Abstract
This study investigated the transformative potential of Compressive Sensing (CS) for optimizing multimodal biomedical signal fusion in Wireless Body Sensor Networks (WBSN), specifically targeting challenges in data storage, power consumption, and transmission bandwidth. Through a Systematic Mapping Study (SMS) and Systematic Literature Review (SLR) following the PRISMA protocol, significant advancements in adaptive CS algorithms and multimodal fusion have been achieved. However, this research also identified crucial gaps in computational efficiency, hardware scalability (particularly concerning the complex and often costly adaptive sensing hardware required for dynamic CS applications), and noise robustness for one-dimensional biomedical signals (e.g., ECG, EEG, PPG, and SCG). The findings strongly emphasize the potential of integrating CS with deep reinforcement learning and edge computing to develop energy-efficient, real-time healthcare monitoring systems, paving the way for future innovations in Internet of Medical Things (IoMT) applications.
1. Introduction
The rapid advancement of wireless body sensor networks (WBSN) technology has revolutionized healthcare monitoring applications, enabling the continuous, real-time acquisition of physiological data from patients [1]. One-dimensional (1D) biomedical signals, such as electrocardiograms (ECG) for heart, photoplethysmogram (PPG) for blood flow, seismocardiogram (SCG) for heart vibrations, and electroencephalograms (EEG) for brain waves, are crucial for monitoring and diagnosing health issues [2]. However, the substantial volume of biomedical signal data collected and transmitted by WBSN devices presents significant challenges in terms of data storage, power consumption, and transmission bandwidth. Efficient data compression techniques are essential to address these issues and facilitate the practical and sustainable deployment of WBSNs [2,3]. Compressive Sensing (CS) has emerged as a particularly promising approach owing to its ability to efficiently acquire and reconstruct signals using significantly fewer samples than traditional methods [4]. This offers a potential solution for reducing the load on data storage and transmission [5].
One particularly promising technique to address these data challenges is Compressive Sensing (CS). Unlike traditional Nyquist-Shannon sampling, which requires sampling at least twice the highest frequency component of a signal, CS leverages the sparsity or compressibility of a signal to acquire and reconstruct it from significantly fewer measurements. This paradigm shift means that instead of first acquiring a dense signal and then compressing it, CS performs both sensing and compression simultaneously during the acquisition phase. The fundamental principle behind CS relies on two main conditions: the signal must be sparse or compressible in some transform domain (e.g., wavelet, Fourier), and the measurement matrix used for acquisition must be incoherent with this sparsity basis. By exploiting this inherent signal structure, CS offers a powerful framework for reducing data acquisition time, storage requirements, and transmission bandwidth, making it highly suitable for resource-constrained environments like Wireless Body Sensor Networks (WBSN). Recent research on CS has demonstrated significant advancements in optimizing compression methods for biomedical signals [2]. Several studies have explored the use of a dynamic sensing matrix to enhance the compression efficiency of ECG signals in Internet of Thing (IoT) applications [6]. Furthermore, a self-adaptive compression ratio approach, utilizing Optimized Discrete Cosine Transform (ODCT) reconstruction, has been proposed for compressing physiological voice data [7]. Deep learning techniques are increasingly being integrated with CS to improve the performance of biomedical signal compression and reconstruction. An example is the development of a deep compressive sensing framework for ECG signals, which employs multiscale feature fusion, along with squeeze-and-excitation (SE) blocks and modified Inception and Long Short-Term Memory (LSTM) blocks [8,9]. Despite these developments, the improvement of CS methods that are more adaptive, efficient, and robust to noise and artifacts in one-dimensional biomedical signals remains an active area of research [10].
