Rasmussen, B.; Bosecke, P.; Diat, O.; Evans, P.F. How coenzyme B12 radicals are generated: The crystal structure of methylmalonyl-coenzyme A mutase at 2 Å resolution. Structure 1996, 4, 339–350. [Google Scholar] [CrossRef]
- Koutmos, M.; Datta, S.; Pattridge, K.A.; Smith, J.L.; Matthews, R.G. Insights into the reactivation of cobalamin-dependent methionine synthase. Proc. Natl. Acad. Sci. USA 2009, 106, 18527–18532. [Google Scholar] [CrossRef]
- Hagemeier, C.H.; Kruer, M.; Rudolf, K.; Thauer, R.K.; Eberhard, W.; Ermler, U. Insight into the mechanism of biological methanol activation based on the crystal structure of the methanol-cobalamin methyltransferase complex. Proc. Natl. Acad. Sci. USA 2006, 103, 18917–18922. [Google Scholar] [CrossRef]
- Reitzer, R.; Gruber, K.; Jogl, G.; Wagner, U.G.; Bothe, H.; Buckel, W.; Kratky, C. Glutamate mutase from Clostridium cochlearium: The structure of a coenzyme B12-dependent enzyme provides new mechanistic insights. Structure 1999, 7, 891–902. [Google Scholar] [CrossRef]
- Jensen, K.P.; Ryde, U. Conversion of Homocysteine to Methionine by Methionine Synthase: A Density Functional Study. J. Am. Chem. Soc. 2003, 125, 13970–13971. [Google Scholar] [CrossRef]
- Kozlowski, P.M.; Kuta, J.; Galezowski, W. Reductive Cleavage Mechanism of Methylcobalamin: Elementary Steps of Co-C Bond Breaking. J. Phys. Chem. B 2007, 111, 7638–7645. [Google Scholar] [CrossRef]
- Kozlowski, P.M.; Kamachi, T. Reductive elimination pathway for homocysteine to methionine conversion in cobalamin-dependent methionine synthase. J. Biol. Inorg. Chem. 2012, 17, 611–619. [Google Scholar] [CrossRef]
- Alfonso-Prieto, M.; Biarnes, X.; Kumar, M.; Rovira, C.; Kozlowski, P.M. Reductive Cleavage Mechanism of Co-C Bond in Cobalamin-Dependent Methionine Synthase. J. Phys. Chem. B 2010, 114, 12965–12971. [Google Scholar] [CrossRef]
- Spataru, T.; Birke, R.L. Carbon-Cobalt Bond Distance and Bond Cleavage in OneElectron Reduced Methylcobalamin: A Failure of the Conventional DFT Method. J. Phys. Chem. A 2006, 110, 8599–8604. [Google Scholar] [CrossRef]
- Spataru, T.; Fernandez, F. The nature of the Co-C bond cleavage processes in the methylcob(II)alamin and adenosylcob(III)alamin. Chem. J. Mold. 2016, 11, 10–20. [Google Scholar] [CrossRef]
- Birke, R.L.; Huang, Q.; Spataru, T.; Gosser, D.K., Jr. Electroreduction of an of Alkylcobalamins: Mechanism of Stepwise Reductive Cleavage of the Co-C Bond. J. Am. Chem. Soc. 2006, 128, 1922–1936. [Google Scholar] [CrossRef]
- Spataru, T.; Birke, R.L. The effect of solvent on the electrode process of methylcobalamin as studied by cyclic voltammetry. J. Electroanal. Chem. 2006, 593, 74–86. [Google Scholar] [CrossRef]
- Lexa, D.; Savéant, J.-M. Electrochemistry of vitamin B12. 3. One-electron intermediates in the reduction of methylcobalamin and methylcobinamide. J. Am. Chem. Soc. 1978, 100, 3220–3222. [Google Scholar] [CrossRef]
- Bersuker, I.B. Limitations of Density Functional Theory in Application to the Degenerate States. J. Comp. Chem. 1997, 2, 260–267. [Google Scholar] [CrossRef]
- Chen, S.-L.; Blomberg, M.R.A.; Siegbahn, P.E.M. How Is a Co-Methyl Intermediate Formed in the Reaction of Cobalamin-Dependent Methionine Synthase? Theoretical Evidence for a Two-Step Methyl Cation Transfer Mechanism. J. Phys. Chem. B 2011, 115, 4066–4077. [Google Scholar] [CrossRef]
- Spataru, T. The complete electronic structure and mechanism of the methionine synthase process as determined by the MCSCF method. J. Organomet. Chem. 2021, 942, 121811. [Google Scholar] [CrossRef]
- James, T.; Drummond, J.T.; Sha, H.; Blumenthal, R.M.; Matthews, R.G. Assignment of Enzymatic Function to Specific Protein Regions of Cobalamin-Dependent Methionine Synthase from Escherichia coli. Biochemistry 1993, 32, 9290–9295. [Google Scholar]
- Spataru, T. The Electronic Structure and Mechanism of the AdenosylcobalaminDependent Bio-processes as Determined by the MCSCF Method. J. Med. Chem. 2021, 11, 595. [Google Scholar]
- Valiev, M.; Bylaska, E.J.; Govind, N.; Kowalski, K.; Straatsma, T.P.; van Dam, H.J.J.; Wang, D.; Nieplocha, D.; Apra, E.; Windus, T.L.; et al. “NwChem”: A comprehensive and scalable open-source solution for large scale molecular simulations. Comput. Phys. Commun. 2010, 181, 1477. [Google Scholar] [CrossRef]
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the author. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).