Improving YOLO-Based Plant Disease Detection Using αSILU: A Novel Activation Function for Smart Agriculture
Abstract
1. Introduction
- We introduce αSiLU, a novel parameterized and adaptive activation function tailored to enhance gradient flow and convergence stability in YOLO-based object detectors. Beyond proposing the function, we conduct a systematic evaluation of αSiLU against standard SiLU and other prevalent activation functions across multiple YOLO architectures. Our results reveal consistent improvements in detection accuracy—quantified by precision, recall, F1-score, mAP@0.5, and mAP@0.5:0.95—while preserving computational efficiency, thereby demonstrating both algorithmic and practical advantages.
- To validate its applicability in real-world agricultural settings, we benchmark αSiLU on two plant disease datasets—tomato and cucumber leaves—demonstrating its robustness and effectiveness under domain-specific conditions.
2. Materials and Methods
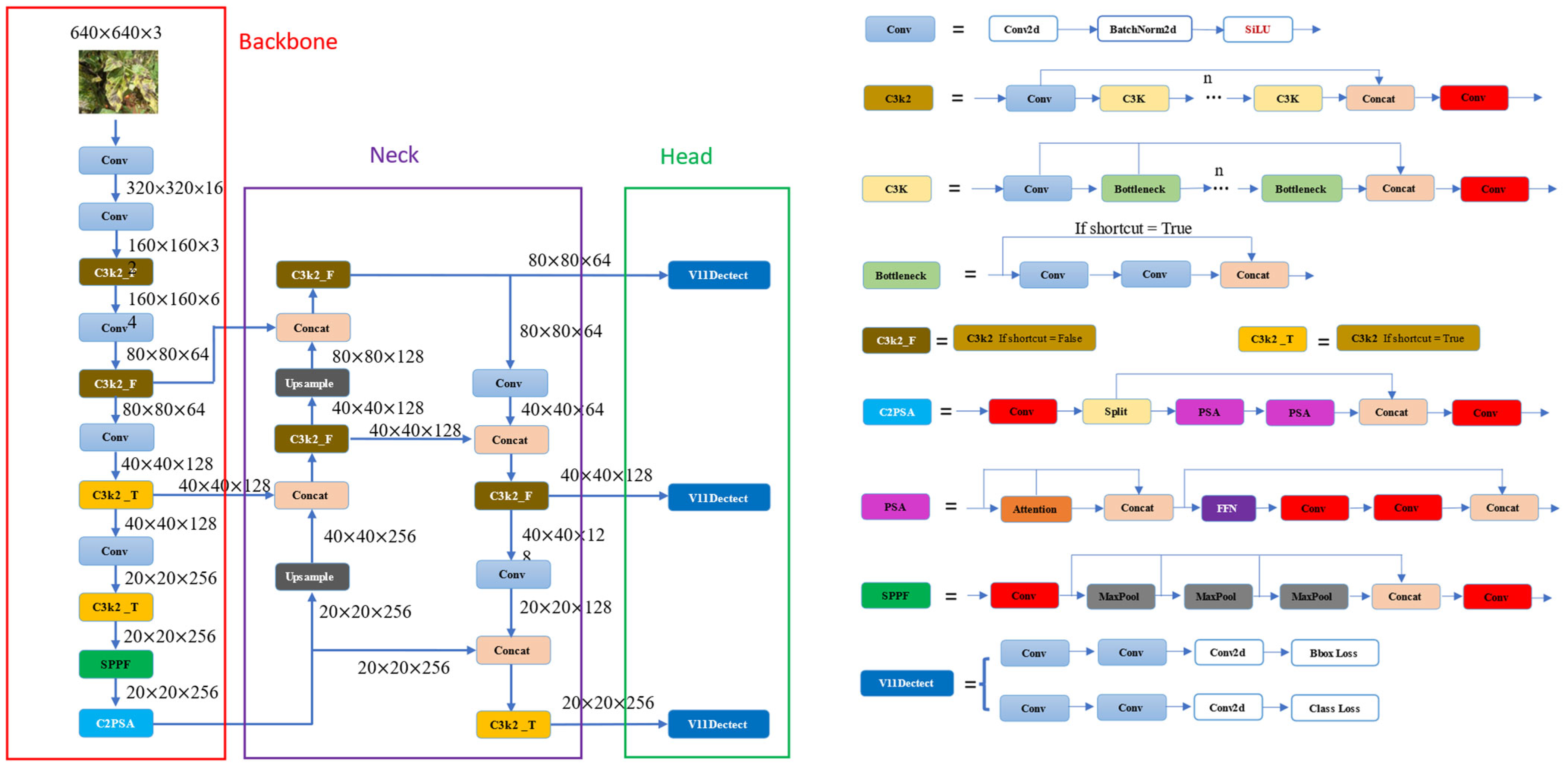
2.1. YOLOv11 Models
- Backbone: The backbone of YOLOv11 is meticulously designed to efficiently extract feature representations at multiple scales. A key advancement is the introduction of the C3k2 block (Cross Stage Partial block with a kernel size of 2), which replaces the previous C2f block. This block employs two smaller convolutions rather than a single large convolution, facilitating faster processing while maintaining the model’s representational capacity. Additionally, YOLOv11 continues to utilize the Spatial Pyramid Pooling—Fast (SPPF) module for multi-scale feature aggregation, enhanced by the integration of the novel C2PSA (Cross Stage Partial with Parallel Spatial Attention) block. The C2PSA block leverages spatial attention mechanisms to direct the model’s focus toward salient regions of an image, thereby improving detection accuracy, particularly for objects with varying sizes or those that are partially occluded.
- Neck: In YOLOv11, the neck serves as the stage where features from different scales are fused and refined before being passed to the head. Similar to the backbone, YOLOv11 substitutes the C2f block with the more computationally efficient C3k2 block at this stage. Moreover, the incorporation of spatial attention mechanisms through the C2PSA module enables the model to prioritize and emphasize critical spatial information, thereby enhancing the robustness and precision of the detection process.
- Head: YOLOv11’s head consists of multiple C3k2 blocks that process the refined feature representations at various depths, effectively balancing parameter efficiency and model expressiveness. It also incorporates CBS blocks (Convolution-BatchNorm-SiLU), which stabilize the training process through normalization and apply the SiLU activation function to enhance non-linearity and improve feature-extraction quality. The final prediction is generated through convolutional layers coupled with a Detect layer, which produces bounding box coordinates, objectness confidence scores, and class probabilities.