Fusion, or merging of multimodal biomedical signals, is a process that integrates diverse information from various modalities (different biomedical signal acquisition devices) [3]. Multimodal feature fusion helps to generate more robust and accurate predictions because information from different modalities (data types) complements each other [2,11]. This approach addresses the limitations of incomplete information obtained from a single modality, thereby strengthening the feature representation and enriching information within a single biomedical signal data stream [12]. Each medical signal has unique characteristics that reflect different aspects of the human body. For instance, heart rate variability exhibits both low- and high-frequency components, indicating parasympathetic and sympathetic nervous system activities, respectively. Integrating various signals can provide a more comprehensive and accurate representation than relying solely on a single signal, leading to richer and more complete understanding. Researchers are increasingly interested in combining different medical signals (multimodal fusion) to gain more comprehensive insights, as a single signal type often proves to be insufficient for differentiating diseases and their symptoms. The appropriate selection of unimodal biomedical signal data and the chosen multimodal fusion strategy are two crucial components of health and affective analysis systems, often outperforming unimodal health detection and emotion recognition systems [13].
Research on multimodal biomedical signal fusion is still in its early stages, yet several studies have already demonstrated its significance in medical research [14]. However, fusion algorithms and strategies require further refinement. Biomedical signal research can not only identify physiological diseases in living beings (humans and animals), but also psychological conditions, such as mental illness [15]. Emotions are complex phenomena that have a profound impact on the quality of life, influencing drive, perception, cognition, creativity, focus, attention, learning, and decision-making [16]. To observe a person’s mental state, research continues to explore the fusion of bioelectric (such as EEG) and non-bioelectric signals (such as respiratory acoustic signals or movement data) [17]. This is because unimodal approaches are often insufficiently informative and biomedical signal fusion has been proven to increase diagnostic accuracy. Beyond these medical applications, multimodal signal fusion can be applied to Body Sensor Networks (BSN) [1]. A real-time respiration pattern diagnostic system can potentially be developed by fusing sensor data from lung sounds and cardiograms [18]. BSNs represent a revolutionary technology across various domains, including healthcare, fitness, smart cities, and numerous other compelling Internet of Things (IoT) applications, enabling a single device to monitor a vast amount of user information [19]. Multimodal biomedical signal fusion generates extremely large datasets, which necessitate effective compression methods [20]. Selecting an appropriate and effective compression technique for biomedical signal data is a key aspect of the problem addressed in this study. Furthermore, the integration of deep reinforcement learning into the compression process is crucial to ensure that no vital information within biomedical signals is lost [21].
The field of compressive sensing applied to multimodal biomedical signals presents a broad and promising scope for exploration, encompassing novel multimodal signal combinations (e.g., integrating electrophysiological signals with motion data), the development of adaptive algorithms for heterogeneous and asynchronous data streams, and the establishment of robust evaluation frameworks for combined datasets. The unique combinations and characteristics of datasets gathered from diverse modalities will yield thousands of distinctive dataset variations, each requiring different treatments. This significantly increases the probability of discovering novel findings. CS methods still offer extensive potential for development, especially in optimizing data storage, computation, and transmission when combined with Deep Reinforcement Learning (DRL) [22,23,24]. This study aims to provide future research analysis and identify research gaps in the current work related to one-dimensional biomedical signal compressive sensing. This analysis was based on existing methodologies, specifically using a systematic mapping study (SMS) and systematic literature review (SLR), following the PRISMA protocol [25].
The remainder of this paper is organized as follows: Section 2 outlines the methodology employed for the systematic literature review, including the article selection process and data extraction. Section 3 presents the results of our comprehensive analysis, encompassing the systematic classification of methods and a comparative performance evaluation. Section 4 discusses the key findings, identified research trends, and existing limitations. Finally, Section 5 concludes the paper and highlights promising directions for future research.
2. Methods
This section details the approach to paper extraction and literature study, encompassing both the Systematic Mapping Study (SMS) and Systematic Literature Review (SLR) stages [26,27]. As illustrated in Figure 1, the methodology was executed in two distinct phases: SMS first, followed by SLR. A Systematic Mapping Study (SMS) serves as a quantitative method designed to provide a broad overview of a specific research area. This includes examining publication demographics, identifying key contributors, understanding research trends, pinpointing promising research topics, and analyzing topic models [28,29,30]. The insights gained from this SMS serve as crucial inputs for subsequent SLR processes. The Systematic Literature Review (SLR) involves an in-depth examination of papers identified as relevant during the SMS phase [31]. This comprehensive methods andto thoroughly understand various research methodologies and objectives related to the topic of interest, including exploring performance metrics, identifying existing issues, analyzing proposed methods, and uncovering potential gaps in previous research concerning compressive sensing and biomedical signal fusion.
