1. Introduction
As the Earth’s climate changes, its population grows, and environments and weather patterns become increasingly unpredictable, problems in efficient crop production are among the most important to solve. Alfalfa, a particularly important livestock feed, has been called “Queen of the forage crops”, and it is the crop of focus for the current study [
1]. Alfalfa is crucial to global food security because it is a valuable, sustainable, and protein-rich livestock feed as well as a nutritious food for people worldwide. In other words, alfalfa is not just food for people, but food for people’s food. Meanwhile, climate change directly threatens alfalfa’s efficient and sustainable cultivation [
2]. The importance of alfalfa is evidenced by the attention it receives from many land-grant universities, which cultivate alfalfa and collect and publish data on it in the form of variety trials. These variety trials, along with weather data, are the data underlying the current work.
Meanwhile, in 2015, the United Nations (U.N.) agreed on and published the “Sustainable Development Goals”, 17 goals that “are the blueprint to achieve a better and more sustainable future for all. They address the global challenges we face, including poverty, inequality, climate change, environmental degradation, peace and justice.” Most specifically, the current work attempts to contribute toward “Goal 2: Zero Hunger,” and “Goal 13: Climate Action” [
3]. Sadly, we are not yet on track to achieve Goal 2 [
4], but it is our fervent hope to contribute towards this goal. Existing methods, such as the ARIMA family of algorithms and deep neural networks are not sufficient for the goals of this paper. One of the end goals of this research is to provide a lightweight and accessible application for farmers as end-users. ARIMA family algorithms and neural networks both require large datasets for training. This is problematic as agricultural datasets are often not large, as we highlight later in this paper. We can sidestep this using synthetic data generated via our novel SITS algorithm and XGBoost. Deep neural networks often require a high quantity of expensive computational resources on which to train, while our models do not require access to GPUs. ARIMA algorithms are also somewhat subjective in progress evaluations, and parameters [
5]. The methods proposed here are more objective and less limited. The current work’s technical focus is forecasting alfalfa yield data as time series. Domain adaptation (DA) was also researched along the way as a steppingstone between crop yield estimation and forecasting. This work is presented as three phases. The first phase is DA, the second is ML-based forecasting, and the third phase is combining ML-based forecasting with DA.
To offer intuitive definitions of univariate and multivariate, time series data may consist of only one feature, which is known as univariate time series, or it may have several features, which is known as multivariate time series [
6]. When the features do not directly cause one another, they may be known as exogenous features or variables, and when they do cause one another, they may be referred to as endogenous features or variables, roughly speaking. For example, the current work focuses on precipitation, solar radiation, temperature, and yield, none of which directly cause each other, though solar radiation affects temperature somewhat. On the other hand, yield is directly affected by the size of the growing site, so that would be more of an endogenous variable. Lütkepohl offers a formal mathematical definition of exogenous and endogenous variables in his book on time series [
7]. The current work proposes both univariate and multivariate forecaster versions and compares them to the traditional family of autoregressive integrated moving average (ARIMA) statistical forecasting models. The proposed univariate ML-based forecaster was compared to the univariate ARIMA and seasonal ARIMA (SARIMA) models, and the multivariate ML-based forecaster was compared to the multivariate SARIMA with exogenous variables model (SARIMAX). ARIMA models are widely used in time series forecasting due to their ability to model trends and seasonality through autoregressive and moving average components [
8]. However, our results show that the proposed univariate ML-based forecaster produced more accurate forecasts than the ARIMA family, as detailed in the Results Section, with symmetric mean absolute percent errors (sMAPEs) as low as 16.94% with the proposed forecaster versus a best of 34.67% with SARIMA. Univariate and multivariate models were compared, showing an improvement in sMAPE from 16.94% to 12.81% when exogenous variables were included in the ML-based forecaster, while SARIMAX produced a sMAPE of 28.08%.
The final phase of this work combines the proposed ML-based forecasting technique with DA using SITS, and shows the most promising sMAPE scores of all, as low as 9.81% at best. Experiments in earlier work showed that DA may be helpful when target data are scarce, and they focused on the very difficult problem of using data with one distribution to estimate that of a different distribution. As research progressed to the current work, results became very promising, but the problem became easier in that local training was used after the initial pretraining step, so even though forecasting is more challenging than estimating the past and present, the local training helps it produce good results.
The current work makes four significant contributions. First, a novel DA technique is proposed that combines data synthesis with pretraining an XGBoost model and substantially increases estimation accuracy. Second, a novel data synthesis algorithm is proposed that shows improvement over established models. Third, a novel ML-based forecasting algorithm is proposed that exploits stationarity. Fourth, the proposed ML-based forecasting technique is combined with the proposed DA pipeline to produce more accurate forecasters.
2. Related Work
Prior approaches in agricultural forecasting often relied on isolated data streams, mostly utilizing either textual data from reports or satellite imagery [
9,
10] in isolation to predict crop yields. A survey by Satore et al. focused on a hybrid approach combining NASA remote sensing data, built administrative boundaries to separate areas of interest, and used a combination of historical satellite imagery and county-level National Agricultural Statistics Service (NASS) to build a dataset for training and validation of the model. While these approaches have made valuable contributions, they faced challenges in capturing all trends present in textual data, leading them to incorporate additional data sources into their methodologies. To address these challenges, they introduced empirical density functions derived from remote sensing data, capturing the distribution of spectral signatures across different crop types. Utilizing multi-dimensional scaling, they transformed these high-dimensional density functions into a lower-dimensional space, generating artificial covariates that retain essential information while simplifying analysis. These artificial covariates, representing distilled, critical features of the agricultural landscape, were then used to train machine learning models. The models then underwent training and validation, leveraging the constructed dataset that integrates the artificial covariates with NASS official statistics. This innovative approach, while effective, still encountered limitations in capturing the entirety of textual data trends [
11].