2.2. αSiLU Activation Function—Proposed Improvement
2.2.1. Introduction to Activation Functions
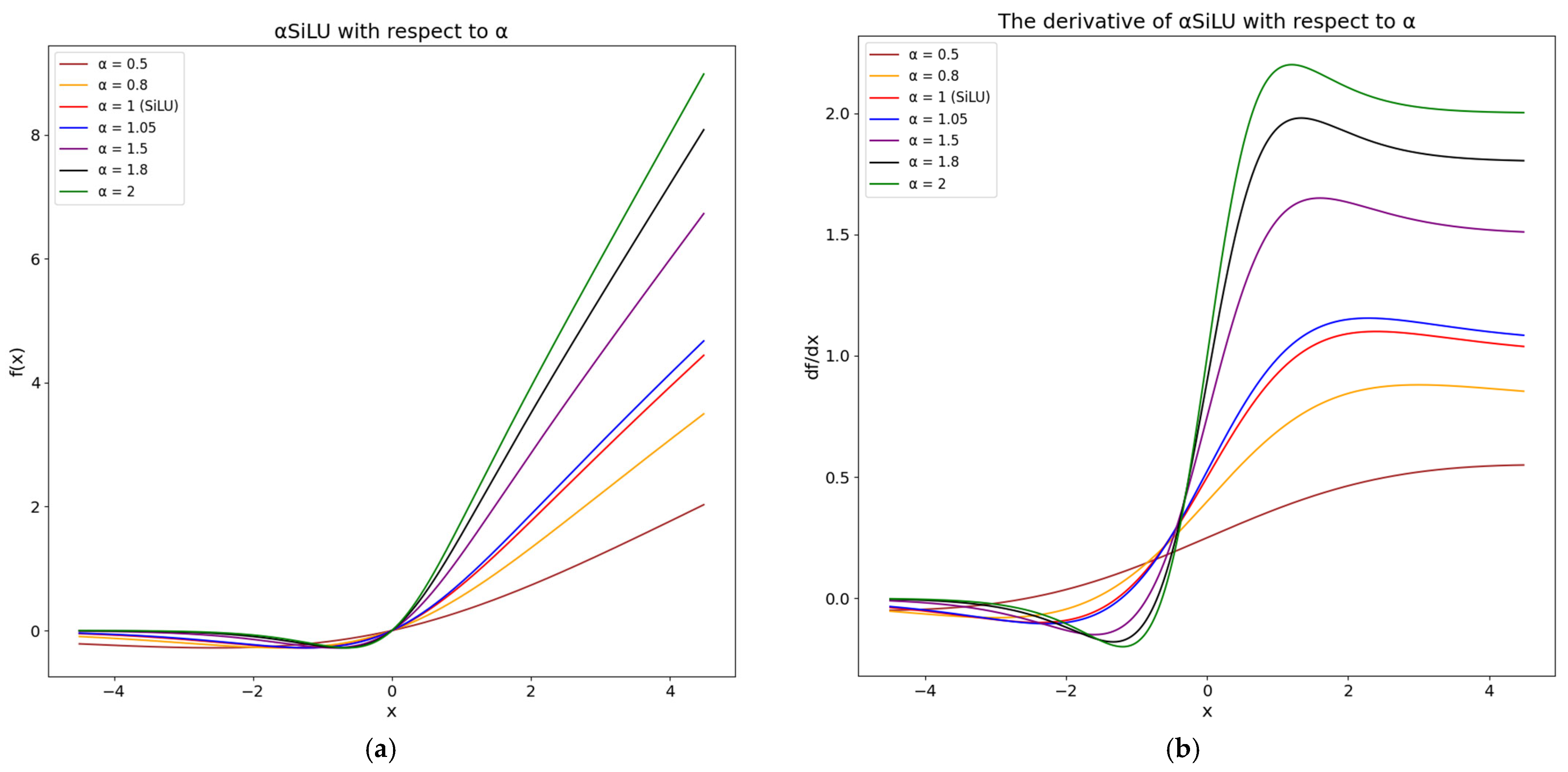
2.2.2. Proposed αSiLU Activation Function