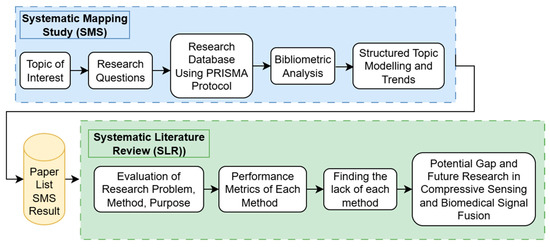
Figure 1.
Research methodology.
The entirety of this systematic literature review, from article metadata collection to data analysis and visualization, was conducted using a dedicated computational environment to ensure methodological rigor and reproducibility. Metadata for the selected publications were systematically obtained from prominent academic databases, including Scopus, ScienceDirect, and IEEE Xplore. These metadata were downloaded in various formats, with CSV files primarily utilized for subsequent processing and analysis. For bibliometric analysis and network visualization, VOSviewer (1.6.20 version) was employed. Further quantitative analyses, including Latent Dirichlet Allocation (LDA) for topic modeling and subsequent statistical analyses for trend identification, were performed in Python (3.12.7 version). Key Python libraries utilized included pandas for data handling and manipulation, gensim for LDA implementation, and scipy for statistical tests. Data visualizations (such as trend plots) were generated using matplotlib and seaborn. This computational environment facilitated the systematic and reliable processing of the extensive literature corpus.
- A.
- Systematic Mapping Study (SMS) Method
A Systematic Mapping Study (SMS) was conducted to gain an understanding of the research landscape surrounding CS and Biomedical Signal Fusion. This included identifying the latest methods or technologies from prior research and analyzing trends in topic modelling. The results of the topic model and trend analysis from the SMS process indicate that CS and Biomedical Signal Fusion constitute a promising research area.
The SMS process is illustrated in Figure 1. The initial stage involved defining the Topic of Interest and outlining the Research Questions (RQs) for the SMS. In this study, three research questions were formulated. RQ1: What is the publication population regarding CS and Biomedical Signal Fusion research between 2014 and 2025? RQ2: What areas are encompassed in this topic? RQ3: What are the trends in this topic within this subject of interest?
The subsequent stage involved paper searching, study selection, and data extraction from research databases, utilizing bibliometric analysis and adhering to the PRISMA protocol (Figure 2). Papers were searched across three prominent research databases: ScienceDirect, Scopus, and IEEE. The search query employed the keywords “Compressive Sensing Biomedical Signal” or “Compressed Sensing Biomedical Signal”. The study selection criteria included publications from 2014 to 2025, encompassing conferences, journals, books, review papers, and book chapters. The query is (title-abs-key (compressive and sensing and biomedical and signal) or title-abs-key (compressed and sensing and biomedical and signal)) and pubyear > 2013 and pubyear < 2026 and (limit-to (subjarea, “engi”) or limit-to (subjarea, “comp”) or limit-to (subjarea, “math”)) and (limit-to (doctype, “ar”) or limit-to (doctype, “cp”) or limit-to (doctype, “re”) or limit-to (doctype, “ch”) or limit-to (doctype, “cr”) or limit-to (doctype, “bk”)).