Building upon this, Gro Intelligence has implemented a real-time application of crop yield predictions to predict corn yields in the United States from diverse data sources. Cai et al.’s approach is a practical execution of the strategies proposed by Satore et al., taking the integration of diverse data sources to the next level by integrating a multi-level machine learning model and leveraging an extensive range of data sources to predict end-of-season corn yields in the United States from 2001 to 2016. Similarly to previous approaches, they faced challenges in fully capturing all trends present in textual data, prompting them to adopt a hybrid approach. This model showcases a sophisticated blend of algorithms, applying knowledge of physiological processes for temporal feature selection, achieving great precision in intra-season forecasts, even in years with anomalous growing conditions. It was back tested between 2000 and 2015, demonstrating the model’s capacity to predict national yield within approximately 2.69% of the actual yield by mid-August. In its first operational year during 2016, the model performed on par with the USDA’s forecasts and commercially available private yield models. At the county level, it could predict 77% of the variation in final yield using data through the beginning of August, with this figure improving to 80% by the beginning of October. Moreover, Gro Intelligence’s methodology enhances the original concept by focusing on the temporal aspects of data collection and analysis, allowing for intra-season adjustments to forecasts based on newly acquired data points. This real-time adaptability ensures that the predictive models are sensitive to sudden changes in weather patterns, crop conditions, and other critical factors affecting yield outcomes. Despite these advancements, the approach still encounters challenges in fully capturing the complexity of textual data trends [
12].
In previous work, the current authors presented results of DA with synthesis classification experiments performed using a conditional tabular generative adversarial network (CTGAN) and tabular variational autoencoder (TVAE) proposed by Park et al. [
13], and that work provided a more detailed description of those networks [
14]. While these synthesizers motivated the current use of data synthesis in precision agriculture, SITS produced the best results in the current work.
Kastens et al. studied crop yield time series forecasting using computer vision with masking, and while they report relative success, they do not explore any forecasting models beyond linear regression, and they do not compare their results to other forecasting models like the ARIMA family. They forecast metric tons per acre for corn, wheat, and soybean, but not tons per acre of alfalfa, and their metric of choice is mean absolute error (MAE), so it is difficult to compare the quality of their results to the current work’s. Kasten et al. claim to require at least 11 years of training data to train a strong forecaster, while the current work produces very low sMAPE scores with as few as six years of training data. Finally, their work is another example of computer vision, which involves extra complexity and expensive equipment beyond the needs of the current work [
15]. Choudhury and Jones compared statistical forecasting techniques to forecast maize yields in Ghana, and they report a significant contribution in this domain with mean squared errors (MSEs) as low as 0.03 metric tons per hectare using an autoregressive (AR) model, though that text does not appear to address or explain the table that features this metric. Furthermore, they emphasize coefficient of determination (R
2) scores in that paper’s discussion, which experiments in the current work have shown to not be a very reliable metric for measuring the quality of time series forecasts. The Results and Discussion Sections herein elaborate on this. Choudhury and Jones also limit their results to those from AR models and a few varieties of exponential smoothing, though they refer to it as an autoregressive and moving average (ARMA) model in their discussions, and they do not explore multivariate models [
16]. Bose et al. propose a spiking neural network (SNN) to forecast winter wheat crop yields from multispectral imaging time series data, and the SNN “encodes temporal information by transforming input data into trains of spikes that represent time-sensitive events” into a positive or negative binary value called a spike. That work reports high average prediction accuracies and R scores, and it reports beating linear regression (LR), k-nearest neighbors (KNNs), and support vector regression (SVR), but they do not compare their model to SARIMAX or other non-ML multivariate models. Most importantly, that work forecasts only one yield per year, six weeks into the future, while the current work forecasts three or four yields per year usually at least nine months into the future. Also, their R score appears to be calculated over the 14 years of predictions, so it bears little relevance to any R scores reported in the current work, which are calculated from a year’s forecast of several time points and the true values for that year. Also, although Bose et al. report that their SNN produces MAE scores as low as 0.24 tons per hectare, the current work’s ML-based technique produces MAE scores as low as 0.15 tons per acre, which is competitive even though the current work tackles an arguably more difficult forecast [
17]. Pavlyshenko proposed a model stacking technique that showed good results in predicting future sales and that their approach beat ARIMA and other ML models [
18]. However, that work does not report being effective for forecasting crop yields.
The traditional forecasting models, against which the current work’s ML-based forecasting technique is compared, are the family of autoregressive integrated moving average (ARIMA). These are ARIMA, seasonal ARIMA (SARIMA), and SARIMA with exogenous variables (SARIMAX). These popular models appear frequently in time series literature and are formally defined in many texts including one by Vishwas and Patel. In layman’s terms, ARIMA uses values in previous time points, called lags, to predict future values, and this is the autoregressive (AR) part. For the moving average (MA), ARIMA uses error lags, or differences between predictions and true values to predict future errors. The integration (I) step involves calculating differences between related time points to reduce seasonality and other statistical issues, called enforcing stationarity, on the time series.
For example, in the current work, alfalfa yields tend to be much higher at the beginning of the season, then lower toward the end, creating a seasonal element that challenges the use of autoregressive (AR) and moving average (MA) components alone. AR and MA models perform best on stationary data, defined as data with constant statistical properties (e.g., mean and variance) over time, such that any two equal-sized subsets of the time series have the same statistical characteristics. Seasonal data, like alfalfa yields, typically lack this stationarity. To address this, ARIMA employs differencing, the “I” step, which calculates differences between consecutive time series values to mitigate seasonality and induce stationarity. This differencing is the primary method by which the ARIMA family (ARIMA/SARIMA) achieves stationarity, with the goal of stabilizing the data’s statistical properties. In contrast, the proposed ML-based forecasting technique predicts differences between annual yields at corresponding time points (e.g., yield 1 in 2000 vs. yield 1 in 2001) to exploit stationarity, directly forecasting changes rather than transforming the series for AR/MA modeling. This approach allows the ML model to capture trends in yield differences without requiring the series to be fully stationary, offering a more flexible forecasting strategy.
Any meaningful description of an ARIMA model should be followed by its parameters in parentheses, such as ARIMA (1, 0, 0), where 1, 0, and 0 represent values for the parameters p, d, and q respectively. The number of AR lags is p, the number of MA lags is q, and the number of times differencing is performed in order to achieve stationarity is d. SARIMA is designed to further address seasonality, and it accepts a second set of seasonal parameters as in SARIMA(p, d, q)(P, D, Q, m), where P is the number of seasonal AR lags, Q is the number of seasonal MA lags, D is the number of times seasonal differences are taken, and m is the number of time points per season. Finally, and again roughly speaking, SARIMAX accepts one more parameter, a vector of exogenous variables from which cross-correlations are exploited to improve forecasts. For precise mathematical definitions of these models, and even Python implementation details, one may refer to the above-mentioned text by Vishwas and Patel [
19].
