Abstract
Crown rot disease is caused by Fusarium pseudograminearum and is one of the major stubble-soil fungal diseases threatening the cereal industry globally. It causes failure of grain establishment, which brings significant yield loss. Screening crops affected by crown rot is one of the key tools to manage crown rot, because it is necessary to understand disease infection conditions, identify the severity of infection, and discover potential resistant varieties. However, screening crown rot is challenging as there are no clear visible symptoms on leaves at early growth stages. Hyperspectral imaging (HSI) technologies have been successfully used to better understand plant health and disease incidence, including light absorption rate, water and nutrient distribution, and disease classification. This suggests HSI imaging technologies may be used to detect crown rot at early growing stages, however, related studies are limited. This paper briefly describes the symptoms of crown rot disease and traditional screening methods with their limitations. It, then, reviews state-of-art imaging technologies for disease detection, from color imaging to hyperspectral imaging. In particular, this paper highlights the suitability of hyperspectral-based screening methods for crown rot disease. A hypothesis is presented that HSI can detect crown-rot-infected plants before clearly visible symptoms on leaves by sensing the changes of photosynthesis, water, and nutrients contents of plants. In addition, it describes our initial experiment to support the hypothesis and further research directions are described.
1. Introduction
Crown rot is a significant stubble-soil fungal disease that affects the cereal industry worldwide as it has been reported in the U.S., China’s southeast coast region, Africa, central Europe, and Australasia [1,2,3,4]. Fusarium pseudograminearum is the major contributing pathogen causing crown rot that is commonly found in Australian wheat [5]. Although wheat can be influenced by other fungal diseases, F. pseudograminearum has been identified and detected in 48% of fungal disease isolates from wheat in Australia [1]. Crown rot may cause the appearance of prematurely senescing culms, leading to the failure of grain/seed establishment, which can result in yield losses up to 89% [1]. F. pseudograminearum almost straddles the Australian wheat belt, due to the relatively warm and dry climate, with reports of crown rot in Queensland, New South Wales, Victoria, South Australia, and Western Australia [6]. In addition, F. pseudograminearum has a rich genetic diversity and adapts to various types of environments [7]. In 2009, the wheat industry in Australia suffered from economic losses of approximately AUD80 million as a result of crown rot, and the potential losses may reach AUD434 million [8,9].
Currently, there is no commercial wheat variety that exhibits obvious resistance to crown rot and no effective biological or chemical method to manage crown rot exists [10,11]. This problem is exacerbated since the pathogen can survive in soil and plant residues for at least three years [8,11]. As a result, the most effective way to manage crown rot is to use crop rotation to reduce the level of infested stubble, thereby reducing the pathogen inoculum level in the soil to decrease the risk of crown rot re-infection [12]. If crown rot disease appears in the wheat field, farmers often have to abandon the cultivation of wheat for at least two growing seasons and choose to grow non-host crops, whose economic value may be lower. For instance, after a three-year rotation of sorghum, the re-infection rate can be reduced to 12%, which is much lower than the 60% re-infection rate of the continuous wheat cultivation method for three years [13]. In addition to using crop rotation to reduce pathogen inoculum level, it is necessary to breed crown-rot-resistant varieties to avoid direct economic loss and further re-infection. The detection of crown rot in infected crops is a prerequisite of a successful screening process; however, the symptoms are difficult to detect at an early stage because crown rot disease does not produce clear visible symptoms at early growth stages. F. pseudograminearum is a soil fungi disease that attacks the crown and root tissues of wheat by colonizing the xylem and phloem after the disease infection [14], thus there is no significant increase in fungal biomass and observable symptoms on the plants during the first four weeks of the disease process [15]. Following this, the crown rot fungus begins to colonize the plant from the root and crown to the lower stem [16,17], causing brown discoloration on the stem tissue above the soil surface [3,18]. This symptom only appears on the lower stem of the plant, so it is difficult to identify in the field [19]. Another typical crown rot symptom is whiteheads that occur at the late growth stages and are more visible than brown stem discoloration. Whiteheads are usually present during early grain fill and the infected wheat will mature earlier, but grain filling is severely inhibited in the shriveled spikelets [20]. The occurrence of whiteheads is exacerbated in certain cultivars and growing conditions. For example, if wheat, at the milk stage, experiences reduced rainfall, the probability of whitehead appearance in the disease-infected wheat crops will greatly increase [21,22]. The pathogen colonizes and damages the plants’ vascular system, which is responsible for water and nutrient uptakes in the plant [5], and water stress during or shortly after the wheat flowering period will increase the symptoms of the disease [23]. Once wheat is infected with crown rot, the accumulation of a high level of the mycotoxin deoxynivalenol (DON) will make the grains inedible [24,25]. Additionally, the accumulation of DON will also aggravate grain development and cause grain shrivel [26,27].
An efficient screening method would not only assist agronomists in better evaluating crown rot infection to avoid excessive pathogen loads remaining in the soil as early as possible, but also support breeders to measure the resistance of different varieties of wheat to crown rot so as to select genotypes with superior resistance [28,29]. There are two common scoring methods to quantify crown rot severity: one is commonly used by breeders to score plants at maturity to identify resistant candidates, and the other is used by researchers to score the disease severity level at early growth stages. In certain breeding programs, researchers collect the main stems of wheat at maturity and use a crown rot screening scale to determine the resistance levels through observation of stem browning [30] on the first internode [2,31]. As infected wheat plants do not have evident symptoms on upper stems and leaves in the early stage of infection, they have to wait four months until maturity. Although current screening methods provide a measure of the resistance levels of individual wheat varieties, they are limited by late detection as well as high associated costs and their time-consuming and subjective nature. In one well-established early growth stage crown rot severity scoring method, ‘0’ represents no symptoms of infection; ‘1’ indicates necrotic lesions on the first leaf sheath; ‘2’ represents the first leaf sheath and the below sub-crown internode have completely changed to the brown color; ‘3’ reflects that the second leaf sheath also has partial necrosis symptoms; ‘4’ means that the brown color area has completely occupied the second leaf sheath and the sub-nodes of the sub-crown; ‘5’ and ‘6’ depict the necrosis of the leaf sheath [32]. By adopting this scale, pathologists are able to quantify the severity of plant infection 30 days after infection. However, this screening method cannot be conveniently applied by the agronomist to screen large field trials because data collection and manual scoring is labor intensive and subjective. Therefore, it is crucial to develop an accurate, objective, and rapid screening approach for general breeders to screen crown rot symptoms in wheat at early stages. Recently, fast-developing computer vision and hyperspectral imaging (HSI) technologies have demonstrated potential to detect crown rot disease at an early stage. To speed up breeding and efficiently manage crown rot disease, this paper reviews advanced imaging technologies for disease detection with an emphasis on hyperspectral imaging technologies and provides a new prospect in imaging-based crown rot disease detection.
Our major contributions in this review are:
- Through the review, we provide evidence that the crown can change photosynthesis (Section 3.2.2) and water and nutrients uptake (Section 3.2.1) in plants, which are major factors to influence wheat growth and cause yield loss.
- We developed a hypothesis that hyperspectral imaging can detect the changes of photosynthesis, water, and nitrogen uptake before visible symptoms on the upper stems and leaves.
- We conducted an initial experiment to support the hypothesis.
- We point out further research directions of using HSI for crown rot detection.
2. Digital Color Imaging
Color imaging is one of the most widely used technologies for plant disease detection. Digital color cameras capture images at three broadbands: red, green, and blue (Figure 1). The center wavelengths are about 450 nm, 550 nm, and 650 nm for blue, green, and red light, respectively [33]. Digital color images contain much less spectral information than hyperspectral images; however, they usually have much higher spatial resolution. In the analysis of the digital color image, the color space of hue, saturation, lightness (HSL), hue, saturation, value (HSV), and lightness, the greens and magentas channel, and blues and yellows channel (LAB) are most often used. Hue refers to the different colors that the human eye perceives, and saturation represents the purity of the colors. Lightness or value represents the intensity of light [34]. Several studies have shown that infected or uninfected areas of plants can be distinguished through color or textural features extracted from color images [35,36]. The general process of analyzing digital color images for crown rot could be: (1) segmentation of region-of-interest: (2) color or textural features extraction; and (3) classification. The digital color image has great potential for assessing Fusarium wheat disease at a late growth stage. For example, digital imaging technology can distinguish between wheat infected with Fusarium head blight and healthy wheat in the late flowering stage (coefficient of determination (R2) > 0.8) [37]. It is worth noting that because, the digital color image itself contains limited image information and is similar to the human eye, it may not produce sufficient data collection to detect the crown rot infection and severity in its early stage. However, hyperspectral imaging may be able to improve detection and screening for this disease.