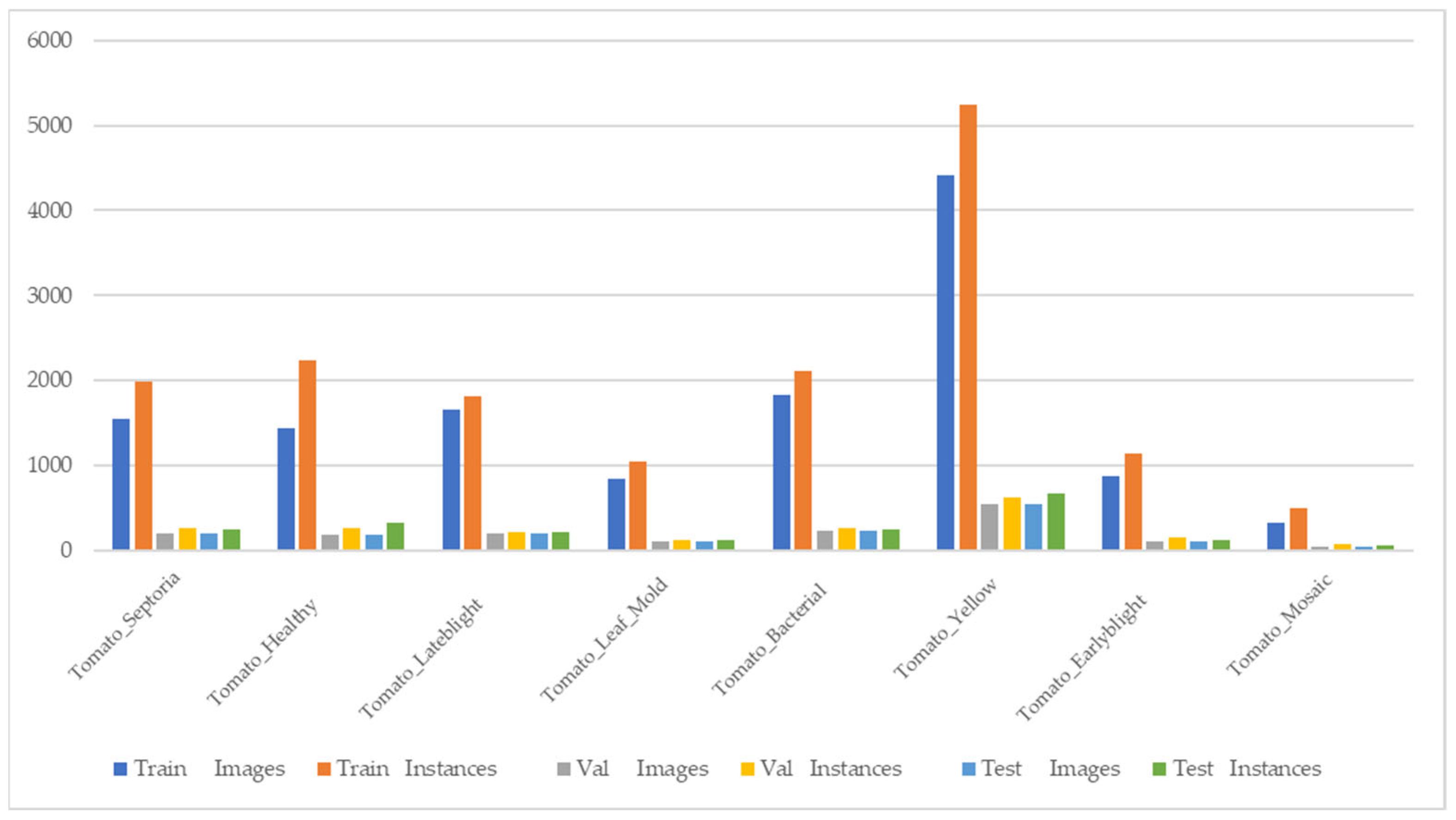
2.3. Dataset
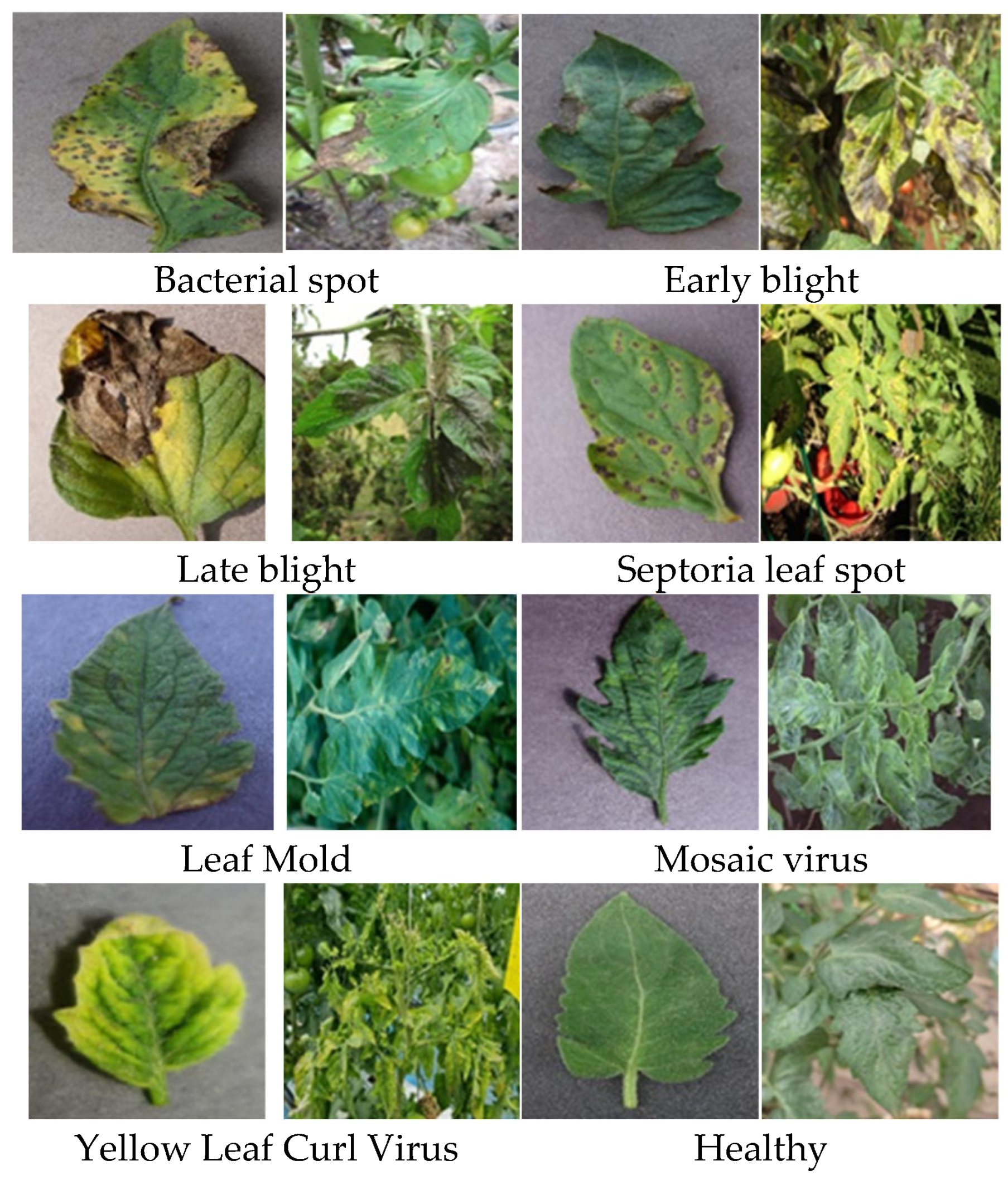
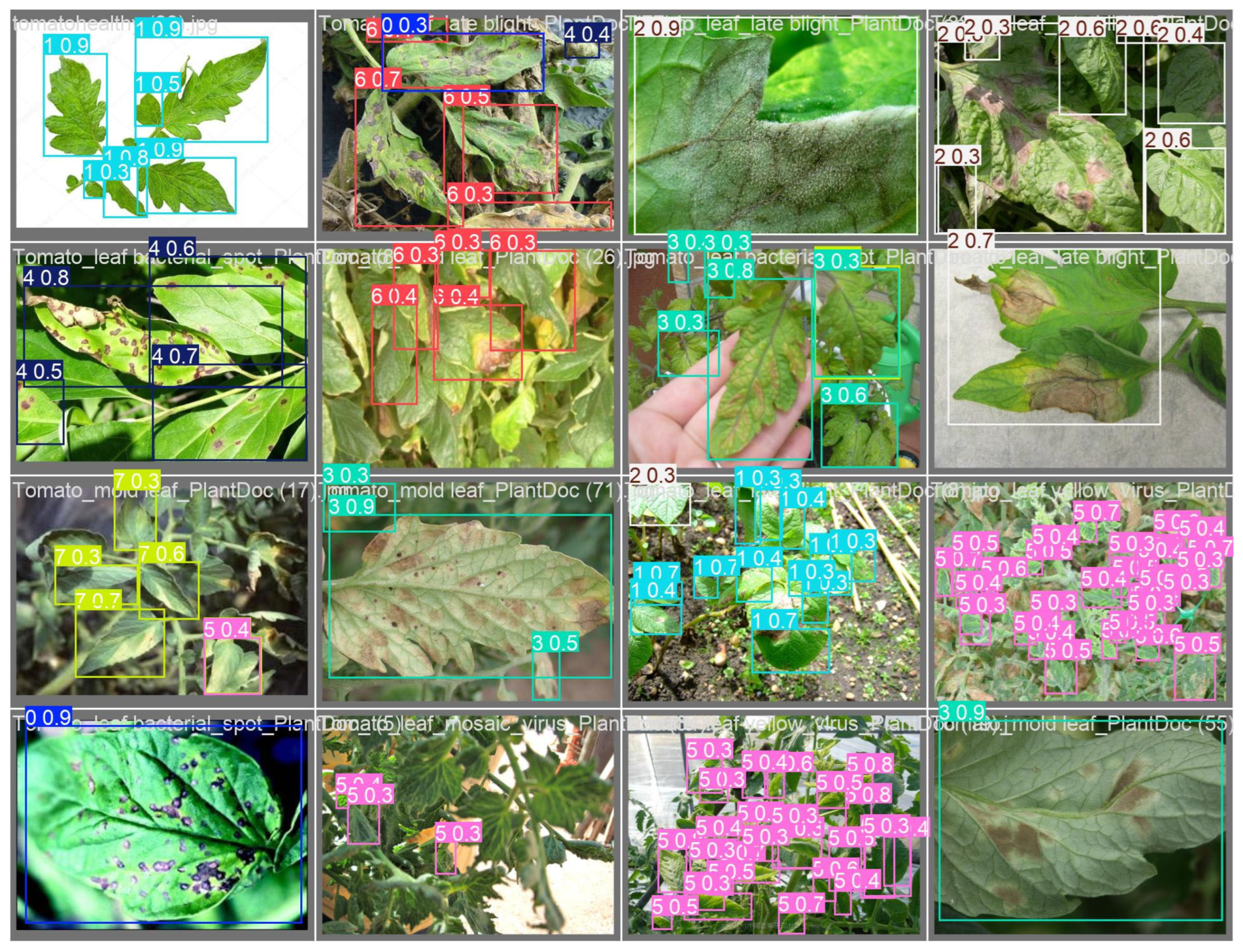
2.3.1. Tomato Disease Dataset
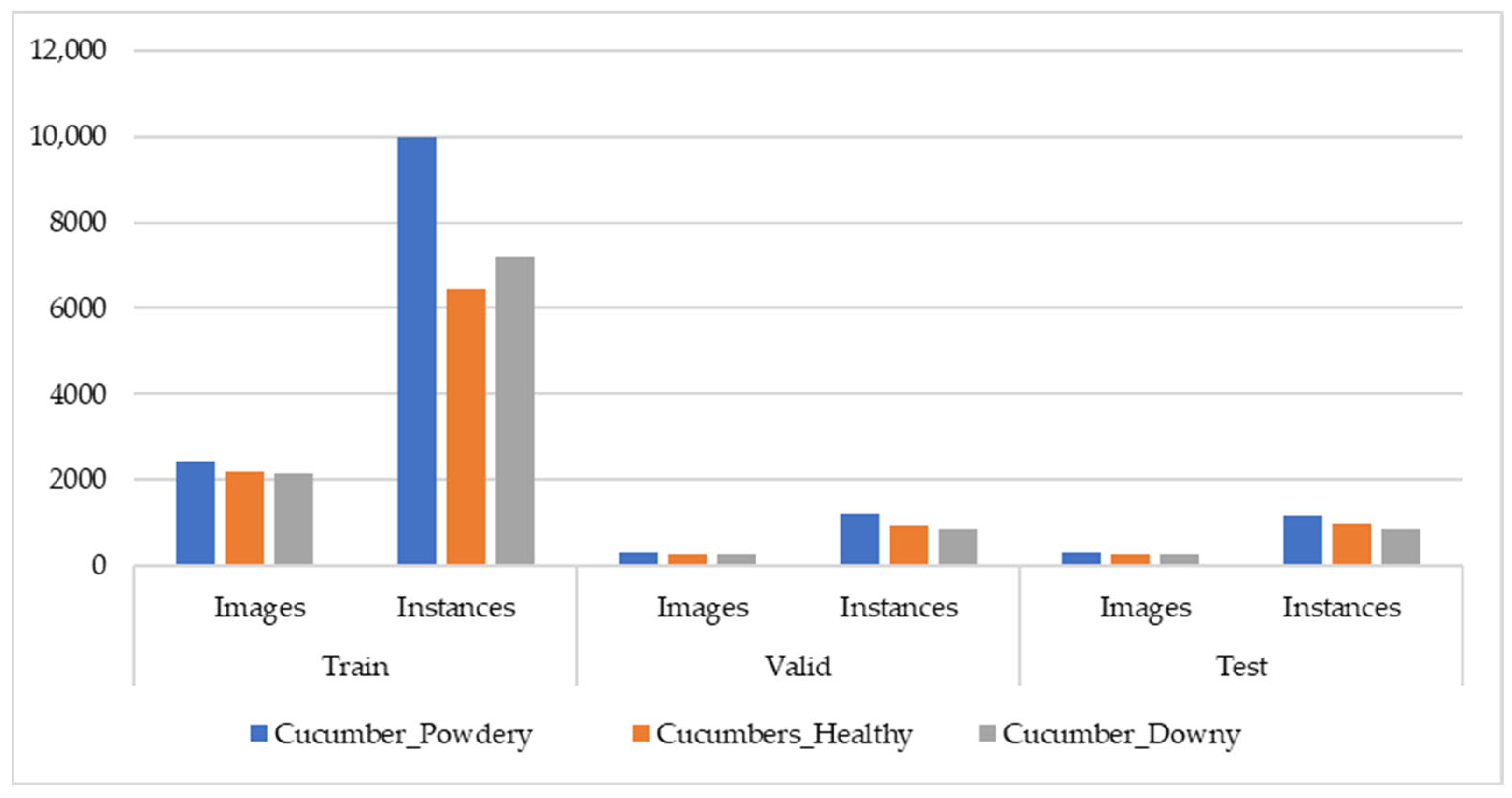
2.3.2. Cucumber Disease Dataset
2.4. Evaluation Method
2.4.1. Performance Metrics
- True Positives (TP): Correctly detected and classified objects that actually exist in the image.