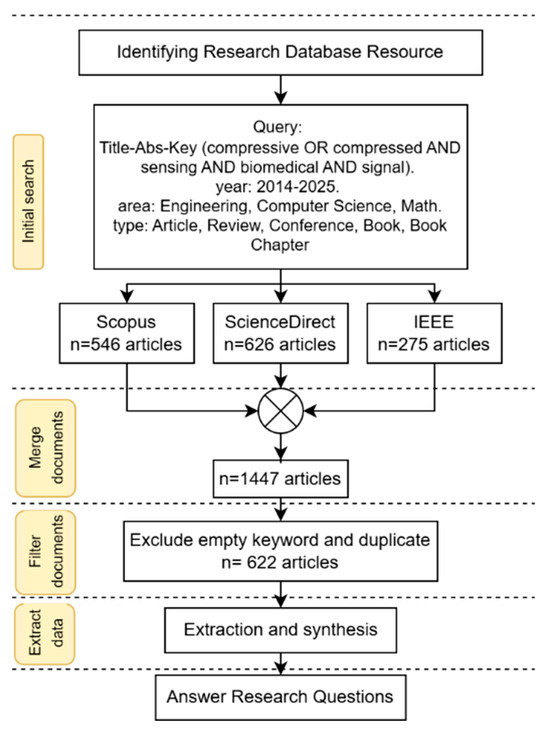
Figure 2.
PRISMA protocol.
The articles were obtained from three different sources: 546 articles from Scopus, 626 articles from ScienceDirect, and 275 articles from IEEE. Following the merging of these results, a manual screening process was performed to remove duplicate titles and entries that did not contain specified keywords. This resulted in a final dataset comprising 622 articles. The last stage involved performing structured topic modelling and trend analysis, which are explained in Section 3.
- B.
- Systematic Literature Review (SLR) Method
The subsequent phase of SMS involves conducting a Systematic Literature Review (SLR). The initial step of the SLR is to define the Research Questions (RQs), which comprise of the three questions detailed in Table 1. These RQs guide more in-depth research pertaining to CS problems in biomedical signals, CS parameter metrics, and biomedical signal fusion methods. Consequently, the RQs facilitate an evaluation of issues related to the methods and techniques employed in CS and biomedical signal fusion, ultimately identifying opportunities for research gaps based on the literature review conducted.

Table 1.
RQs for SLR.
The SLR conducts a more focused search for articles performed (from SMS) to gain a deeper understanding of CS and biomedical signal fusion. Before commencing the detailed review, articles were further explored using more specific queries, such as “CS optimization,” “Adaptive CS,” and “Biomedical signal fusion.” The articles retrieved from this targeted search formed an augmented version of the dataset, merged with the initial SMS results, and were moved to the exclusion stage. Articles were selected from both the SMS process and the augmented set based on specific exclusion criteria, including accessibility, publication type, and content suitability. The articles that remained after this rigorous selection process were subsequently analyzed to answer predefined Research Questions (RQs). The combined outcome of the SMS and SLR analyses is the State of The Art (SOTA), which comprises key articles that serve as critical references for exploring potential research gaps and future research [32].
3. Results
The findings from both the SMS and SLR are presented in relation to their respective Research Questions (RQs). The RQs guiding the SMS will inform the results regarding trends in CS within biomedical signals, along with bibliometric analyses, topic modelling, and their corresponding trends. Conversely, the RQs of the SLR have clarified issues, methodologies, and performance metrics related to CS, identified limitations and proposed solutions, and detailed various biomedical signal fusion methods.
3.1. The SMS Result of RQ1, RQ2, and RQ3: Trend, Bibliometric and Topic Modelling
For bibliometric analysis, VOSviewer was used to construct and visualize bibliometric networks. VOSviewer is a powerful tool that aids researchers in understanding the structure and dynamics of a research field [33]. It functions by analyzing and visualizing various types of bibliometric networks, including co-authorship, citation, and co-occurrence networks. The analysis of keyword (or term) co-occurrence is central to how VOSviewer identifies topical trends. In this study, metadata from various sources, namely Scopus, ScienceDirect, and IEEE, were used and stored in the CSV file format. This data contained information such as title, abstract, keywords, authors, affiliations, publication year, references, source, and document type [34]. The threshold applied was the minimum number of occurrences of a keyword, meaning that only keywords appearing at least a specified number of times were considered in the analysis. The method used for calculating co-occurrence was Full Counting. In this method, if keywords A and B appear together in one document, they receive a score of one. If they appear in 10 documents, their score is 10. This represents a direct count of the co-occurrences. For example:
- Document 1: A, B, C
- Document 2: A, B, D
- Document 3: A, C
Then:
- Co-occurrence (A, B) = 2 (because they appear in Documents 1 and 2)
- Co-occurrence (A, C) = 2 (because they appear in Documents 1 and 3)
- Co-occurrence (B, C) = 1 (because they appear only in Document 1)
VOSviewer employs normalization techniques to calculate the link strength between keywords, because some keywords may appear more frequently in general. The formula for the Link Strength between two items (e.g., keywords) i and j is:
- Sij = Link strength between item i and j.