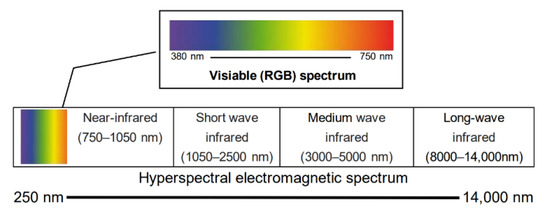
Figure 1.
Depiction of the different wavelength ranges of visible color imaging and hyperspectral imaging. The wavelength range of visible light is from approximately 380 to 750 nm, a small portion of the range of HSI.
3. HSI Technologies for Plant Phenotyping
3.1. Hyperspectral Imaging
As a modern and advanced approach, the applications of HSI technologies in agricultural research have become increasingly prevalent. In contrast to digital color imaging that only captures images at the three broad bands in the visible light range, HSI collects and processes the spectrum in the visible light as well as in the infrared range, including hundreds of narrow bands (Figure 1). Hyperspectral images contain significant amounts of spectral information at different bands, which describe the unique ‘fingerprints’ of objects and may reveal the composition of the objects [38]. When light hits the surface of plants, reflection, absorption, or transmission occurs, all of which are influenced by the cellular structure and chemical and biological properties of the leaves [39]. For example, the cell structure of a leaf controls the absorption of near-infrared light, and short-wave infrared light is affected by the water and nitrogen content (Figure 2) [40]. Therefore, HSI is currently one of the most promising tools for quantifying plant parameters with less human bias [41].
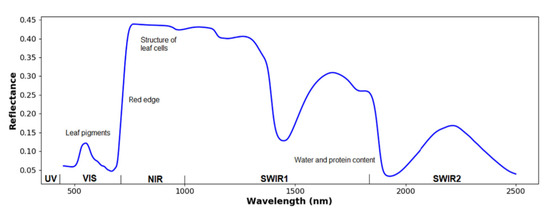
Figure 2.
Illustration of a typical reflectance spectrum of a green leaf. UV, VIS, NIR, and SWIR refers to ultraviolet, visible spectrum, near-infrared, and short-wave infrared, respectively [41]. Near-infrared light is affected by the structure of leaf cells, while the SWIR light is dominated by water and protein content.
HSI data are especially sensitive to the light environment, so laboratory-based imaging and field-based remote sensing are quite different procedures. Laboratory-based imaging is usually relatively easy to manage and control the lighting environment, while outdoor remote sensing is more dependent on sunlight [42,43]. Relatively uncontrollable sunlight density, the appearance of clouds, and the angle and height of the sun are factors that affect the collection of hyperspectral image data. Laboratory imaging usually has better resolution because sensors can photograph plants more closely and there are more pixels for representing a single plant, which can provide pathologists with more information about disease development [44]. Although laboratory imaging usually has controllable illumination, the self-shadow and the leaf curvature of plants could cause errors in hyperspectral images that need to be carefully addressed [41]. The standard normal variates (SNV) [45] and hypercube-to-hyper-hue, -saturation, and -intensity (HC2HHSI) [46,47] algorithms have proven to be useful to reduce the effects of shadow and leaf angle. In the pre-processing of HSI, the first step is to reduce the impact of scatter effects and bad pixels. Scatter effects are usually caused by physical change (e.g., cell, leaf or canopy structure, size, or shape) [39]. Different scatter-correction techniques can be used to reduce the influence of scattering effects. For example, multiplicative scatter calibration is a common scatter correction method that reduces additive and multiplicative effects by regressing the measured spectrum with a spectral reference [48]. SNV transformation uses a normalization method to reduce scatter effects [49,50]. The derivative methods calculate the first or higher derivative of reflectance to improve the resolution and correct the baseline shift in the hyperspectral data [39]. Savitzky–Golay (SG) derivative [51] and Norris–Williams (NW) derivative [52] are the two common methods used in this process [53,54]. Bad pixels, including dead pixels and spikes, are errors of hyperspectral images that need to be removed. Bad pixels are permanent and usually caused by the damaged cells on the detectors [55]. Signal smoothing algorithms are usually used to remove the effects of dead pixels or spikes. In addition, field-based remote sensing can collect plant data in a large area, which can provide large-scale information of plants for both breeders and agronomists.
HSI usually collects hundreds of images in different wavelengths, providing considerable spectral and spatial information [37,56]. Therefore, HSI data must be analyzed with reasonable computer software that supports state-of-art machine learning methods. As a popular and well-developed computing analysis platform and programing language, Python is an ideal option for data analysis. Compared with other programing languages, Python is an interpreted, advanced, and open-source programing language supporting multiple programing paradigms, including functional, instructional, structured, object-oriented, and reflective programing [57]. Moreover, Python is more user-friendly when it comes to non-programing researchers and free to access. Throughout the past 30 years, machine learning has developed into a multi-field interdisciplinary subject, involving a vast number of statistical theories. In particular, there is a strong relationship between machine learning and computational statistics. Machine learning theory is mainly used to design and analyze some algorithms that allow computers to automatically ‘learn’. This is especially helpful in the sense that computers can automatically analyze and acquire rules from data that can be used to predict unknown data [58]. Notably, Scikit-learn is a free machine-learning library based on Python, and it can be used for various classification, regression, and clustering algorithms [59]. Generally speaking, machine learning is divided into two categories: supervised learning and unsupervised learning. Supervised learning achieves a function from a given training data set that includes input and output data. When new data is input, it can predict the result based on the trained function. Compared with supervised learning, unsupervised learning algorithms do not need training data and algorithms can learn by themselves. Specifically, the process of analyzing HSI data roughly consists of four parts: pre-processing, segmentation, feature extraction, and data analysis [41]. The purpose of pre-processing is to improve the image quality and eliminating noise [39]. Once pre-processing is completed, the region of interest is segmented from the entire image. In terms of feature extraction, researchers need to interpret the hyperspectral data into feature vectors that are more descriptive.
In data analysis, statistical analysis, regression, and classification will be utilized to obtain the relationship between image information and the plant parameters of the target crop. For example, the support vector machine (SVM) model, a non-probabilistic supervised machine learning classifier, can classify a large dataset to the greatest extent while simulating linear and non-linear relationships depend on the kernel functions [60]. SVM also can carry out multiple classification and regression. Its decision boundary is the maximum margin hyperplane that is solved by learning samples. The algorithm can analyze the objective’s maximum distance between the decision boundary and the training sample (support vectors) closest to the hyperplane to achieve HSI imaging classification purposes [61,62]. For example, SVM was used to predict the nutrient and water contents of wheat with R2 above 0.5 [63]. As a kind of unsupervised learning method, k-means can group objects with similar properties together without human guidance. In plant disease detection, the k-means clustering algorithm can be used to cluster infected pixels of leaves from uninfected pixels [64]. The reasonable application of k-means to pre-process data to reduce the size of the data set can provide more support for SVM analysis in training data [44]. In addition, a principal component analysis (PCA) also effectively reduces data dimensions to improve analysis accuracy and interpretability, and minimize information loss [44,65]. PCA as statistical analysis can be used in the simplification of data sets; it uses an orthogonal transformation to linearly transform a series of variable values, so as to obtain the principal components of the sample library data to reduce the complexity of the data. As one of the simplest approaches in the hyperspectral imaging analysis method, it has a strength of common method of eigen-decomposition of the covariance matrix for solving problems in construction of soft classification rules, dealing with outlying samples and missing data [66]. For the PCA in hyperspectral image analyses, first a few spectra of images has been extracted to provide principal components, and then the principal components were used to classify images with an accuracy of 70% [67]. Compared to classification, regression analysis is a quantitative method that associates spectral response with specific parameters (e.g., yield loss). The most common regression techniques applicable to HSI data include principal component regression and partial least squares regression [39].
3.2. HSI and Crown Rot Related Plant Traits
Traditional plant phenotyping methods present challenges in crown rot resistance plant breeding because the visual symptoms of crown rot generally present at late growth stages, leading to late decision making and selection. In contrast, the biological and morphological changes at early growth stages caused by the disease can affect the reflectance of spectra beyond visible light range; thus, HSI might be a potential method for breeders to detect crown rot at an early stage in a more efficient manner. Notably, numerous studies have shown that HSI is an efficient and non-destructive method that can accurately detect traits related to crown rot diseases before human experts are able to observe the symptoms [68,69,70].