4. Results
First, results from DA with data synthesis experiments are presented, comparing several ML models in a set of target and source locations with leave-one-out cross-validation.
Table 1,
Table 2,
Tables S2 and S3 present these DA results. Second, results from experiments with the ML-based forecaster compared to ARIMA family models are presented.
Table 3,
Table 4,
Tables S4 and S5 present these time series forecast results. Finally,
Table 5,
Table 6,
Table 7,
Table 8 and
Table 9 present results which generally demonstrate that ForDA can produce better forecasters than ML-based forecasting without DA, and that SITS beats established synthesizers in this task.
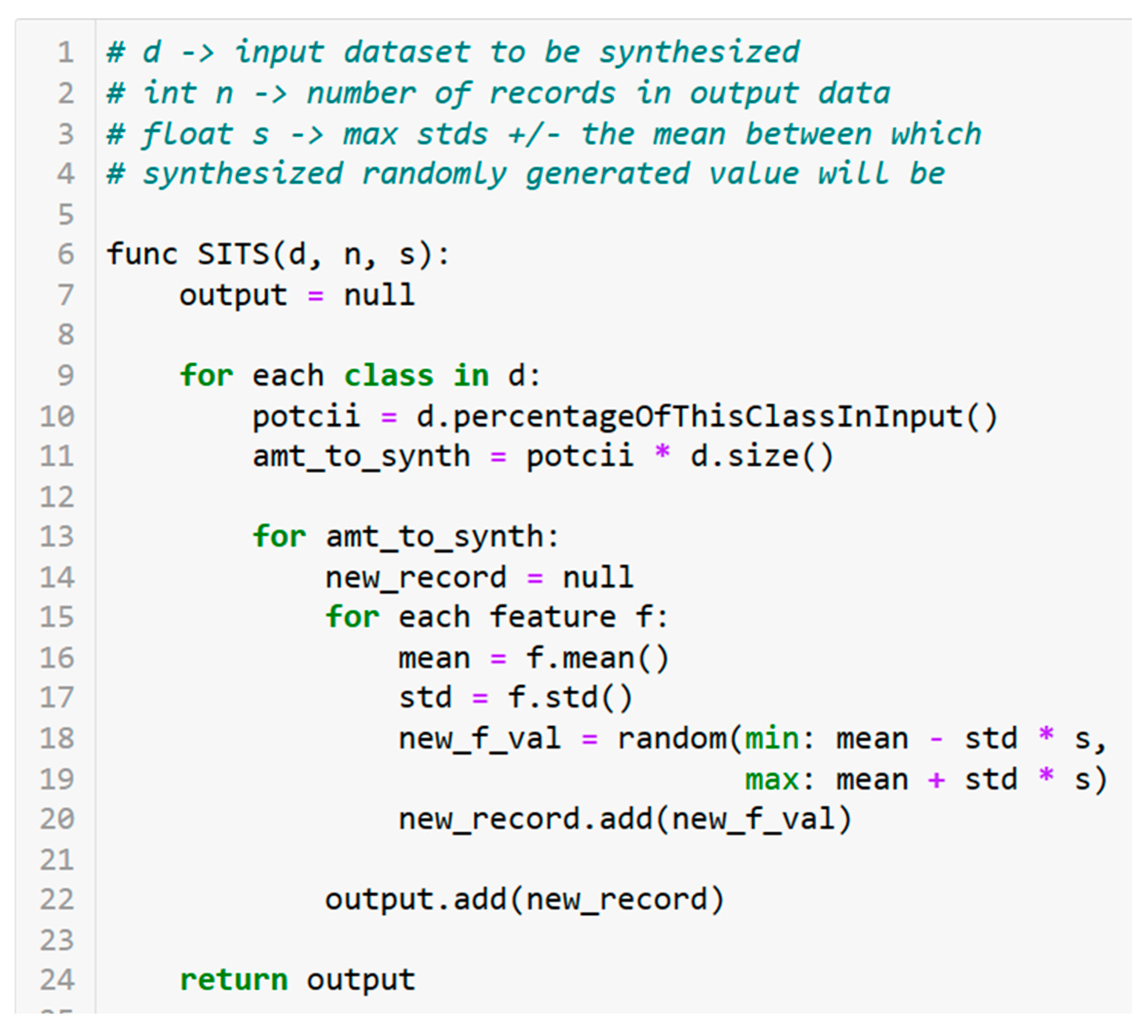
Table 1 depicts preliminary results using DASP and synthesizing training datasets of 20,000 samples generated from an initial dataset of about 2000 records from SD. We chose 20,000 because experiments showed improving results up to around that point but diminishing results after.
Table 1 shows that SITS is competitive with or superior to CTGAN and TVAE in this domain. While CTGAN sometimes beats SITS anecdotally, SITS on average generates datasets that train more accurate estimators than the others.
Table 1 does not reflect forecasting or use time series data, but trains on a separate source and estimates unseen values anywhere in the timeline of the target data. Estimating these past and current crop yields can help reveal which states may be good candidates for ForDA and provide a steppingstone toward designing that technique.
Table 1 is approximately a 66/33 training/test split, with pretraining in all of SD followed by the boost step in XGBoost with a subset of Wooster, OH data.
Table 1.
SITS vs. TVAE, CTGAN. XGBoost pretraining, 20 k samples, source: SD, target: Wooster, OH, 32 samples. Estimating past and current yields. SITS has best average R (bold).
Table 1.
SITS vs. TVAE, CTGAN. XGBoost pretraining, 20 k samples, source: SD, target: Wooster, OH, 32 samples. Estimating past and current yields. SITS has best average R (bold).
| Synth | Avg R | Avg MAE | Avg sMAPE |
|---|
| SITS | 0.68 | 0.48 | 30.24 |
| TVAE | 0.53 | 0.42 | 25.86 |
| CTGAN | 0.54 | 0.47 | 29.41 |
| TDA | 0.25 | 0.60 | 36.87 |
The next round of experiments trained and tested on locations that are within-state, but still non-local, and the boosting step was skipped, because early experiments showed no benefit from boosting with further within-state data versus training with it all at once. Also, a comparison of TDA along with the same three flavors of synthesizers as before was included. In these experiments, a sample size of 10,000 was chosen because early tests showed increasing gains up to that point but diminishing results after on this dataset. Source training data came from all of SD except the town of Highmore, and target test data came from Highmore, SD. The results from these closer source and target neighbors were also promising, but not quite as strong as more remote SD to OH results with the boosting step. As
Table 2 shows, SITS again generates datasets that train more accurate estimators than CTGAN or TVAE, not only on average but the best overall as well. CTGAN and TVAE beat TDA, however. SITS average R was 0.63, more than a 100% improvement over CTGAN, and the average MAE was 0.43.