- False Positives (FP): Predictions that either do not match any real object or are assigned an incorrect class.
- False Negatives (FN): Ground-truth objects that the model fails to detect, either due to omission or low IoU overlap.
- Precision (P) measures the proportion of predicted bounding boxes that are correct, indicating how well the model avoids false detections.
- Recall (R) quantifies the model’s ability to retrieve all relevant instances, reflecting its sensitivity to missed objects.
- F1-score is the harmonic mean of precision and recall, offering a balanced performance indicator, particularly in scenarios with class imbalance.
- Mean Average Precision (mAP) summarizes overall detection accuracy across all object categories by averaging the Average Precision (AP) scores:
- +
- mAP@50: Computed at a fixed IoU threshold of 0.50.
- +
- mAP@50:95: Calculated by averaging AP over IoU thresholds ranging from 0.50 to 0.95 in 0.05 increments, following the COCO evaluation protocol.
2.4.2. Experimental Setup
3. Results and Discussion
3.1. Experimental Results
3.1.1. YOLOv11-αSiLU Performance
- Tomato Dataset:
- B.
- Cucumber Dataset:
- C.
- Discussion:
3.1.2. Comparison with Other YOLO Versions
3.2. Comparative Analysis of Activation Functions
4. Discussion and Future Work
- Dataset Diversity, Annotation Granularity, and Multi-Disease Detection
- Computational Constraints, Activation Expressiveness, and Model Scalability
- Statistical Robustness and Experimental Reproducibility
- Practical Deployment, System Cost, and Edge-AI Profiling
- Multimodal Sensing and Hardware-Level Enhancement
5. Conclusions
Author Contributions
Funding
Data Availability Statement
Conflicts of Interest
References
- Oerke, E.C. Crop losses to pests. J. Agric. Sci. 2006, 144, 31–43. [Google Scholar] [CrossRef]
- Savary, S.; Ficke, A.; Aubertot, J.N.; Hollier, C. Crop losses due to diseases and their implications for global food production losses and food security. Food Secur. 2012, 4, 519–537. [Google Scholar] [CrossRef]
- Bock, C.H.; Chiang, K.S.; Del Ponte, E.M. Plant disease severity estimated visually: A century of research, best practices, and opportunities for improving methods and practices to maximize accuracy. Trop. Plant Pathol. 2022, 47, 25–42. [Google Scholar] [CrossRef]
- Savary, S.; Teng, P.S.; Willocquet, L.; Nutter, F.W., Jr. Quantification and modeling of crop losses: A review of purposes. Annu. Rev. Phytopathol. 2006, 44, 89–112. [Google Scholar] [CrossRef]
- Bouhouch, Y.; Esmaeel, Q.; Richet, N.; Barka, E.A.; Backes, A.; Steffenel, L.A.; Hafidi, M.; Jacquard, C.; Sanchez, L. Deep Learning-Based Barley Disease Quantification for Sustainable Crop Production. Phytopathology® 2024, 114, 2045–2054. [Google Scholar] [CrossRef]
- Fang, Y.; Ramasamy, R.P. Current and prospective methods for plant disease detection. Biosensors 2015, 5, 537–561. [Google Scholar] [CrossRef]
- Ngongoma, M.S.; Kabeya, M.; Moloi, K. A review of plant disease detection systems for farming applications. Appl. Sci. 2023, 13, 5982. [Google Scholar] [CrossRef]
- Singla, A.; Nehra, A.; Joshi, K.; Kumar, A.; Tuteja, N.; Varshney, R.K.; Gill, S.S.; Gill, R. Exploration of machine learning approaches for automated crop disease detection. Curr. Plant Biol. 2024, 40, 100382. [Google Scholar] [CrossRef]
- Jafar, A.; Bibi, N.; Naqvi, R.A.; Sadeghi-Niaraki, A.; Jeong, D. Revolutionizing agriculture with artificial intelligence: Plant disease detection methods, applications, and their limitations. Front. Plant Sci. 2024, 15, 1356260. [Google Scholar] [CrossRef]
- El Sakka, M.; Ivanovici, M.; Chaari, L.; Mothe, J. A Review of CNN Applications in Smart Agriculture Using Multimodal Data. Sensors 2025, 25, 472. [Google Scholar] [CrossRef] [PubMed]
- Mao, M.; Hong, M. YOLO Object Detection for Real-Time Fabric Defect Inspection in the Textile Industry: A Review of YOLOv1 to YOLOv11. Sensors 2025, 25, 2270. [Google Scholar] [CrossRef]
- Lee, Y.S.; Patil, M.P.; Kim, J.G.; Seo, Y.B.; Ahn, D.H.; Kim, G.D. Hyperparameter Optimization for Tomato Leaf Disease Recognition Based on YOLOv11m. Plants 2025, 14, 653. [Google Scholar] [CrossRef]
- Sangaiah, A.K.; Yu, F.N.; Lin, Y.B.; Shen, W.C.; Sharma, A. UAV T-YOLO-rice: An enhanced tiny YOLO networks for rice leaves diseases detection in paddy agronomy. IEEE Trans. Netw. Sci. Eng. 2024, 11, 5201–5216. [Google Scholar] [CrossRef]
- Kumar, D.; Malhotra, A. Fast and Precise: YOLO-based Wheat Spot Blotch Recognition. In Proceedings of the 2024 5th IEEE Global Conference for Advancement in Technology (GCAT), Bangalore, India, 4–6 October 2024; pp. 1–5. [Google Scholar]
- Xie, J.; Xie, X.; Xie, W.; Xie, Q. An Improved YOLOv8-Based Method for Detecting Pests and Diseases on Cucumber Leaves in Natural Backgrounds. Sensors 2025, 25, 1551. [Google Scholar] [CrossRef]