This section can be broadly classified into two categories: (1) water and nutrient distribution map in wheat, and (2) hyperspectral and chlorophyll fluorescence imaging interaction in photosynthesis and deoxynivalenol (DON) screening.
3.2.1. Water and Nutrient Distribution Maps of Wheat
The colonization of F. pseudograminearum of a plant restricts water and nutrient movement in the vascular system of the diseased plant. Although disease symptoms at early growth stages are not obvious, the mycelium have already occupied the xylem and phloem of the lower stem of the plant up to the first node [8,71]. At the beginning of the disease process, F. pseudograminearum enters the leaf sheath tissue through the stomata of the infected seedling leaf sheath. The eventual abundant growth of the hyphae will occur in vascular bundles of the whole plant [72]. After the pathogen enters the plant vasculature, it will first colonize the plant’s xylem vessels, and then spread to the phloem tissues as the severity of the disease increases [73]. The proportion of vascular bundles colonized in the prematurely senescent culms is usually greater than non-senescent culms [73]. Around 36 to 99% of the xylem and phloem tissue in prematurely senescent culms are colonized, while the percentage of vasculature occupied by the fungus in non-senescent culms is relatively low [74]. There is an established relationship between the extent of F. pseudograminearum vessel colonization and appearance of visual discoloration, but the extent of tissue discoloration is different between varieties, as well as at different infection/incubation dates [75]. In addition, although both xylem and phloem are responsible for the transfer of substances within plants, the xylem is mainly responsible for transporting water and nutrients from the roots to the aerial parts of the plant, while phloem transports nutrients (such as sugar and amino acids) from the leaves to the growing tissue or root storage [76,77]. As such, while the initial colonization of the fungus is usually within the xylem tissue or vascular bundles, the severity of phloem colonization is more related to the appearance of premature senescence. Moreover, F. pseudograminearum is more willing to grow in the nutrient-rich areas, such as the phloem, thus can quickly restrict the nutrient flow to the plant [73]. As the infection progresses, the fungus will form a dense hypha network in the epidermal cells and then extend to the mesophyll tissue and other asymptomatic parts of the plant [74]. Therefore, when wheat is infected with crown rot, understanding the water and nutrient distribution in a plant could be an important surrogate trait for phenotyping infection level. This could not only predict the severity of the disease earlier, but also improve support for the screening of different varieties at resistant levels. Because the transportation of water and nutrients is affected by the colonization of crown rot, this trait may differ between varieties and growth environments.
The analysis of HSI with a partial least square regression method has shown great potential to predict the distribution of nitrogen and water in plants, as shown in Figure 3 [63,78]. If water and nitrogen distribution maps of wheat can be created through HSI, the maps could help quantify the potential impact and yield penalty of crown rot on different wheat varieties. Some resistance genes may only be expressed at certain disease developmental stages in the plant, thereby reducing the fungus’s influence on available nutrients and water transport in the plants’ vascular system [26]. Therefore, a water and nutrient distribution analysis of young plant at the early stage of infection may be able to provide phenotyping information to support breeders to find the expression of some resistance genes and plant phenotypes by genetic mapping. The severity of the crown rot infection is highly likely to impact the water and nutrient movement within infected plants differently since the phloem and xylem occupied by a bundle of hyphae are not able to transport water and nutrients normally leading to serious symptoms and premature senescence [22,79].
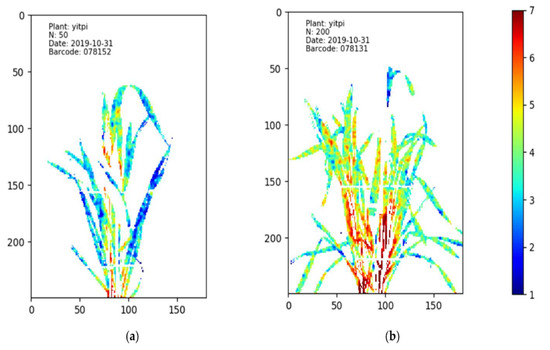
Figure 3.
The HSI nitrogen (N) distribution maps of wheat cultivar Yitpi under different N treatments. Different levels of N% are presented using different colors, with red referring to the highest N% while blue refers to the lowest N% [78]. In image (a), the plant treated with 50 mg/kg N has more blue pixels than the plant treated with 200 mg/kg N, which is shown in image (b). This means that the leaves of the plant in image (a) contain lower N content than the leaves in the plant of image (b). The imaging date was 31 October 2019.
Hyperspectral imaging-based high-throughput phenotyping has been employed to quantify the water content, micronutrients, and macronutrient concentrations of plants in a non-destructive way [80,81]. A combination analysis of visible light and near-infrared (VNIR) (400–1000 nm) and short-wave infrared (SWIR) (1000–2500 nm) can improve accuracy in water and nutrient content predictions (validated R2 = 0.69 in water and R2 = 0.66 in nitrogen, respectively), while a poor accuracy is obtained by using VNIR alone to predict the water and nitrogen content of plants (both R2 values are lower than 0.6) [63]. The water distribution map also provides an intuitive representation of the concentration and distribution of water in plants [63], which might be helpful to analyze the extent of fungal colonization of plants that are water-restricted as a result of the infection. The prediction of the severity of crown rot disease based on changes in nutrient and water transport should be thoroughly evaluated.
It should be noted that leaf morphological features and shadows have a significant influence on HSI analysis [82]. Wheat leaves are narrow, and the structure is relatively irregular and distorted, which can make it more difficult to accurately predict a nitrogen distribution map [63]. In contrast, corn leaves are wider and relatively regular, which makes it easier to predict nitrogen distribution. Separate photographic analyses of various parts of the plant may be a potential solution. Compared with the photograph of the whole plant, in partial photographs of plant organs (e.g., leaf, stem, and root) it is easier to predict the nitrogen content [83]. The possible reason for this is that sensors can photograph plants more closely, providing sufficient pixel resolution for a small region of the plant. Thus, increased resolution may provide pathologists with the required information about disease development [44]. Crown rot also gradually spreads from the bottom to the top of the plant during colonization. Therefore, rather than using the whole plant photo, partial photos of each plant’s basic parts from root to stem will be more beneficial to the pathological conditions of screening crown rot at each stage, as well as to observe the response of plants to disease resistance. In addition, in the process of analyzing plant nutrient content, plants at different growth stages may need to be analyzed with different small-range specific wavelengths. The control group and nitrogen stressed plants could be identified by different ranges of specific wavelengths at 20 days, 30 days, and 70 days after sowing [84]. For example, after 30 days sowing, stressed and control plants can be distinguished by reflectance at 350–415 nm and 706–934.5 nm, while at grain filling (70 days) they can be separated by reflectance at 355–515.5 nm, 617–695 nm, and 726–1075 nm [84]. Similarly, another study indicated that Fusarium-infected plants and healthy plants whose phenological growth stage is at approximately 70 days are successfully distinguished under similar wavelengths (682–733 nm and 927–931 nm); the potential reason is the distinct water loss in the plant’s spike [29]. The nutrient stress in plants caused by crown rot will change with the growth of the plants as fungal hyphae proliferate in nutrient rich plant organs. Studies have shown that if, pathogens consume nutrients in a certain area of the plant, it may reduce the colonization of that area [74]. Further research is required to determine whether specific wavelengths identifying nutrient and water content of plants are also useful for screening crown rot infection levels and how dynamic this relationship is over the development of wheat plants through time.
3.2.2. Hyperspectral and Chlorophyll Fluorescence Imaging Interaction in Photosynthesis and DON Screening
The interaction of HSI system and chlorophyll fluorescence imaging technology represents another potential screening approach for crown rot in wheat. The HSI combined with chlorophyll fluorescence imaging technology has been successfully employed to identify the ears of Fusarium-infected wheat and healthy wheat with high accuracy, while also helping to assess the severity of the disease infection [29]. Although the screening of wheat infection occurred at the flowering or early milk growth stage, the interaction of the hyperspectral imaging system and chlorophyll fluorescence imaging technology still has considerable advantages to assist agronomists and pathologists in the analysis of the severity and region of wheat infection in the field. Chlorophyll fluorescence imaging is able to assess chlorophyll fluorescence, which is re-emitted by chlorophyll molecules during the return from excited to non-excited states [85], and most fungal diseases will impact on plant’s photosynthetic metabolism negatively, such as chlorophyll degradation. Therefore, chlorophyll fluorescence imaging is an effective way of analyzing plant diseases and is also considered a non-destructive screening method. For instance, another hemibiotrophic fungi, Colletotrichum lindemuthianum, can colonize plant leaves and inhibit plant photosynthesis. Previous research successfully used chlorophyll fluorescence imaging to distinguish healthy and infected plants, also found that the effect of plant photosynthesis on necrotic lesions is more evident [86]. Consequently, chlorophyll fluorescence imaging can be used to evaluate and better understand the physiological state of the leaf or plant—especially the photosynthesis process.