Table 2.
SITS vs. TVAE, CTGAN, TDA. XGBoost w/no pretraining, 10 K samples. Source: SD, target: Highmore, SD. Estimating past and current yields. SITS beats others (bold).
Table 2.
SITS vs. TVAE, CTGAN, TDA. XGBoost w/no pretraining, 10 K samples. Source: SD, target: Highmore, SD. Estimating past and current yields. SITS beats others (bold).
| Synth | Avg R | Avg MAE | Avg sMAPE |
|---|
| SITS | 0.63 | 0.43 | 31.26% |
| TVAE | 0.36 | 0.48 | 32.65% |
| CTGAN | 0.30 | 0.47 | 32.50% |
| TDA (none) | 0.34 | 0.70 | 41.76% |
The final and most systematic round of DA experiments used leave-one-out cross-validation (LOOCV) to demonstrate average estimation accuracies over many locations, to identify any locations that stood out as significantly weak or strong trainers or targets, and to identify the best and worst models and synthesizers. A total of 20,000 samples were generated for all leave-one-out tests.
Table S2 details LOOCV test results for OH, and
Table S3 shows the averages. Though TVAE with LR had the highest anecdotal R score of 0.61, overall SITS results in slightly better R and MAE scores than TVAE, while CTGAN came in last. The three OH locations tested in
Table S2 are Wooster (W), North Baltimore (NB), and South Charleston (SC).
The remaining results are from time series experiments, where forecasters are trained only on historical data to predict yields in future years that the model has not previously seen.
Figure 2,
Figure 3,
Figure 4,
Figure 5 and
Figure 6 highlight select time plots.
Table S4 depicts results from our preliminary univariate time series experiments comparing our ML-based technique to ARIMA. We used one long time window from 1999 to 2010 for training and forecasted 2011 alfalfa yields. The resulting sMAPE scores show that our ML-based model produced more accurate forecasts than ARIMA or SARIMA, with BRR producing the best average score of sMAPE = 16.94%.
Tables S6 and S7 depict preliminary experiments with univariate and multivariate time series, respectively, in Beresford, SD.
Table S4 compares the results of the univariate version of the proposed ML-based technique with ARIMA and SARIMA, and
Table S4 compares the multivariate version of our technique with the multivariate version of ARIMA called SARIMAX. Again, one long training time window of 1999 to 2010 is used, and the resulting sMAPE scores, as low as 12.81%, indicate that the ML-based approach produces more accurate forecasts than ARIMA, SARIMA, and SARIMAX. For the ARIMA(p,d,q) and SARIMA(p, d, q)(P, D, Q, m) runs reported in each table the values of d and D were selected based on stationarity testing, m=4 is representative of the 4-month seasonal harvesting cycle, and the other values were selected based on grid search and experimentation. The multivariate approaches produce better results with almost every model, which is expected since they are provided with more training information. These experiments omit years 2002, 2005, and 2006, as these years report three cuts instead of the normal four. Neither the dataset for Univariate time series trial, nor the dataset for the Multivariate time series trials described in
Tables S4 and S5,
Table 3 and
Table 4 contain synthesized data. For these experiments we used exclusively original data.
Table 3 summarizes the results of a more systematic approach to univariate time series experiments that uses a sliding window, or rolling validation with multiple forecast horizons, which is a common validation approach in the time series literature [
3,
34]. This sliding window of training data was used to compare several ML models trained on only one feature, alfalfa yield in tons per acre. Arguably, the yield’s position in the time series may be thought of as a second feature. The results of the univariate approach were compared to the results of ARIMA and its seasonal counterpart SARIMA, and
Table 3 shows that the ML-based technique almost always produces more accurate forecasts than ARIMA and SARIMA. This table presents symmetrical mean absolute percent errors (sMAPEs) from a six-year sliding window in Beresford, SD, with a forecast horizon (FH) of 1 year at a time. It depicts forecasts starting with source 1999 to 2007 and target 2008, then the source years’ window slides forward one year at a time up to forecasting 2011. Training windows of six years are used because that is the width required before the ML-based forecasting technique begins to show an advantage over ARIMA and SARIMA. With training windows smaller than six years, ARIMA models usually forecasted more accurately in these experiments. The p and q and seasonal P and Q parameters were optimized through experimentation for each step of the sliding windows, so these parameters are different for each table row. As
Table 7 shows, the proposed ML-based forecasting technique is consistently competitive with or more accurate than ARIMA and SARIMA as measured by sMAPE, across every sliding window tested. Highlights include SVR with sMAPE = 13.77%, RF with sMAPE = 19.97%, and KNN with sMAPE = 18.53% on training window 2003 to 2010.
Table 3 shows sMAPE scores on top and R on bottom.
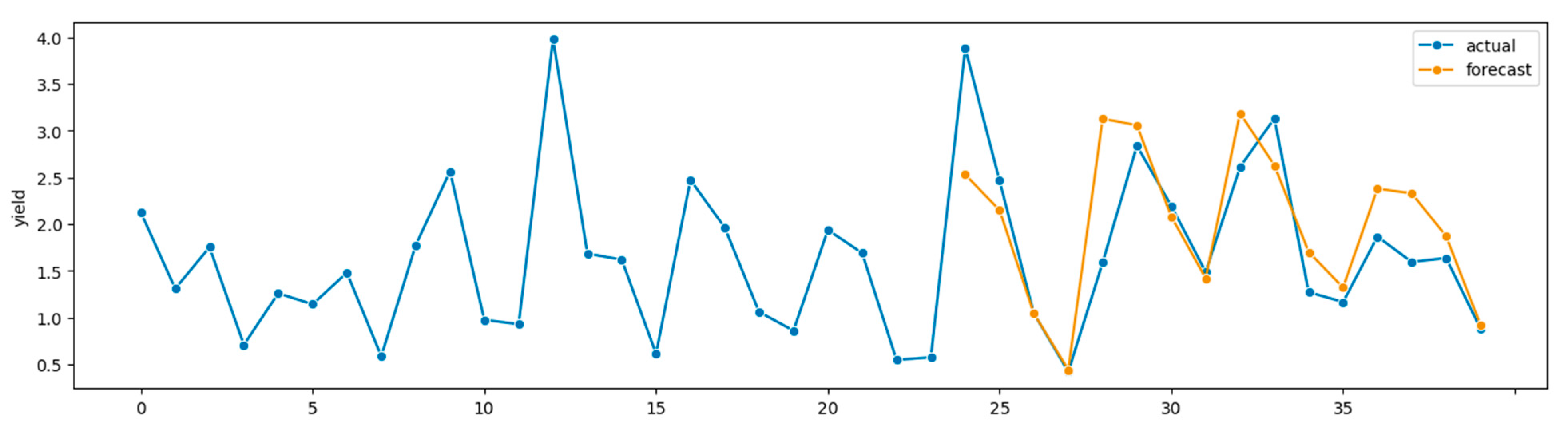
Figure 2 depicts a time plot for the SVR forecaster.