- Mamun, S.B.; Payel, I.J.; Ahad, M.T.; Atkins, A.S.; Song, B.; Li, Y. Grape Guard: A YOLO-based mobile application for detecting grape leaf diseases. J. Electron. Sci. Technol. 2025, 23, 100300. [Google Scholar] [CrossRef]
- Singh, Y.; Shukla, S.; Mohan, N.; Parameswaran, S.E.; Trivedi, G. Real-time plant disease detection: A comparative study. In Proceedings of the International Conference on Agriculture-Centric Computation, Chandigarh, India, 11–13 May 2023; Springer Nature: Cham, Switzerland, 2023; pp. 210–224. [Google Scholar]
- Lou, Y.; Hu, Z.; Li, M.; Li, H.; Yang, X.; Liu, X.; Liu, F. Real-time detection of cucumber leaf diseases based on convolution neural network. In Proceedings of the 2021 IEEE 5th Information Technology, Networking, Electronic and Automation Control Conference (ITNEC), Xi’an, China, 15–17 October 2021; Volume 5, pp. 1040–1046. [Google Scholar]
- Wang, Y.; Zhang, P.; Tian, S. Tomato leaf disease detection based on attention mechanism and multi-scale feature fusion. Front. Plant Sci. 2024, 15, 1382802. [Google Scholar] [CrossRef]
- Ding, J.; Jeon, W.; Rhee, S. DM-YOLOv8: Cucumber disease and insect detection using detailed multi-intensity features. In Proceedings of the 2024 International Conference on Artificial Intelligence in Information and Communication (ICAIIC), Osaka, Japan, 19–22 February 2024; IEEE: New York, NY, USA, 2024; pp. 199–204. [Google Scholar]
- Abulizi, A.; Ye, J.; Abudukelimu, H.; Guo, W. DM-YOLO: Improved YOLOv9 model for tomato leaf disease detection. Front. Plant Sci. 2025, 15, 1473928. [Google Scholar] [CrossRef] [PubMed]
- He, Z.; Tong, M. LT-YOLO: A Lightweight Network for Detecting Tomato Leaf Diseases. Comput. Mater. Contin. 2025, 82, 3. [Google Scholar] [CrossRef]
- Rajamohanan, R.; Latha, B.C. An optimized YOLO v5 model for tomato leaf disease classification with field dataset. Eng. Technol. Appl. Sci. Res. 2023, 13, 12033–12038. [Google Scholar] [CrossRef]
- Liu, Z.; Guo, X.; Zhao, T.; Liang, S. YOLO-BSMamba: A YOLOv8s-Based Model for Tomato Leaf Disease Detection in Complex Backgrounds. Agronomy 2025, 15, 870. [Google Scholar] [CrossRef]
- Tian, Y.; Ye, Q.; Doermann, D. Yolov12: Attention-centric real-time object detectors. arXiv 2025, arXiv:2502.12524. [Google Scholar]
- Li, Y.; Yan, H.; Li, D.; Wang, H. Robust Miner Detection in Challenging Underground Environments: An Improved YOLOv11 Approach. Appl. Sci. 2024, 14, 11700. [Google Scholar] [CrossRef]
- Cheng, S.; Han, Y.; Wang, Z.; Liu, S.; Yang, B.; Li, J. An Underwater Object Recognition System Based on Improved YOLOv11. Electronics 2025, 14, 201. [Google Scholar] [CrossRef]
- Ye, T.; Huang, S.; Qin, W.; Tu, H.; Zhang, P.; Wang, Y.; Gao, C.; Gong, Y. YOLO-FIX: Improved YOLOv11 with Attention and Multi-Scale Feature Fusion for Detecting Glue Line Defects on Mobile Phone Frames. Electronics 2025, 14, 927. [Google Scholar] [CrossRef]
- Gao, Y.; Xin, Y.; Yang, H.; Wang, Y. A Lightweight Anti-Unmanned Aerial Vehicle Detection Method Based on Improved YOLOv11. Drones 2024, 9, 11. [Google Scholar] [CrossRef]
- Li, Y.; Guo, Z.; Sun, Y.; Chen, X.; Cao, Y. Weed Detection Algorithms in Rice Fields Based on Improved YOLOv10n. Agriculture 2024, 14, 2066. [Google Scholar] [CrossRef]
- Wang, D.; Tan, J.; Wang, H.; Kong, L.; Zhang, C.; Pan, D.; Li, T.; Liu, J. SDS-YOLO: An improved vibratory position detection algorithm based on YOLOv11. Measurement 2025, 244, 116518. [Google Scholar] [CrossRef]
- Liao, Y.; Li, L.; Xiao, H.; Xu, F.; Shan, B.; Yin, H. YOLO-MECD: Citrus Detection Algorithm Based on YOLOv11. Agronomy 2025, 15, 687. [Google Scholar] [CrossRef]
- Shah, V.; Youngblood, N. Leveraging continuously differentiable activation functions for learning in quantized noisy environments. arXiv 2024, arXiv:2402.02593. [Google Scholar] [CrossRef]
- Kaseb, Z.; Xiang, Y.; Palensky, P.; Vergara, P.P. Adaptive Activation Functions for Deep Learning-based Power Flow Analysis. In Proceedings of the 2023 IEEE PES Innovative Smart Grid Technologies Europe (ISGT EUROPE), Grenoble, France, 23–26 October 2023; pp. 1–5. [Google Scholar]
- Rajanand, A.; Singh, P. ErfReLU: Adaptive activation function for deep neural network. Pattern Anal. Appl. 2024, 27, 68. [Google Scholar] [CrossRef]
- Chen, J.; Pan, Z. Competition-based Adaptive ReLU for Deep Neural Networks. arXiv 2024, arXiv:2407.19441. [Google Scholar]
- Lee, M. Gelu activation function in deep learning: A comprehensive mathematical analysis and performance. arXiv 2023, arXiv:2305.12073. [Google Scholar] [CrossRef]