The severity of the Fusarium disease symptoms is highly correlated with the efficiency of photosynthesis, and the photosynthetic efficiency of infected plants is significantly decreased [87,88]. For instance, DON produced by F. pseudograminearum can inhibit protein synthesis in both stem and leaf cells. When a small piece of mesophyll has been exposed to a DON solution, DON will whiten portions of tissue after a 48-to-96-h period and is due to a decline in the content of chlorophyll a and b and total carotenoids [89]. After the chloroplast begins to lose pigment, the leaf segments begin to lose electrolytes as well. The light absorption within the chlorophyll zone decreases rapidly with the destruction of chloroplasts and with the gradual decomposition of chlorophyll in cells that are affected by fungi. Photosynthesis has been shown to be inhibited after the seedlings are infected by Fusarium [90]. In this study, the maximum photochemical efficiency in the photosystem I reaction decreased more significantly compared with that in the photosystem II, and the oxidation process of the photosystem I reaction was also inhibited. However, as the infection period increased, the donor and acceptor sides of the photosystem II reaction center also began to suffer from damage, thus further inhibiting the performance and coordination of the plant’s photosystem I and II reaction centers. Under such circumstances, Fusarium will inevitably cause a decrease in tissue photosynthetic activity, which provides a basis for the application of chlorophyll fluorescence and HSI. Notably, there is a high possibility to combine chlorophyll fluorescence imaging and HSI to detect Fusarium disease, since the reduction in chlorophyll content in these cells reduces the potential for internal photon reflection and reabsorption processes in the relevant HSI wavelength range [91].
When a plant is infected with Fusarium, the epidermis, cell wall, and epidermal cells inside the plant will undergo changes that can be detected by NIR hyperspectral imaging [92,93]. There is a significant shift in reflectance between healthy and infected plants at 680–730 nm [87]. In this paper, it was observed that there is a considerable drop at this wavelength range for the infected plant as the chlorophyll degrades in the plant.
When the green reflection peak of wheat leaves becomes smooth due to disease infection, the reflection rate in the near-infrared region also generally decreases [94]. The decrease in chlorophyll content in cells can not only be detected by chlorophyll fluorescence imaging, but it also affects the potential of the photon remission and re-absorption processes of plant leaves in the wavelength range of 690 nm to 735 nm [95]. Consequently, it can be inferred that HSI technology can be combined with chlorophyll fluorescence images to improve accuracy in the screening analysis of plant pathology.
The chlorophyll of Fusarium resistant plants exposed to DON decreased less than in Fusarium-sensitive plants [96]. For example, when a gene encoding ethylene insensitive 2 (EIN2) in wheat was silenced, the wheat leaves were less affected by DON, and the probability of programmed cell death was significantly reduced [97]. Therefore, hyperspectral imaging technology is able to interact with chlorophyll fluorescence images to quickly identify change in leaf greenness area, which could provide a tool for breeders to identify wheat varieties with strong tolerance to DON. However, compared with other broadleaf crops (e.g., canola, beans, and peas), wheat leaves, as a grass-like crop, possess smaller leaf surfaces to absorb radiation at the seedling stage, which poses challenges for the analysis of chlorophyll content at early growth stages. Notably, neither of the two imaging techniques can distinguish infected plants at maturity, which is attributed to the fact that the chlorophyll of the plant head is completely degraded and the tissue water content is reduced [91]. Therefore, the interaction of hyperspectral imaging system and chlorophyll fluorescence imaging technology has considerable advantages to help agronomists and pathologists in analyzing the severity of wheat infection in the field and in predicting the disease spread region before maturity stage.
Previous work has employed HSI to identify wheat heads affected by F. pseudograminearum and people can apply HSI to understand the water and nutrient content in plant. In addition, by applying the PCA analysis method, researchers successfully distinguished diseased and healthy wheat ear tissues in the wavelength ranges of 500–533 nm, 560–675 nm, 682–733 nm, and 927–931 nm [29]. As a popular machine learning method, the artificial neural network (ANN) is a mathematical model or computational model that imitates the structure and function of biological neural networks. ANN can analyze and process image data through a neural network with input vectors and output vectors (neurons/nodes) [98]. The application ANN distinguished healthy versus infected wheat plant in near-infrared (900–1700 nm) images and obtained an accuracy of 74.14%, after nine weeks of infection [99]. Such several studies indicate the crown rot could be detected by the HSI as summarized in Table 1, and there is high probability to develop a rapid method to detection crown by HSI at infection early stage.

Table 1.
Summary information of previous studies using HSI for measurement of water and nutrient content or Fusarium disease related trait discovery. Included are the sensors, related image types, or special wavelength and further comments.
3.3. Preliminary Evaluation of HSI to Detect Crown Rot in Wheat
The literature shows that HSI combined with machine learning technologies can be successfully applied to sense photosynthetic activity, water, and nutrient content. Thus, we made a hypothesis that HSI can be applied to detect crown rot infected wheat plants at an early stage without visible symptoms on leaves by sensing photosynthesis or water and nutrient contents in plants, and conducted a preliminary experiment to support the hypothesis. The experiment was conducted in The Plant Accelerator located in the Waite campus of The University of Adelaide. Four widely grown wheat varieties (Aurora, Yipti, Trojan, and Emu Rock), which have different levels of susceptibility to crown rot disease [100], were chosen for study. The plants were grown in soil substrate under 2 disease treatments of “controlled” and “infected” with 26 replicates for 4 varieties, resulting in a total of 208 pots of wheat plants for the study. To infect the plants, we placed 10 F. pseudograminearum incubated seeds around the base of the seedling’s shoot 14 days after sowing, defined as the first day after infection (1 DAI). On the same day, all seedlings were transported to the lanes of the LemnaTec plant phenotyping platform (LemnaTec GmbH, Aachen Germany) for hyperspectral imaging. Once on the phenotyping platform, the plants were automatically watered daily to a set target weight. Two cameras were located in the hyperspectral imaging chamber (WIWAM, Ghent, Netherlands) of the phenotyping platform for whole-plant imaging, as shown in Figure 4. The FX10 camera (Specim, Oulu, Finland) captured the visible and near infrared (VNIR) data from 400 nm to 1000 nm with 5.5 nm full wavelength at half maximum (FWHM), while the short-wavelength infrared (SWIR) camera (Specim, Oulu, Finland) operated in the range of 1000 nm to 2600 nm with 12 nm FWHM. Hyperspectral imaging was conducted weekly starting on 15 July (11 DAI), when the shoots were large enough for side-view imaging, to 2 September 2021 (60 DAI), when the plants were in the early milk stage.

Figure 4.
Present the process of hyperspectral image acquisition by two cameras (FX 10 and SWIR cameras) in WIWAM hyperspectral imaging system.
After the data collection was completed, all HSI data were firstly calibrated and then the Savitzky–Golay filter [54] was applied to complete the data smoothing. The least squares method was used to fit two adjacent spectrums for linear regression to estimate continued value [78,101]. In the calibrated hyperspectral images, the backgrounds were segmented from the plants using a crop segmentation method proposed by a previous study [78]. After the final plant region segmentation, the original spectral signatures of the pixels for whole plant in each pot were extracted and averaged. As the aim of this study was to determine if hyperspectral imaging technologies can detect crown rot disease in wheat plants in an early stage without obvious symptoms on leaves, the dead and rotting plants removed from further analysis and therefore only healthy plants and infected plants without clear visible symptoms on leaves remained for further processing.
The processed data of each individual day was input into a SVM for binary classification, and the kernel method was radial basis function (RBF) kernel. The cross-validation technique was used to divide the data samples into several smaller sub-datasets, so that the initial sub-dataset was used as the training set, and the other sub-datasets were used to follow-up confirmation and verification of this analysis. Five-fold cross-validation with three repetitions was performed to train and validate the models. We used weighted accuracy, precision, recall, and F1 score to evaluate the performance of classification.