Table 3.
Univariate sliding window validation results. Beresford, SD. Four training windows of 6 years with each forecast of the following year, from 2008 to 2011. Top three average results in bold.
Table 3.
Univariate sliding window validation results. Beresford, SD. Four training windows of 6 years with each forecast of the following year, from 2008 to 2011. Top three average results in bold.
| Target Year | ARIMA | SARIMA | KNN | DT | SVR | XGB | MLP | RF | LR | BRR |
|---|
| 2008 | 49.47
0.62 | 39.05
0.88 | 42.22
0.99 | 36.62
0.99 | 37.04
0.74 | 39.38
0.79 | 43.18
0.99 | 41.22
0.93 | 41.01
0.99 | 37.46
0.99 |
| 2009 | 34.96
0.28 | 50.64
0.13 | 37.45
0.18 | 36.92
0.51 | 46.08
0.16 | 54.01
0.36 | 32.86
0.16 | 33.80
0.26 | 43.81
0.16 | 46.20
0.16 |
| 2010 | 29.80
0.86 | 14.43
0.90 | 27.71
0.85 | 59.04
0.44 | 25.89
0.82 | 71.04
0.34 | 26.83
0.85 | 39.34
0.65 | 23.65
0.85 | 22.56
0.85 |
| 2011 | 29.52
0.59 | 25.91
0.79 | 18.53
0.91 | 24.25
0.48 | 13.77
0.85 | 24.14
0.48 | 31.04
0.85 | 19.97
0.76 | 24.10
0.85 | 20.83
0.86 |
| Average | 35.94
0.59 | 32.51
0.68 | 31.48
0.73 | 39.21
0.48 | 30.70
0.64 | 47.14
0.49 | 33.48
0.71 | 33.58
0.65 | 33.14
0.71 | 31.76
0.71 |
Figure 2.
Time plot,
Table 3 SVR. Forecasts 2008–2011, true alfalfa yields 1999–2011.
Figure 2.
Time plot,
Table 3 SVR. Forecasts 2008–2011, true alfalfa yields 1999–2011.
Table 4 shows the results of the multivariate version of the sliding window validation experiments, where models trained on more than one feature were compared. The established model against which we compared the multivariate version of the ML-based forecasting technique is SARIMA with exogenous variables, or SARIMAX. Results from the same ML models as in the univariate version were compared. These data include precipitation, solar radiation, and temperature, and the location is Beresford, SD from 1999 to 2011, omitting 2002, 2005, and 2006 because those years did not feature four cuts.
Table 4 shows that the ML-based technique with stationarity results in more accurate forecasts than SARIMAX. The top row scores are sMAPE and the bottom row scores are R. RF was the top scorer in these tests, with an average sMAPE of 22.38% over all windows, and
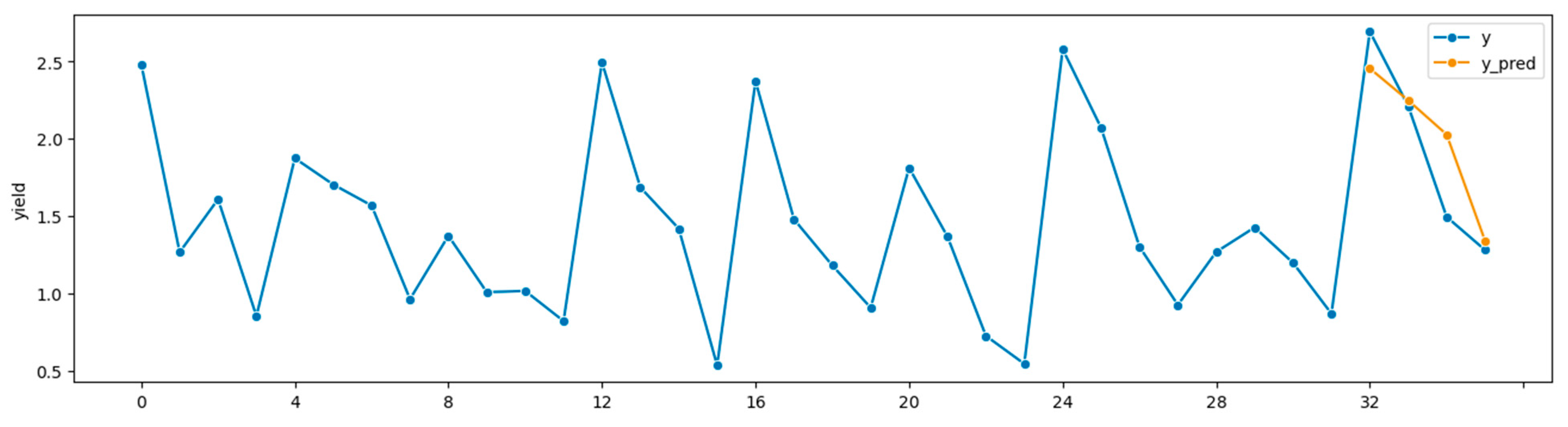
Figure 3 depicts its time plot.
Table 4.
Multivariate sliding window validation results. Beresford, SD. Four training windows of 6 years with each forecast of the following year, from 2008 to 2011. Top three average results in bold.
Table 4.