- Cij = Number of co-occurrences of items i and j (i.e., how many times they appear together).
- Wi = Total weight (e.g., total number of occurrences) of item i.
- Wj = Total weight (e.g., total number of occurrences) of item j.
Figure 3 presents a visualization of research trend clustering derived from the metadata generated in the SMS. This analysis, powered by VOSviewer, served as a crucial initial step in our systematic literature review, aiming to map the intellectual landscape of compressive sensing in multimodal biomedical signals over the past decade and to systematically guide our article selection process. Based on the co-occurrence data and calculated link strengths, VOSviewer builds network. Network Visualization then displays nodes (representing individual keywords) and connecting lines (representing co-occurrence relationships) with different colors indicating different clusters. The clustering algorithm employed was based on modularity optimization, ensuring a robust identification of thematic groups.
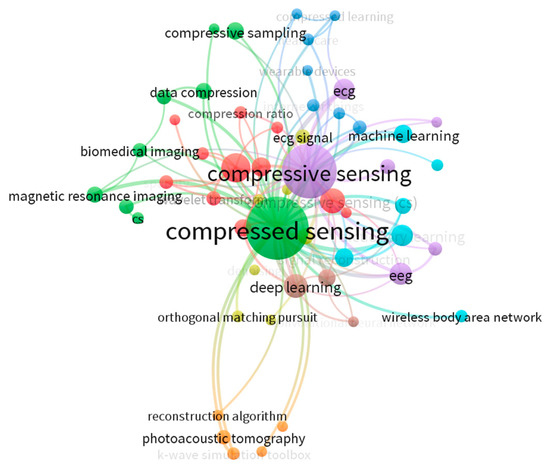
Figure 3.
VOS Viewer Analysis.
This SMS contains 1436 keywords, of which 54 meet the threshold. The minimum number of occurrences of a keyword was 5. For each of the 54 keywords, the total strength of the co-occurrences link with other keywords was calculated. The visualization clearly identifies 8 distinct clusters, each represented by a different color, signifying a specific thematic area within the research domain. In this map, the size of each node is proportional to its total number of occurrences, highlighting its overall prominence, while the thickness of the links indicates the strength of the co-occurrence relationship between connected keywords, illustrating their strong thematic connections.
As observed in Figure 3, “compressed sensing” (and its variant “compressive sensing”) emerges as the central and most prominent keyword, with the highest number of occurrences (168) and a total link strength of 195, unequivocally confirming its centrality to our review topic. The following keywords were used in this analysis: compressive sensing, deep learning, ECG, EEG, compressed sensing (cs), dictionary learning, electrocardiogram, signal reconstruction, classification, and so on. The clustering revealed several key thematic areas that define the field’s current research fronts. For instance, the purple cluster prominently features ‘deep learning’, ‘machine learning’, and ‘EEG’, indicating a strong convergence of advanced computational techniques with neurophysiological signal processing. Similarly, the red and orange clusters highlight the significant application of compressive sensing in biomedical imaging, including ‘magnetic resonance imaging’ and ‘photoacoustic tomography’. Other notable themes include specific signal applications like ‘ECG’ (light blue cluster) and fundamental ‘reconstruction algorithms’ (orange/yellow cluster). The insights gained from this VOSviewer analysis were instrumental in refining our literature selection. By visualizing the connections and thematic clusters from thousands of initial search results, we were able to validate the core relevance of our research focus, identify the most active and central sub-fields, and efficiently filter the vast corpus of literature.