In the VNIR region, F1 score and accuracy have a clear increasing trend until 32 DAI, at 0.790 in F1 score and 0.790 in accuracy (Figure 5). In the contrast, F1 score and accuracy in SWIR increased considerably from 11 to 39 DAI and became stable from 39 DAI to 53 DAI, as shown in Figure 6. Notably, although the F1 score and accuracy values at 32 DAI are 0.756 for accuracy and 0.750 for F1 scores, the peak values of both factors at 60 DAI are around 0.83. The results showed that both VNIR and SWIR have F1 classification scores over 0.75 after 32 DAI, which provides support for the early detection of crown rot in the future by both sensors.
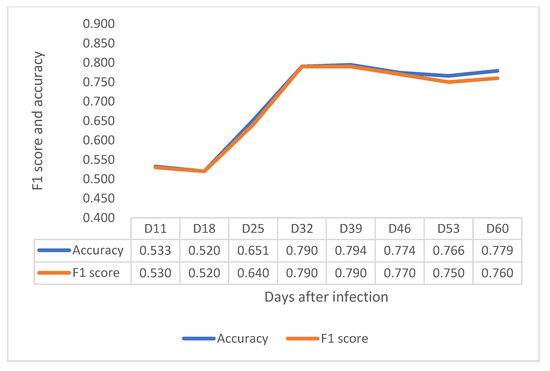
Figure 5.
Depiction of F1 score and accuracy of the VNIR sensor from 11 to 60 DAI.
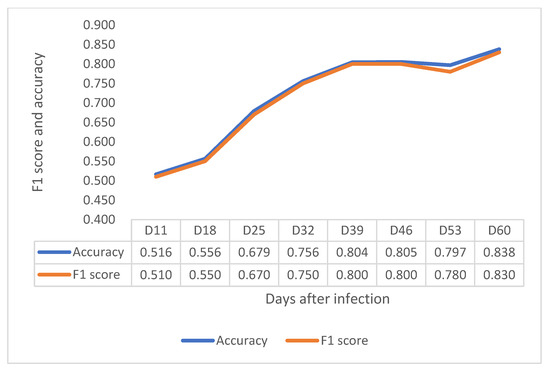
Figure 6.
Depiction of F1 score and accuracy of the SWIR sensor from 11 to 60 DAI.
The pathogen does not spread up to the stem immediately after infecting the roots, as it waits four weeks before it starts to increase biomass [15]. The HSI result suggest that, once the crown rot starts to colonize the stem region, and even though the leaves do not show obvious visible symptoms, HSI identifies some difference between infected and control plants. As previously hypothesized, the VNIR camera might distinguish the infected plants by detecting differences in photosynthetic capacity in the vegetative stage (18–32 DAI), while the SWIR cameras identified the infected plants probably by detecting the changing of water and nutrient content with increasing accuracy through development (11–60 DAI). However, further investigation should be conducted to find out which wavelengths play the most important roles for detection.
4. Conclusions
The harmful impact of crown rot on the wheat industry is well-established, especially in areas where drought events are increasing due to global warming. Unfortunately, there is currently no effective chemical or biological method to control this disease. Farmers rely on rotation to reduce the residual level of pathogens in the soil to reduce the risk of infection. An efficient and economical screening method is essential to help wheat breeders effectively select varieties with resistance. HSI is a high-throughout, non-invasive screening technique that has been successfully used for detection of other diseases and to measure water and nutrient in plants. We believe that HSI has significant promise to detect crown rot disease at an early growth stage when plants do not have visible symptoms. According to the results of our preliminary experiment, either VNIR or SWIR HSI can distinguish the difference between the infected and healthy plants after 32 DAI with an F1 score up to 0.790 in VNIR and 0.750 in SWIR even though there are no visible symptoms on the leaves or upper stems. It is not only more convenient than the current method, which requires people to observe the plant’s crown and root section, but also it can be employed earlier in the cropping season than using whitehead symptoms to identify the crown rot disease at harvest time.
Despite the success of the preliminary experiment, a number of further investigations should be considered:
- The wavelengths range of the spectrum mentioned in this study is very wide. The key-wavelengths that play the most important role in the classification need to be further studied. Determining key-wavelengths can guide the design of low-cost, light-weight multispectral sensors for field applications.
- In the preliminary experiment, we used the reflectance data alone as input for SVM classification. Different data types, such as SNV, hyper-hue, or principal components, need to be further studied.
- The preliminary experiment supports the hypothesis that HSI can distinguish the difference between infection and healthy plant, which can provide support for early disease detection. However, further research needs to investigate how to use HSI in disease screening to determine symptom severity level and levels of crown rot resistance in diverse varieties.
- It is important to further analyze how the pathogen affects the transport and distribution of water and nutrients in plants, especially at different growth stages.
- The initial experimental results are limited to side-view imaging only. An experiment of top-view imaging should be undertaken in a further study, since top-view images would be easier to obtain via remote sensing in field trials.
- The initial experiment was limited to the greenhouse environment and further investigation needs to be conducted in field trials.
- We conducted a preliminary experiment to demonstrate the feasibility of using HSI for crown rot disease detection. However, different types of sensors, data collection, data processing, and machine learning algorithms need further intensive study.
Author Contributions
Writing—original draft, Y.X.; writing—review and editing, H.L. and D.P.; project administration, H.L. and D.P. All authors have read and agreed to the published version of the manuscript.
Funding
This research was funded by the Yitpi Foundation, grant ID: 0006009868.
Institutional Review Board Statement
Not applicable.
Informed Consent Statement
Not applicable.
Data Availability Statement
Conflicts of Interest
The authors declare no conflict of interest.
References
- Akinsanmi, O.A.; Mitter, V.; Simpfendorfer, S.; Backhouse, D.; Chakraborty, S. Identity and pathogenicity of Fusarium spp. isolated from wheat fields in Queensland and northern New South Wales. Aust. J. Agric. Res. 2004, 55, 97–107. [Google Scholar] [CrossRef]
- Moya-Elizondo, E.; Arismendi, N.; Castro, M.P.; Doussoulin, H. Distribution and prevalence of crown rot pathogens affecting wheat crops in Southern Chile. Chil. J. Agric. Res. 2015, 75, 78–84. [Google Scholar] [CrossRef] [Green Version]
- Shikur Gebremariam, E.; Sharma-Poudyal, D.; Paulitz, T.C.; Erginbas-Orakci, G.; Karakaya, A.; Dababat, A.A. Identity and pathogenicity of Fusarium species associated with crown rot on wheat (Triticum spp.) in Turkey. Eur. J. Plant Pathol. 2018, 150, 387–399. [Google Scholar] [CrossRef]
- Xu, F.; Yang, G.; Wang, J.; Song, Y.; Liu, L.; Zhao, K.; Li, Y.; Han, Z. Spatial distribution of root and crown rot fungi associated with winter wheat in the north China plain and its relationship with climate variables. Front. Microbiol. 2018, 9, 1054. [Google Scholar] [CrossRef] [PubMed]