Multivariate sliding window validation results. Beresford, SD. Four training windows of 6 years with each forecast of the following year, from 2008 to 2011. Top three average results in bold.
| Target Year | SARIMAX | KNN | DT | SVR | XGB | MLP | RF | LR | BRR |
|---|
| 2008 | 29.33
0.97 | 27.47
0.96 | 9.08
0.99 | 29.61
0.99 | 28.24
0.98 | 30.90
0.99 | 17.23
0.98 | 43.79
0.84 | 32.13
0.99 |
| 2009 | 27.29
0.30 | 35.45
0.11 | 34.52
0.85 | 30.37
0.20 | 38.43
0.78 | 26.83
0.27 | 27.16
0.45 | 36.33
0.17 | 45.11
0.16 |
| 2010 | 29.54
0.54 | 16.06
0.87 | 48.18
0.49 | 21.70
0.84 | 36.61
0.92 | 18.93
0.86 | 23.39
0.89 | 31.36
0.86 | 22.16
0.85 |
| 2011 | 40.00
0.79 | 23.80
0.71 | 44.13
0.99 | 21.89
0.86 | 52.26
0.86 | 20.04
0.76 | 21.72
0.94 | 24.99
0.86 | 19.29
0.86 |
| Average | 31.54
0.65 | 25.70
0.66 | 33.98
0.83 | 25.89
0.72 | 38.89
0.89 | 24.18
0.72 | 22.38
0.82 | 34.12
0.68 | 29.67
0.72 |
Figure 3.
Time plot,
Table 4 RF. Forecasts 2008–2011, true alfalfa yields 1999–2011.
Figure 3.
Time plot,
Table 4 RF. Forecasts 2008–2011, true alfalfa yields 1999–2011.
Table 5 and
Table 6 present results from ML-based forecasting with DA (ForDA), which produced the best results, like sMAPE = 9.81%. They show that ForDA beats ML-based forecasting without DA, and that SITS leads to more accurate forecasts than CTGAN or TVAE. The source data in
Table 5 comes from Watertown, SD 1999 to 2011 (no 2003 due to insufficient data), and the target is Highmore, SD 1999 to 2011 (no 2000 to 2003, 2005, 2006, 2009 due to insufficient data). When data synthesis is useful due to data scarcity, these synthesis techniques will likely improve results over forecasting with TDA as previous experiments have demonstrated.
Table 6 presents the results from the same technique, but with source Highmore and target Watertown, for validation.
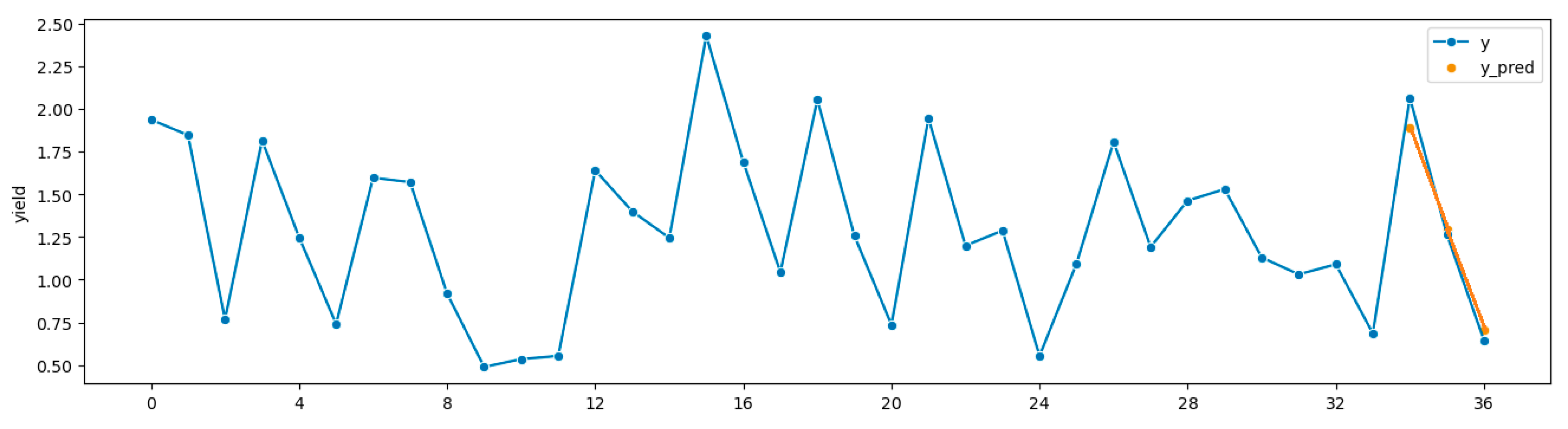
Figure 4 depicts the time plot for the most successful ForDA run in
Table 6.
Table 7 depicts the averages of
Table 5 and
Table 6.
Table 5.
ForDA with SITS vs. CTGAN, TVAE w/XGBoost; 20 k samples, SITS wins (bold). Source: Watertown, SD 1999 to 2011; target: Highmore, SD, 1999 to 2011.
Table 5.
ForDA with SITS vs. CTGAN, TVAE w/XGBoost; 20 k samples, SITS wins (bold). Source: Watertown, SD 1999 to 2011; target: Highmore, SD, 1999 to 2011.
| ForDA Synth | sMAPE | R | MAE |
|---|
| SITS | 10.22 | 0.87 | 0.21 |
| TVAE | 16.75 | 0.74 | 0.33 |
| CTGAN | 15.38 | 0.75 | 0.29 |
Table 6.
ForDA with SITS vs. CTGAN, TVAE w/XGBoost; 20 k samples, SITS wins (bold). Source: Highmore, SD 1999 to 2011; target: Watertown, SD, 1999 to 2011.
Table 6.
ForDA with SITS vs. CTGAN, TVAE w/XGBoost; 20 k samples, SITS wins (bold). Source: Highmore, SD 1999 to 2011; target: Watertown, SD, 1999 to 2011.
| ForDA Synth | sMAPE | R | MAE |
|---|
| SITS | 9.81 | 0.94 | 0.14 |
| TVAE | 19.20 | 0.92 | 0.26 |
| CTGAN | 18.50 | 0.90 | 0.25 |
Table 7.
ForDA with SITS vs. CTGAN, TVAE w/XGBoost averages; 20 k samples, SITS wins (bold). Averages of
Table 5 and
Table 6.
Table 7.