- Chen, Z.; Yang, J.; Li, F.; Feng, Z.; Chen, L.; Jia, L.; Li, P. Foreign Object Detection Method for Railway Catenary Based on a Scarce Image Generation Model and Lightweight Perception Architecture. IEEE Trans. Circuits Syst. Video Technol. 2025. Available online: https://ieeexplore.ieee.org/document/10988810 (accessed on 12 August 2025). [CrossRef]
- Zhao, Z.; Qin, Y.; Qian, Y.; Wu, Y.; Qin, W.; Zhang, H.; Wu, X. Automatic potential safety hazard evaluation system for environment around high-speed railroad using hybrid U-shape learning architecture. IEEE Trans. Intell. Transp. Syst. 2024, 26, 1071–1087. [Google Scholar] [CrossRef]
- Khanam, R.; Hussain, M. Yolov11: An overview of the key architectural enhancements. arXiv 2024, arXiv:2410.17725. [Google Scholar] [CrossRef]
- He, K.; Zhang, X.; Ren, S.; Sun, J. Delving Deep into Rectifiers: Surpassing Human-Level Performance on ImageNet Classification. In Proceedings of the IEEE International Conference on Computer Vision (ICCV), Santiago, Chile, 7–13 December 2015; pp. 1026–1034. [Google Scholar]
- He, K.; Zhang, X.; Ren, S.; Sun, J. Deep Residual Learning for Image Recognition. In Proceedings of the IEEE Conference on Computer Vision and Pattern Recognition (CVPR), Las Vegas, NV, USA, 27–30 June 2016; pp. 770–778. [Google Scholar]
- Ramachandran, P.; Zoph, B.; Le, Q.V. Searching for Activation Functions. arXiv 2017, arXiv:1710.05941v2. [Google Scholar] [CrossRef]
- Misra, D. Mish: A self-regularized non-monotonic activation function. arXiv 2019, arXiv:1908.08681. [Google Scholar]
- Ramachandran, P.; Zoph, B.; Le, Q.V. Swish: A self-gated activation function. arXiv 2017, arXiv:1710.05941v1. [Google Scholar]
- Hussain, M. Yolov5, yolov8 and yolov10: The go-to detectors for real-time vision. arXiv 2024, arXiv:2407.02988. [Google Scholar]
- Elfwing, S.; Uchibe, E.; Doya, K. Sigmoid-weighted linear units for neural network function approximation in reinforcement learning. Neural Netw. 2018, 107, 3–11. [Google Scholar] [CrossRef]
- Mohanty, S.P.; Hughes, D.P.; Salathé, M. Using deep learning for image-based plant disease detection. Front. Plant Sci. 2016, 7, 1419. [Google Scholar] [CrossRef] [PubMed]
- Singh, D.; Jain, N.; Jain, P.; Kayal, P.; Kumawat, S.; Batra, N. PlantDoc: A dataset for visual plant disease detection. In Proceedings of the 7th ACM IKDD CoDS and 25th COMAD, Hyderabad, India, 5–7 January 2020; pp. 249–253. [Google Scholar]
- Uoc, N.Q.; Duong, N.T.; Son, L.A.; Thanh, B.D. A novel automatic detecting system for cucumber disease based on the convolution neural network algorithm. GMSARN Int. J. 2022, 16, 295–301. [Google Scholar]
- Silva, J.P.; Vieira, E.M.F.; Gwozdz, K.; Silva, N.E.; Kaim, A.; Istrate, M.C.; Ghica, C.; Correia, J.H.; Pereira, M.; Marques, L.; et al. High-performance and self-powered visible light photodetector using multiple coupled synergetic effects. Mater. Horiz. 2024, 11, 803–812. [Google Scholar] [CrossRef]
- Su, L. Room temperature growth of CsPbBr3 single crystal for asymmetric MSM structure photodetector. J. Mater. Sci. Technol. 2024, 187, 113–122. [Google Scholar] [CrossRef]



| Activation Funtion | Formula | Derivative | Non- Linearity | Gradient Stability |
|---|---|---|---|---|
| ReLU | Yes | |||
| LeakyReLU | Yes | Yes | ||
| Mish | Yes | Yes, smooth grandient flow | ||
| GELU | Yes | Yes, smooth and adaptive | ||
| ELU | Yes | Yes, avoids vanishing gradients | ||
| SiLU | Yes | Yes | ||
| CAReLU | ) | Yes | Yes (tanh ensures smooth transition and adaptive stability) | |
| αSiLU | Yes | Yes, adjustable by α |
| Model | Activation | α | P (%) | R (%) | F1-Score (%) | mAP@50 (%) | mAP@50–95 (%) |
|---|---|---|---|---|---|---|---|
| YOLOv11n | αSiLU | 0.5 | 95.70 | 83.40 | 89.13 | 91.80 | 82.00 |
| αSiLU | 0.7 | 95.20 | 83.80 | 89.14 | 91.80 | 81.70 | |
| αSiLU | 0.8 | 96.10 | 82.80 | 89.14 | 92.20 | 82.10 | |
| αSiLU | 0.9 | 94.80 | 84.10 | 89.13 | 92.00 | 82.20 | |
| αSiLU | 0.95 | 95.30 | 84.60 | 89.63 | 91.90 | 81.90 | |
| SiLU | 1 | 95.20 | 83.40 | 88.91 | 91.30 | 81.30 | |
| αSiLU | 1.025 | 96.70 | 83.60 | 89.67 | 91.70 | 81.60 | |
| αSiLU | 1.05 | 95.70 | 84.40 | 89.70 | 92.40 | 82.00 | |
| αSiLU | 1.055 | 95.30 | 83.50 | 89.01 | 91.70 | 81.70 | |
| αSiLU | 1.1 | 95.20 | 83.30 | 88.85 | 91.60 | 81.80 | |
| αSiLU | 1.5 | 94.40 | 84.60 | 89.23 | 91.30 | 81.30 | |
| αSiLU | 1.8 | 95.60 | 83.80 | 89.31 | 91.50 | 81.40 | |
| αSiLU | 2 | 96.30 | 83.40 | 89.39 | 91.60 | 81.50 |
| Model | Activation | α | P (%) | R (%) | F1-Score (%) | mAP@50 (%) | mAP@50–95 (%) |
|---|---|---|---|---|---|---|---|
| YOLOv11n | αSiLU | 0.5 | 87.40 | 87.50 | 87.45 | 94.20 | 80.70 |
| αSiLU | 0.7 | 87.40 | 87.10 | 87.25 | 94.30 | 80.90 | |
| αSiLU | 0.85 | 87.10 | 87.30 | 87.20 | 94.10 | 80.60 | |
| αSiLU | 0.9 | 87.60 | 86.30 | 86.95 | 94.00 | 80.50 | |
| αSiLU | 0.95 | 89.10 | 85.90 | 87.47 | 94.20 | 80.70 | |