- Knight, N.L.; Macdonald, B.; Percy, C.; Sutherland, M.W. Disease responses of hexaploid spring wheat (Triticum aestivum) culms exhibiting premature senescence (dead heads) associated with Fusarium pseudograminearum crown rot. Eur. J. Plant Pathol. 2021, 159, 191–202. [Google Scholar] [CrossRef]
- Chakraborty, S.; Liu, C.J.; Mitter, V.; Scott, J.B.; Akinsanmi, O.A.; Ali, S.; Dill-Macky, R.; Nicol, J.; Backhouse, D.; Simpfendorfer, S. Pathogen population structure and epidemiology are keys to wheat crown rot and Fusarium head blight management. Australas. Plant Pathol. 2006, 35, 643–655. [Google Scholar] [CrossRef]
- Khudhair, M.; Obanor, F.; Kazan, K.; Gardiner, D.M.; Aitken, E.; McKay, A.; Giblot-Ducray, D.; Simpfendorfer, S.; Thatcher, L.F. Genetic diversity of Australian Fusarium pseudograminearum populations causing crown rot in wheat. Eur. J. Plant Pathol. 2021. [Google Scholar] [CrossRef]
- Kazan, K.; Gardiner, D.M. Fusarium crown rot caused by Fusarium pseudograminearum in cereal crops: Recent progress and future prospects. Mol. Plant Pathol. 2018, 19, 1547–1562. [Google Scholar] [CrossRef] [Green Version]
- Murray, G.M.; Brennan, J.P. Estimating disease losses to the Australian wheat industry. Australas. Plant Pathol. 2009, 38, 558–570. [Google Scholar] [CrossRef]
- Moya-Elizondo, E.A.; Jacobsen, B.J. Integrated management of Fusarium crown rot of wheat using fungicide seed treatment, cultivar resistance, and induction of systemic acquired resistance (SAR). Biol. Control 2016, 92, 153–163. [Google Scholar] [CrossRef]
- Simpfendorfer, S. Evaluation of the Seed treatment Rancona Dimension as a Standalone Option for Managing Crown Rot in Wheat–2015; DAN00175; NSW DPI Northern Grains Research Results; Department of Primary Industries NSW: Tamworth, Australia, 2016; pp. 121–124. [Google Scholar]
- Wildermuth, G.; Thomas, G.; Radford, B.; McNamara, R.; Kelly, A. Crown rot and common root rot in wheat grown under different tillage and stubble treatments in southern Queensland, Australia. Soil Tillage Res. 1997, 44, 211–224. [Google Scholar] [CrossRef]
- Backhouse, D. Modelling the behaviour of crown rot in wheat caused by Fusarium pseudograminearum. Australas. Plant Pathol. 2014, 43, 15–23. [Google Scholar] [CrossRef]
- Erginbas-Orakci, G.; Poole, G.; Nicol, J.M.; Paulitz, T.; Dababat, A.A.; Campbell, K. Assessment of inoculation methods to identify resistance to Fusarium crown rot in wheat. J. Plant Dis. Prot. 2016, 123, 19–27. [Google Scholar] [CrossRef]
- Stephens, A.E.; Gardiner, D.M.; White, R.G.; Munn, A.L.; Manners, J.M. Phases of infection and gene expression of Fusarium graminearum during crown rot disease of wheat. Mol. Plant-Microbe Interact. 2008, 21, 1571–1581. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Knight, N.L.; Sutherland, M.W. Histopathological assessment of Fusarium pseudograminearum colonization of cereal culms during crown rot infections. Plant Dis. 2016, 100, 252–259. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Summerell, B.A.; Burgess, L.W.; Backhouse, D.; Bullock, S.; Swan, L.J. Natural occurrence of perithecia of Gibberella coronicola on wheat plants with crown rot in Australia. Australas. Plant Pathol. 2001, 30, 353–356. [Google Scholar] [CrossRef]
- Saad, A.; Macdonald, B.; Martin, A.; Knight, N.L.; Percy, C. Comparison of disease severity caused by four soil-borne pathogens in winter cereal seedlings. Crop Pasture Sci. 2021, 72, 325–334. [Google Scholar] [CrossRef]
- Smiley, R.W. Fusarium Crown Rot Whitehead Symptom as Influenced by Wheat Crop Management and Sampling Date. Plant Dis. 2019, 103, 2612–2623. [Google Scholar] [CrossRef]
- Alahmad, S.; Simpfendorfer, S.; Bentley, A.R.; Hickey, L.T. Crown rot of wheat in Australia: Fusarium pseudograminearum taxonomy, population biology and disease management. Australas. Plant Pathol. 2018, 47, 285–299. [Google Scholar] [CrossRef]
- Alahmad, S. Accelerating the Development of Durum Wheat Adapted to Drought and Crown Rot Conditions. Ph.D. Thesis, The University of Queensland, Brisbane, Australia, 2019. [Google Scholar]
- Hollaway, G.J.; Exell, G.K. Survey of wheat crops for white heads caused by crown rot in Victoria, 1997–2009. Australas. Plant Pathol. 2010, 39, 363–367. [Google Scholar] [CrossRef]
- Moya-Elizondo, E.A. Fusarium crown rot disease: Biology, interactions, management and function as a possible sensor of global climate change. Int. J. Agric. Nat. Resour. 2013, 40, 235–252. [Google Scholar] [CrossRef]
- Pestka, J.J.; Smolinski, A.T. Deoxynivalenol: Toxicology and potential effects on humans. J. Toxicol. Environ. Health Part B 2005, 8, 39–69. [Google Scholar] [CrossRef] [PubMed]
- Sobrova, P.; Adam, V.; Vasatkova, A.; Beklova, M.; Zeman, L.; Kizek, R. Deoxynivalenol and its toxicity. Interdiscip. Toxicol. 2010, 3, 94. [Google Scholar] [CrossRef]
- Mudge, A.M.; Dill-Macky, R.; Dong, Y.; Gardiner, D.M.; White, R.G.; Manners, J.M. A role for the mycotoxin deoxynivalenol in stem colonisation during crown rot disease of wheat caused by Fusarium graminearum and Fusarium pseudograminearum. Physiol. Mol. Plant Pathol. 2006, 69, 73–85. [Google Scholar] [CrossRef]
- Yli-Mattila, T. Ecology and evolution of toxigenic Fusarium species in cereals in northern Europe And Asia. J. Plant Pathol. 2010, 92, 7–18. [Google Scholar]
- Ozdemir, F.; Koc, N.K.; Paulitz, T.; Nicol, J.M.; Schroeder, K.L.; Poole, G. Determination of fusarium crown rot resistance in wheat to Fusarium culmorum and Fusarium pseudogramineaum using real time PCR. Crop Prot. 2020, 135. [Google Scholar] [CrossRef]
- Bauriegel, E.; Giebel, A.; Herppich, W. Hyperspectral and chlorophyll fluorescence imaging to analyse the impact of Fusarium culmorum on the photosynthetic integrity of infected wheat ears. Sensors 2011, 11, 3765–3779. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Davies, P. Resistance and Tolerance Where We Are with Crown Rot Breeding; GRDC, ACT 2604: Kingston, Australia, 2016. [Google Scholar]
- Tapia, M.P.C.; Burrows, R.P.M.; Sepúlveda, B.R.; Concha, M.V.; Palma, C.V.; Moya-Elizondo, E.A. Antagonistic activity of chilean strains of pseudomonas protegens against fungi causing crown and root rot of wheat (Triticum aestivum L.). Front. Plant Sci. 2020, 11, 951. [Google Scholar] [CrossRef] [PubMed]
- Shi, S.; Zhao, J.; Pu, L.; Sun, D.; Han, D.; Li, C.; Feng, X.; Fan, D.; Hu, X. Identification of new sources of resistance to crown rot and Fusarium head blight in Wheat. Plant Dis. 2020, 104, 1979–1985. [Google Scholar] [CrossRef] [PubMed]
- Nassau, K. Colour. Available online: https://www.britannica.com/science/color (accessed on 21 September 2021).