ForDA with SITS vs. CTGAN, TVAE w/XGBoost averages; 20 k samples, SITS wins (bold). Averages of
Table 5 and
Table 6.
| ForDA Synth | sMAPE | R | MAE |
|---|
| SITS | 10.01 | 0.90 | 0.18 |
| TVAE | 17.98 | 0.83 | 0.30 |
| CTGAN | 16.94 | 0.83 | 0.27 |
Figure 4.
Best results from ForDA. Source: Highmore, SD 1999 to 2011; target: Watertown, SD, 1999 to 2011 sMAPE = 6.72%.
Figure 4.
Best results from ForDA. Source: Highmore, SD 1999 to 2011; target: Watertown, SD, 1999 to 2011 sMAPE = 6.72%.
Table 8 shows average results from round-robin style experiments with ForDA in OH in Wooster (2011 to 2018), North Baltimore (2010 to 2018 without 2015 or 2017 due to insufficient data), and South Charleston (2010 to 2019 without 2017 due to insufficient data); each location is given a turn at being the source and another its target. These experiments showed the best results on much smaller synthesized datasets, so a sample size of 30 was settled on, as was using SITS for data synthesis, since earlier experiments suggested that it produces better results than CTGAN or TVAE for these purposes. The bottom row of
Table 8 shows averages of each metric over all round-robin experiments, so it represents the average forecast accuracy in OH.
Table 8 forecasts are not as striking as those where SD is the source and Beresford, SD is the target, but they are better than the ML-based forecasts without DA, and they are relatively strong forecasts overall. Also,
Table 8 depicts four-point seasons.
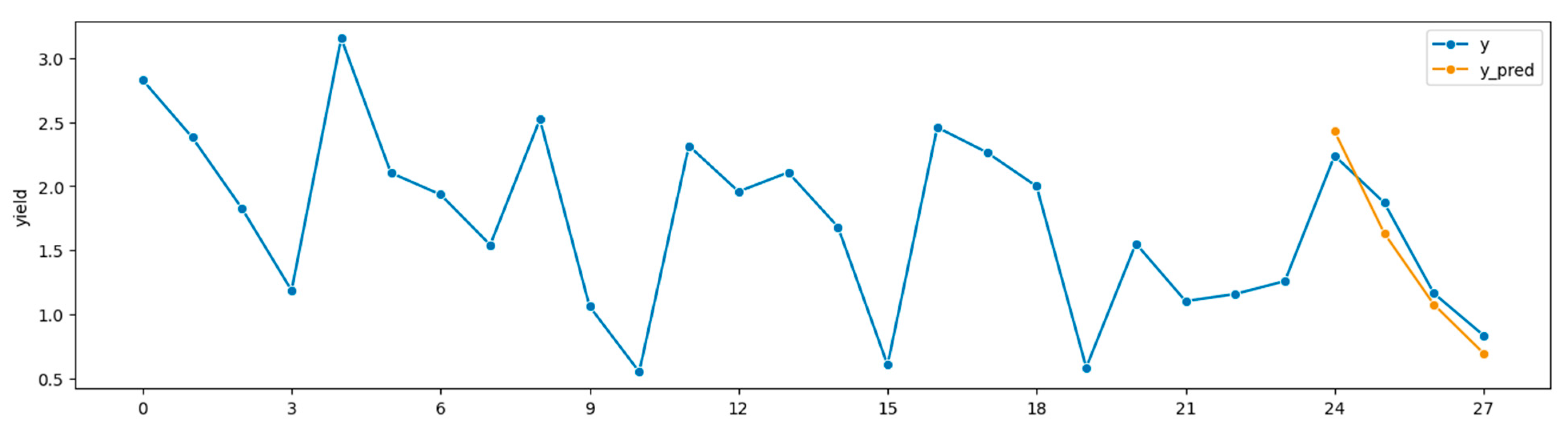
Figure 5 shows the best run, which produced sMAPE = 11.45%, MAE = 0.22 tons/acre, and R = 0.88 with source Wooster and target South Charleston. Since
Table 8 presents ForDA results with pretraining, all these source/target pairs use XGBoost; however, to determine whether pretraining helps, these same targets and the other seven models were also experimented with, using synthesis only and no pretraining. Those results and their corresponding time plot generally show that pretraining almost always helps, and skipping it produces mostly inferior results, with overall average sMAPE = 25.70% and R = 0.66 as shown in
Table 4.
Table 8.
ForDA results using SITS. Round-robin experiments in OH. Best two in bold; 2011 to 2018 in Wooster, 2010 to 2018 in North Baltimore, 2010 to 2019 in South Charleston.
Table 8.
ForDA results using SITS. Round-robin experiments in OH. Best two in bold; 2011 to 2018 in Wooster, 2010 to 2018 in North Baltimore, 2010 to 2019 in South Charleston.
| Source: Target | SITS(s) Parameter | sMAPE | MAE | R |
|---|
| Wooster: North Baltimore | 0.5 | 16.47 | 0.19 | 0.98 |
| Wooster: South Charleston | 1.5 | 16.55 | 0.30 | 0.82 |
| North Baltimore: Wooster | 1.5 | 26.51 | 0.51 | 0.21 |
| North Baltimore: South Charleston | 1.0 | 18.33 | 0.55 | 0.76 |
| South Charleston: Wooster | 2.0 | 22.84 | 0.44 | 0.26 |
| South Charleston: North Baltimore | 0.5 | 16.54 | 0.23 | 0.86 |
| Average | - | 19.54 | 0.37 | 0.65 |
Figure 5.
Best run in OH round-robin tests. Source: Wooster 2011 to 2018, target: South Charleston 2010 to 2019, forecasting 2019, sMAPE = 11.45%, MAE = 0.22 tons/acre, R = 0.88.
Figure 5.
Best run in OH round-robin tests. Source: Wooster 2011 to 2018, target: South Charleston 2010 to 2019, forecasting 2019, sMAPE = 11.45%, MAE = 0.22 tons/acre, R = 0.88.
As
Table 9 shows, LOOCV was attempted next, at the state level for sources and the local level for targets, since it is not feasible to forecast per-cut at the state level, as cut dates, weather, and number of cuts vary within the state. Three states were combined for each source and again experiments were conducted with and without synthesis, rotating among MI, OH, SD, and KY. Those results were promising in at least the case of target North Baltimore, OH, which produced a sMAPE = 15.06%, MAE = 0.16 tons/acre, and R = 0.98. SITS data synthesis clearly improved results in these tests, with an average sMAPE = 25.85%, MAE = 0.36 tons/acre, and R = 0.75 with synthesis versus sMAPE = 33.62, MAE = 0.49 tons/acre, and R = 0.65 without synthesis.