| SiLU | 1 | 87.80 | 87.00 | 87.40 | 94.10 | 80.80 | |
| αSiLU | 1.025 | 88.00 | 86.00 | 86.99 | 94.10 | 80.60 | |
| αSiLU | 1.05 | 87.80 | 87.60 | 87.70 | 94.30 | 81.00 | |
| αSiLU | 1.06 | 88.00 | 87.30 | 87.65 | 94.20 | 80.80 | |
| αSiLU | 1.08 | 89.60 | 85.60 | 87.55 | 94.30 | 80.70 | |
| αSiLU | 1.1 | 88.40 | 86.30 | 87.34 | 94.20 | 80.70 | |
| αSiLU | 1.5 | 88.10 | 87.90 | 88.00 | 94.20 | 80.90 | |
| αSiLU | 1.8 | 86.60 | 88.10 | 87.34 | 93.90 | 80.80 | |
| αSiLU | 2 | 87.80 | 86.70 | 87.25 | 94.10 | 80.80 |
| Model | Precision | Recall | F1-Score | mAP@50 | mAP@50–95 |
|---|---|---|---|---|---|
| YOLOv5n (SiLU) | 96.90% | 83.10% | 89.47% | 91.60% | 80.80% |
| YOLOv5n (α = 1.05) | 95.60% | 83.50% | 89.14% | 92.20% | 80.80% |
| YOLOv8n (SiLU) | 97.10% | 82.80% | 89.38% | 91.90% | 81.40% |
| YOLOv8n (α = 1.05) | 95.80% | 82.60% | 88.71% | 92.00% | 81.20% |
| YOLOv10n (SiLU) | 94.70% | 83.60% | 88.80% | 91.00% | 80.90% |
| YOLOv10n (α = 1.05) | 96.60% | 82.80% | 89.17% | 91.60% | 80.90% |
| YOLOv11n(SiLU) | 95.20% | 83.40% | 88.91% | 91.30% | 81.30% |
| YOLOv11n (α = 1.05) | 95.70% | 84.40% | 89.70% | 92.40% | 82.00% |
| Model | Precision | Recall | F1-Score | mAP@50 | mAP@50–95 |
|---|---|---|---|---|---|
| YOLOv5n (SiLU) | 88.10% | 84.50% | 86.26% | 93.50% | 79.30% |
| YOLOv5n (α = 1.05) | 88.40% | 85.00% | 86.67% | 93.60% | 79.30% |
| YOLOv8n (SiLU) | 85.70% | 87.00% | 86.35% | 93.40% | 79.70% |
| YOLOv8n (α = 1.05) | 86.50% | 87.20% | 86.85% | 93.50% | 79.80% |
| YOLOv10n (SiLU) | 88.80% | 84.60% | 86.65% | 93.50% | 79.40% |
| YOLOv10n (α = 1.05) | 89.70% | 84.90% | 87.23% | 94.00% | 79.80% |
| YOLOv11n(SiLU) | 87.80% | 87.00% | 87.40% | 94.10% | 80.80% |
| YOLOv11n (α = 1.05) | 87.80% | 87.60% | 87.70% | 94.30% | 81.00% |
| Model | Activation Functions | mAP@50 (%) | mAP@50–95 (%) | Inference (ms) |
|---|---|---|---|---|
| YOLOv11n | LeakyReLU | 91.00 | 80.60 | 5.9 |
| YOLOv11n | ReLU | 91.2 0 | 80.80 | 5.8 |
| YOLOv11n | Mish | 91.20 | 81.30 | 5.9 |
| YOLOv11n | GELU | 91.40 | 81.30 | 5.9 |
| YOLOv11n | ELU | 91.70 | 81.60 | 5.9 |
| YOLOv11n | SiLU | 91.30 | 81.30 | 6.1 |
| YOLOv11n | CAReLU | 91.50 | 80.90 | 12.1 |
| YOLOv11n (α = 1.05) | αSiLU | 92.40 | 82.00 | 6.6 |
| Model | Activation Functions | mAP@50 (%) | mAP@50–95 (%) | Inference (ms) |
|---|---|---|---|---|
| YOLOv11n | LeakyReLU | 94.00 | 80.10 | 6.4 |
| YOLOv11n | ReLU | 94.20 | 80.20 | 6.3 |
| YOLOv11n | Mish | 94.10 | 80.80 | 6.2 |
| YOLOv11n | GELU | 94.20 | 80.70 | 6.4 |
| YOLOv11n | ELU | 94.10 | 80.60 | 6.3 |
| YOLOv11n | SiLU | 94.10 | 80.80 | 6.6 |
| YOLOv11n | CAReLU | 93.80 | 80.10 | 12.4 |
| YOLOv11n (α = 1.05) | αSiLU | 94.30 | 81.00 | 7.0 |
| Class ID | Object Class | P | R | F1-Score | mAP@50 | mAP@50–95 |
|---|---|---|---|---|---|---|
| 0 | Tomato_Septoria | 91.1% | 83.8% | 87.3% | 92.3% | 79.8% |
| 1 | Tomato_Healthy | 95.7% | 84.2% | 89.6% | 95.5% | 81.0% |
| 2 | Tomato_Lateblight | 96.3% | 94.9% | 95.6% | 97.8% | 84.5% |
| 3 | Tomato_Leaf_Mold | 94.2% | 87.8% | 90.9% | 94.2% | 86.6% |
| 4 | Tomato_Bacterial | 96.6% | 86.3% | 91.2% | 91.3% | 88.6% |
| 5 | Tomato_Yellow | 96.1% | 88.4% | 92.1% | 96.8% | 83.1% |
| 6 | Tomato_Earlyblight | 98.1% | 85.5% | 91.4% | 93.1% | 81.6% |
| 7 | Tomato_Mosaic | 97.6% | 64.5% | 77.7% | 78.5% | 70.9% |
| Average | 95.7% | 84.4% | 89.7% | 92.4% | 82.0% |
| Class ID | Object Class | P | R | F1-Score | mAP@50 | Class ID |
|---|---|---|---|---|---|---|
| 0 | Cucumber_Powdery | 88.1% | 88.3% | 88.2% | 95.4% | 81.2% |
| 1 | Cucumbers_Healthy | 92.9% | 80.2% | 86.1% | 93.8% | 80.9% |
| 2 | Cucumber_Downy | 88.2% | 86.1% | 87.1% | 92.7% | 77.2% |
| Average | 89.7% | 84.9% | 87.2% | 94.0% | 79.8% |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Nguyen, D.T.; Bui, T.D.; Ngo, T.M.; Ngo, U.Q. Improving YOLO-Based Plant Disease Detection Using αSILU: A Novel Activation Function for Smart Agriculture. AgriEngineering 2025, 7, 271. https://doi.org/10.3390/agriengineering7090271
Nguyen DT, Bui TD, Ngo TM, Ngo UQ. Improving YOLO-Based Plant Disease Detection Using αSILU: A Novel Activation Function for Smart Agriculture. AgriEngineering. 2025; 7(9):271. https://doi.org/10.3390/agriengineering7090271
Chicago/Turabian StyleNguyen, Duyen Thi, Thanh Dang Bui, Tien Manh Ngo, and Uoc Quang Ngo. 2025. "Improving YOLO-Based Plant Disease Detection Using αSILU: A Novel Activation Function for Smart Agriculture" AgriEngineering 7, no. 9: 271. https://doi.org/10.3390/agriengineering7090271
APA StyleNguyen, D. T., Bui, T. D., Ngo, T. M., & Ngo, U. Q. (2025). Improving YOLO-Based Plant Disease Detection Using αSILU: A Novel Activation Function for Smart Agriculture. AgriEngineering, 7(9), 271. https://doi.org/10.3390/agriengineering7090271