- Moroney, N.; Fairchild, M.D.; Hunt, R.W.; Li, C.; Luo, M.R.; Newman, T. The CIECAM02 color appearance model. In Proceedings of the Color and Imaging Conference, Scottsdale, AZ, USA, 12 November 2002; pp. 23–27. [Google Scholar]
- Ahmed, N.; Asif, H.M.S.; Saleem, G. Leaf Image-based Plant Disease Identification using Color and Texture Features. arXiv 2021, arXiv:2102.04515. [Google Scholar]
- Tian, Y.-W.; LI, C.-h. Color image segmentation method based on statistical pattern recognition for plant disease diagnose. J. Jilin Univ. Technol. (Nat. Sci. Ed.) 2004, 2, 028. [Google Scholar]
- Qiu, R.; Yang, C.; Moghimi, A.; Zhang, M.; Steffenson, B.J.; Hirsch, C.D. Detection of Fusarium Head Blight in wheat using a deep neural network and color imaging. Remote Sens. 2019, 11, 2658. [Google Scholar] [CrossRef] [Green Version]
- Lu, G.; Fei, B. Medical hyperspectral imaging: A review. J. Biomed. Opt. 2014, 19, 10901. [Google Scholar] [CrossRef] [PubMed]
- Bruning, B.; Berger, B.; Lewis, M.; Liu, H.; Garnett, T. Approaches, applications, and future directions for hyperspectral vegetation studies: An emphasis on yield-limiting factors in wheat. Plant Phenome J. 2020, 3, e20007. [Google Scholar] [CrossRef]
- Sahoo, R.N.; Ray, S.S.; Manjunath, K.R. Hyperspectral remote sensing of agriculture. Curr. Sci. 2015, 108, 848–859. [Google Scholar]
- Liu, H.; Bruning, B.; Garnett, T.; Berger, B. Hyperspectral imaging and 3D technologies for plant phenotyping: From satellite to close-range sensing. Comput. Electron. Agric. 2020, 175, 105621. [Google Scholar] [CrossRef]
- Abdulridha, J.; Ampatzidis, Y.; Qureshi, J.; Roberts, P. Laboratory and UAV-based identification and classification of tomato yellow leaf curl, bacterial spot, and target spot diseases in tomato utilizing hyperspectral imaging and machine learning. Remote Sens. 2020, 12, 2732. [Google Scholar] [CrossRef]
- Schut, A.; Ketelaars, J. Monitoring grass swards using imaging spectroscopy. Grass Forage Sci. 2003, 58, 276–286. [Google Scholar] [CrossRef]
- Lowe, A.; Harrison, N.; French, A.P. Hyperspectral image analysis techniques for the detection and classification of the early onset of plant disease and stress. Plant Methods 2017, 13, 80. [Google Scholar] [CrossRef]
- Vigneau, N.; Ecarnot, M.; Rabatel, G.; Roumet, P. Potential of field hyperspectral imaging as a non destructive method to assess leaf nitrogen content in Wheat. Field Crop. Res. 2011, 122, 25–31. [Google Scholar] [CrossRef] [Green Version]
- Liu, H.; Chahl, J.S. A multispectral machine vision system for invertebrate detection on green leaves. Comput. Electron. Agric. 2018, 150, 279–288. [Google Scholar] [CrossRef]
- Liu, H.; Lee, S.H.; Chahl, J.S. Transformation of a high-dimensional color space for material classification. J. Opt. Soc. Am. A Opt. Image Sci. Vis. 2017, 34, 523–532. [Google Scholar] [CrossRef] [PubMed]
- Thennadil, S.; Martens, H.; Kohler, A. Physics-based multiplicative scatter correction approaches for improving the performance of calibration models. Appl. Spectrosc. 2006, 60, 315–321. [Google Scholar] [CrossRef] [PubMed]
- Grisanti, E.; Totska, M.; Huber, S.; Krick Calderon, C.; Hohmann, M.; Lingenfelser, D.; Otto, M. Dynamic Localized SNV, Peak SNV, and Partial Peak SNV: Novel Standardization Methods for Preprocessing of Spectroscopic Data Used in Predictive Modeling. J. Spectrosc. 2018, 2018, 5037572. [Google Scholar] [CrossRef]
- Zhu, H.; Gowen, A.; Feng, H.; Yu, K.; Xu, J.-L. Deep spectral-spatial features of near infrared hyperspectral images for pixel-wise classification of food products. Sensors 2020, 20, 5322. [Google Scholar] [CrossRef]
- Savitzky, A.; Golay, M.J. Smoothing and differentiation of data by simplified least squares procedures. Anal. Chem. 1964, 36, 1627–1639. [Google Scholar] [CrossRef]
- Norris, K.; Williams, P. Optimization of mathematical treatments of raw near-infrared signal in the measurement of protein in hard red spring wheat. I. Influence of particle size. Cereal Chem. 1984, 61, 158–165. [Google Scholar]
- Rinnan, Å.; Van Den Berg, F.; Engelsen, S.B. Review of the most common pre-processing techniques for near-infrared spectra. TrAC Trends Anal. Chem. 2009, 28, 1201–1222. [Google Scholar] [CrossRef]
- Steinier, J.; Termonia, Y.; Deltour, J. Smoothing and differentiation of data by simplified least square procedure. Anal. Chem. 1972, 44, 1906–1909. [Google Scholar] [CrossRef]
- Celestre, R.; Rosenberger, M.; Notni, G. A novel algorithm for bad pixel detection and correction to improve quality and stability of geometric measurements. Proc. J.Phys. Conf.Ser. 2016, 772, 012002. [Google Scholar] [CrossRef] [Green Version]
- Stellacci, A.M.; Castrignanò, A.; Diacono, M.; Troccoli, A.; Ciccarese, A.; Armenise, E.; Gallo, A.; De Vita, P.; Lonigro, A.; Mastro, M.A. Combined approach based on principal component analysis and canonical discriminant analysis for investigating hyperspectral plant response. Ital. J. Agron. 2012, 7, e34. [Google Scholar] [CrossRef] [Green Version]
- Kuhlman, D. A Python Book: Beginning Python, Advanced Python, and Python Exercises; Platypus Global Media: Washington, DC, USA, 2011. [Google Scholar]
- Koza, J.R.; Bennett, F.H.; Andre, D.; Keane, M.A. Automated design of both the topology and sizing of analog electrical circuits using genetic programming. In Artificial Intelligence in Design ’96; Gero, J.S., Sudweeks, F., Eds.; Springer: Dordrecht, The Netherlands, 1996; pp. 151–170. [Google Scholar]
- Pedregosa, F.; Varoquaux, G.; Gramfort, A.; Michel, V.; Thirion, B.; Grisel, O.; Blondel, M.; Prettenhofer, P.; Weiss, R.; Dubourg, V. Scikit-learn: Machine learning in Python. the J. Mach. Learn. Res. 2011, 12, 2825–2830. [Google Scholar]
- Dotto, A.C.; Dalmolin, R.S.D.; ten Caten, A.; Grunwald, S. A systematic study on the application of scatter-corrective and spectral-derivative preprocessing for multivariate prediction of soil organic carbon by Vis-NIR spectra. Geoderma 2018, 314, 262–274. [Google Scholar] [CrossRef]
- Kaur, R.; Kang, S.S. An enhancement in classifier support vector machine to improve plant disease detection. In Proceedings of the 2015 IEEE 3rd International Conference on MOOCs, Innovation and Technology in Education (MITE), Amritsar, India, 1–2 October 2015; pp. 135–140. [Google Scholar]
- Nguyen, C.; Sagan, V.; Maimaitiyiming, M.; Maimaitijiang, M.; Bhadra, S.; Kwasniewski, M.T. Early Detection of Plant Viral Disease Using Hyperspectral Imaging and Deep Learning. Sensors 2021, 21, 742. [Google Scholar] [CrossRef]
- Bruning, B.; Liu, H.; Brien, C.; Berger, B.; Lewis, M.; Garnett, T. The development of hyperspectral distribution maps to predict the content and distribution of nitrogen and water in wheat (Triticum aestivum). Front. Plant Sci. 2019, 10. [Google Scholar] [CrossRef]
- Maity, S.; Sarkar, S.; vinaba Tapadar, A.; Dutta, A.; Biswas, S.; Nayek, S.; Saha, P. Fault area detection in leaf diseases using k-means clustering. In Proceedings of the 2018 2nd International Conference on Trends in Electronics and Informatics (ICOEI), Tirunelveli, India, 11–12 May 2018; pp. 1538–1542. [Google Scholar]
- Jolliffe, I.T.; Cadima, J. Principal component analysis: A review and recent developments. Philos. Trans. R. Soc. A Math. Phys. Eng. Sci. 2016, 374, 20150202. [Google Scholar] [CrossRef] [PubMed]
- Severson, K.A.; Molaro, M.C.; Braatz, R.D. Principal component analysis of process datasets with missing values. Processes 2017, 5, 38. [Google Scholar] [CrossRef] [Green Version]
- Rodarmel, C.; Shan, J. Principal component analysis for hyperspectral image classification. Surv. Land Inf. Sci. 2002, 62, 115–122. [Google Scholar]
- Bock, C.H.; Barbedo, J.G.A.; Del Ponte, E.M.; Bohnenkamp, D.; Mahlein, A.-K. From visual estimates to fully automated sensor-based measurements of plant disease severity: Status and challenges for improving accuracy. Phytopathol. Res. 2020, 2, 9. [Google Scholar] [CrossRef] [Green Version]
- Dong, L.; Na, L.; Dongyan, Z.; Jinling, Z.; Fenfang, L.; Linsheng, H.; Qing, Z.; Yuwan, D. Discrimination of powdery mildew and yellow rust of winter wheat using high resolution hyperspectra and imageries. Infrared Laser Eng. 2017, 46, 50–58. [Google Scholar]
- Guo, A.T.; Huang, W.J.; Ye, H.C.; Dong, Y.Y.; Ma, H.Q.; Ren, Y.; Ruan, C. Identification of wheat yellow rust using spectral and texture features of hyperspectral images. Remote Sens. 2020, 12, 1419. [Google Scholar] [CrossRef]
- Hagerty, C.H.; Irvine, T.; Rivedal, H.M.; Yin, C.; Kroese, D.R. Diagnostic Guide: Fusarium Crown Rot of Winter Wheat. Plant Health Prog. 2021, 22, 176–181. [Google Scholar] [CrossRef]
- Knight, N.L.; Sutherland, M.W. Histopathological assessment of wheat seedling tissues infected by Fusarium pseudograminearum. Plant Pathol. 2013, 62, 679–687. [Google Scholar] [CrossRef]
- Knight, N.L.; Macdonald, B.; Sutherland, M.W. Colonization of durum wheat (Triticum turgidum L. var. durum) culms exhibiting premature senescence (dead heads) associated with Fusarium pseudograminearum crown rot. Plant Dis. 2017, 101, 1788–1794. [Google Scholar] [CrossRef] [Green Version]
- Beccari, G.; Covarelli, L.; Nicholson, P. Infection processes and soft wheat response to root rot and crown rot caused by Fusarium culmorum. Plant Pathol. 2011, 60, 671–684. [Google Scholar] [CrossRef]
- Burgess, L.; Backhouse, D.; Summerell, B.; Swan, L. Crown rot of wheat. In Fusarium: Paul E. Nelson Memorial Symposium; Burgess, L., Backhouse, D., Summerell, B., Swan, L., Eds.; APS Press: St Paul, MN, USA, 2001; pp. 271–294. [Google Scholar]
- Britannica, T.E.o.E. Phloem. Available online: https://www.britannica.com/science/phloem (accessed on 21 August 2021).