Figure 6 shows a time plot from the best run of these experiments.
Table 9.
ForDA LOOCV with MI (1999 to 2022), OH (2010 to 2019), SD (1999 to 2011), and KY (2012 to 2018).
Table 9.
ForDA LOOCV with MI (1999 to 2022), OH (2010 to 2019), SD (1999 to 2011), and KY (2012 to 2018).
| Source: Target | SITS(s) param | samples | sMAPE | MAE | R |
|---|
| MI, OH, SD: KY | 1.2 | 500 | 26.55 | 0.28 | 0.51 |
| MI, OH, SD: KY no synth | - | - | 19.82 | 0.19 | 0.71 |
| MI, SD, KY: NB, OH | 0.3 | 500 | 15.06 | 0.16 | 0.98 |
| MI, SD, KY: NB, OH no synth | - | - | 25.47 | 0.35 | 0.99 |
| OH, SD, KY: MI | 1.5 | 400 | 25.21 | 0.40 | 0.94 |
| OH, SD, KY: MI no synth | - | - | 39.39 | 0.56 | 0.88 |
| MI, OH, KY: SD | 1.2 | 500 | 36.59 | 0.59 | 0.55 |
| MI, OH, KY: SD no synth | - | - | 49.79 | 0.84 | 0.00 |
| Average w/synth | - | - | 25.85 | 0.36 | 0.75 |
| Average no synth | - | - | 33.62 | 0.49 | 0.65 |
Figure 6.
Source: MI, SD, KY; target: OH. SITS w/500 samples synthesized, s = 0.3, sMAPE = 11.89, MAE = 0.16, R = 0.98. Forecasting 2019.
Figure 6.
Source: MI, SD, KY; target: OH. SITS w/500 samples synthesized, s = 0.3, sMAPE = 11.89, MAE = 0.16, R = 0.98. Forecasting 2019.
Our statistical analysis of the experiments presented above can be located in
Table S6.
6. Discussion
Over the course of this work, modest promise was shown with the early DA experiments, but DA was harnessed to power the final ForDA technique, which yields very promising results.
Our results generally show that data diversity presents a serious issue. When we train on one set of locations and test on another, this creates a data diversity challenge, often leading to worse results. In estimation experiments where there was disparity or diversity between training and test data, we used SITS (Scale-Invariant Tabular Synthesizer) to synthesize data based on training datasets, as it would be inappropriate and lead to overfitting to synthesize data based on test sets. The rationale for using SITS is that it is scale-invariant, making it particularly suitable for handling data from different locations with varying scales or distributions, such as environmental conditions across states like Michigan, Ohio, South Dakota, and Kentucky. The purpose is to produce more accurate estimators, and our results confirm its effectiveness, with SITS leading to R scores over 100% higher than TVAE in certain experiments (e.g., improving R from 0.36 to 0.63 when estimating past and current yields). In forecasting, we handle data diversity mainly by using the boosting mechanism in XGBoost. When the datasets for the target location are too small, we often pre-train on a different location; however, this increases diversity between training and testing data. For the purpose of producing more accurate forecasts where data are scarce, we leverage XGBoost’s ability to pre-train on a large dataset (often synthesized by SITS) and then boost training using a smaller, less divergent dataset from the target location. This approach is supported by the literature, which has shown that such methods are effective for transfer learning with XGBoost (Huber et al, 2022). Since the actual test target data remain hidden during training, this boosting step is appropriate and does not lead to overfitting. Most importantly, this approach allows us to handle data diversity when training and testing on disparate datasets while still attaining useful, relatively accurate results, as evidenced by ForDA experiments achieving sMAPE scores as low as 9.81%.
One phenomenon observed is that while models in the ARIMA family provide diminishing returns after considering too many lags in these experiments, the ML-based forecasting technique appears to only improve more as the training window widens. Another interesting observation is that ARIMA sometimes outperforms SARIMA, so trying to account for seasonality is not always helpful. On the other hand, though alfalfa yields display some seasonality, looking at the time plots reveals that the trends are not always consistent.
Until the current work, the authors have focused on estimating past and current yields as tabular data. In that problem, R and coefficient of determination (R2) scores closer to 1 are the best and usually indicate a good model that makes accurate predictions.
Readers may note that the results for forecasting the future are sometimes better than results for estimating past and current yields, which may seem surprising; however, better forecasting results are expected when they are locally trained, while the estimators are trained on non-local data. Even though ForDA pretrains on non-local source data, it ultimately trains on a small subset of data local to the target area. As the current authors showed in previous work, local training to estimate past and current yields is still arguably the most accurate, as one would expect, reporting R2 scores over 0.98 (J. R. Vance).
While the results herein suggest that the proposed multivariate ML-based forecasting technique and ForDA produce more accurate forecasts than the well-established SARIMAX, this may be a little like comparing apples to oranges. The authors hypothesize that this technique wins because it is fitted to the forecast horizon’s weather features during testing to make its predictions, so it has an advantage over SARIMAX, which only looks at exogenous variables in the lags. On the other hand, the vision for the current project has always been to build a what-if tool like PYCS, where the “ifs” are the weather features in the forecast horizon, whether they are hypothetical or known, and these results indicate that the proposed techniques can forecast “what-if” with high accuracy. XGBoost often performs best with several thousand training samples, but it is not usually a top performer when the data size is very limited. On the other hand, when plenty of training data for pretraining is used and the booster function is employed, XGBoost becomes the winner. Overall, it would make this work easier if all the historical yield data was more consistent and tightly controlled, and if it went back further. While six years is enough training for this technique to beat SARIMAX, that small window might not reveal the potential of ML-based forecasting, and more data and consistency would likely help.
When ForDA was pushed to its limits, using non-local and state-level sources to forecast targets in disparate regions, it showed that ForDA is still potentially useful, but not as accurate as when sources are in-state. Therefore, and not too surprisingly, it is best to use in-state or otherwise very nearby sources and targets. While data synthesis and our SITS algorithm led to higher average accuracies than without synthesis in most experiments, synthesis did not demonstrate a clear advantage in those more challenging validation experiments. However, data synthesis continued to produce the anecdotally best models, which may be important to pay attention to, as one can save these pretrained models and reuse them in the final PYCS application, and they will likely keep and reuse those that lead to the anecdotally best results.