- Myburg, A.A.; Sederoff, R.R. Xylem structure and function. eLS 2001. [Google Scholar] [CrossRef]
- Liu, H.; Bruning, B.; Garnett, T.; Berger, B. The performances of hyperspectral sensors for proximal sensing of nitrogen levels in wheat. Sensors 2020, 20, 4550. [Google Scholar] [CrossRef]
- Hogg, A.; Johnston, R.; Dyer, A. Applying real-time quantitative PCR to Fusarium crown rot of wheat. Plant Dis. 2007, 91, 1021–1028. [Google Scholar] [CrossRef] [Green Version]
- Pandey, P.; Ge, Y.; Stoerger, V.; Schnable, J.C. High throughput in vivo analysis of plant leaf chemical properties using hyperspectral imaging. Front. Plant Sci. 2017, 8, 1348. [Google Scholar] [CrossRef] [Green Version]
- Wahabzada, M.; Mahlein, A.-K.; Bauckhage, C.; Steiner, U.; Oerke, E.-C.; Kersting, K. Plant phenotyping using probabilistic topic models: Uncovering the hyperspectral language of plants. Sci. Rep. 2016, 6, 1–11. [Google Scholar] [CrossRef] [Green Version]
- Nguyen, H.D.D.; Pan, V.; Pham, C.; Valdez, R.; Doan, K.; Nansen, C. Night-based hyperspectral imaging to study association of horticultural crop leaf reflectance and nutrient status. Comput. Electron. Agric. 2020, 173, 105458. [Google Scholar] [CrossRef]
- Yu, K.-Q.; Zhao, Y.-R.; Li, X.-L.; Shao, Y.-N.; Liu, F.; He, Y. Hyperspectral imaging for mapping of total nitrogen spatial distribution in pepper plant. PLoS ONE 2014, 9, e116205. [Google Scholar]
- Ansari, M.S.; Young, K.R.; Nicolas, M.E. Determining wavelenth for nitrogen and phosphorus nutrients through hyperspectral remote sensing in wheat (Triticum aestivum L.) plant. Int. J. Bio-Resour. Stress Manag. 2016, 7, 653–662. [Google Scholar]
- Barbagallo, R.P.; Oxborough, K.; Pallett, K.E.; Baker, N.R. Rapid, noninvasive screening for perturbations of metabolism and plant growth using chlorophyll fluorescence imaging. Plant Physiol 2003, 132, 485–493. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Meyer, S.; Saccardy-Adji, K.; Rizza, F.; Genty, B. Inhibition of photosynthesis by Colletotrichum lindemuthianum in bean leaves determined by chlorophyll fluorescence imaging. Plant Cell Environ. 2001, 24, 947–956. [Google Scholar] [CrossRef]
- Bauriegel, E.; Giebel, A.; Geyer, M.; Schmidt, U.; Herppich, W.B. Early detection of Fusarium infection in wheat using hyper-spectral imaging. Comput. Electron. Agric. 2011, 75, 304–312. [Google Scholar] [CrossRef]
- Nogués, S.; Cotxarrera, L.; Alegre, L.; Trillas, M.I. Limitations to photosynthesis in tomato leaves induced by Fusarium wilt. New Phytol. 2002, 461–470. [Google Scholar] [CrossRef]
- Bushnell, W.; Perkins-Veazie, P.; Russo, V.; Collins, J.; Seeland, T. Effects of deoxynivalenol on content of chloroplast pigments in barley leaf tissues. Phytopathology 2010, 100, 33–41. [Google Scholar] [CrossRef] [Green Version]
- Yan, K.; Han, G.; Ren, C.; Zhao, S.; Wu, X.; Bian, T. Fusarium solani infection depressed photosystem performance by inducing foliage wilting in apple seedlings. Front. Plant Sci. 2018, 9, 479. [Google Scholar] [CrossRef] [Green Version]
- Bauriegel, E.; Herppich, W. Hyperspectral and Chlorophyll Fluorescence Imaging for Early Detection of Plant Diseases, with Special Reference to Fusarium spec. Infections on Wheat. Agriculture 2014, 4, 1–26. [Google Scholar] [CrossRef] [Green Version]
- Dammer, K.-H.; Möller, B.; Rodemann, B.; Heppner, D. Detection of head blight (Fusarium ssp.) in winter wheat by color and multispectral image analyses. Crop Prot. 2011, 30, 420–428. [Google Scholar] [CrossRef]
- Kang, Z.; Buchenauer, H. Cytology and ultrastructure of the infection of wheat spikes by Fusarium culmorum. Mycol. Res. 2000, 104, 1083–1093. [Google Scholar] [CrossRef]
- Muhammed, H.H.; Larsolle, A. Feature vector based analysis of hyperspectral crop reflectance data for discrimination and quantification of fungal disease severity in wheat. Biosyst. Eng. 2003, 86, 125–134. [Google Scholar] [CrossRef]
- Baret, F.; Guyot, G.; Major, D. Coupled fluorescence and reflectance measurements to improve crop productivity evaluation. In Applications of Chlorophyll Fluorescence in Photosynthesis Research, Stress Physiology, Hydrobiology and Remote Sensing; Springer: Berlin/Heidelberg, Germany, 1988; pp. 319–324. [Google Scholar]
- Šrobárová, A.; Nadubinská, M.; Čiamporová, M. Relative Efficacy of Fusamotoxins on Young Maize Plants. Cereal Res. Commun. 2004, 32, 241–248. [Google Scholar] [CrossRef]
- Gunupuru, L.R.; Perochon, A.; Doohan, F.M. Deoxynivalenol resistance as a component of FHB resistance. Trop. Plant Pathol. 2017, 42, 175–183. [Google Scholar] [CrossRef] [Green Version]
- Thai, L.H.; Hai, T.S.; Thuy, N.T. Image classification using support vector machine and artificial neural network. Int. J. Inf. Technol. Comput. Sci. 2012, 4, 32–38. [Google Scholar] [CrossRef] [Green Version]
- Humpal, J.; McCarthy, C.; Percy, C.; Thomasson, J.A. Detection of crown rot in wheat utilising near-infrared spectroscopy: Towards remote and robotic sensing. In Proceedings of the SPIE—The International Society for Optical Engineering, Online, 27 April–8 May 2020. [Google Scholar]
- Wallwork, H.; Zwer, P. Cereal Variety Disease Guide 2016; SARDI: Urrbrae, Australia, 2016. [Google Scholar]
- Liu, S.; Tan, X.; Liu, C.Y.; Zhu, C.L.; Li, W.H.; Cui, S.; Du, Y.F.; Huang, D.C.; Xie, F. Recognition of Fusarium Head Blight Wheat Grain Based on Hyperspectral Data Processing Algorithm. Guang Pu Xue Yu Guang Pu Fen Xi/Spectrosc. Spectr. Anal. 2019, 39, 3540–3546. [Google Scholar] [CrossRef]
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).