In the context of analyzing author keywords from the Scopus dataset, the Structure Topic Model (STM) with Latent Dirichlet Allocation (LDA) can effectively identify research topic trends and reveal inter-topic relationships within a research publication database. As a probabilistic topic modeling method, LDA is specifically designed to uncover inherent topic structures, where each topic is characterized by a set of frequently co-occurring words grouped through an unsupervised learning approach. Critically, topic prevalence (how much a topic appears in a document) and/or topic content (words associated with a topic) can vary as a function of external covariates. Fundamentally, LDA operates under the bag-of-words assumption, treating documents as collections of word frequencies where word order is disregarded. Effective topic identification necessitates a sufficiently large corpus with diverse vocabulary, as inadequate data impedes the discovery of clear thematic patterns. Furthermore, the optimal number of topics in LDA is not automatically determined by the algorithm but rather through user experimentation. For the purpose of a systematic mapping study, STM with LDA proved to be quite powerful and capable of analyzing author keywords from a Scopus dataset to help discover patterns and topical trends [35,36]. STM models the prevalence of topics in document d as a function of document metadata Xd:
The θd (topic distribution for document d) is drawn from or distributed according to a Logistic-Normal distribution of metadata Xd to the mean of the logistic-normal distribution for θd with covariance Σ. The results of the STM with LDA for this metadata study are shown in Figure 4.
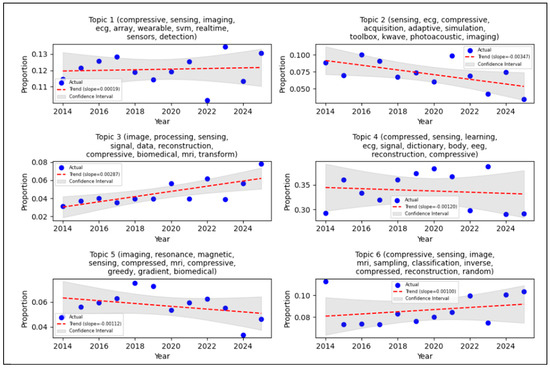
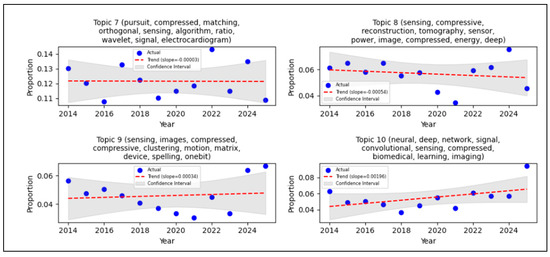
Figure 4.
STM with LDA result.
Figure 4 shows the trends of each topic based on metadata through STM with LDA analysis. These trends are visualized using a linear regression plot, applied to the annual proportion of a topic’s occurrence within the specified start-year and end-year range. Linear regression models the long-term trends of a topic’s proportion within a publication database. In the graph, the blue dots represent the actual data proportion, indicating the real-time proportion of the topic in each given year. This proportion signifies the distribution of a particular topic in the Scopus dataset. For instance, a proportion between 0.01 and 0.02 suggests that approximately 1–2% of the publications in that specific year discuss this topic. If the proportion increases annually, this implies that the topic is gaining more attention in the research database. The red dashed line denotes the trend line derived from the linear regression, illustrating whether the topic’s prevalence generally increased or decreased. The gray shaded area represents the confidence interval, which indicates the range of uncertainty for the regression model, thereby providing an estimate of the model’s confidence in the predicted trend. The interpretation of the slope (gradient) is crucial, a positive slope indicates that the proportion of publications related to a topic increase over time. A very small slope value suggests that the topic’s increase was relatively slow. Table 2 provides a comprehensive summary of the identified topics, their observed trends (trending up or down), and the explicit statistical support derived from the time-series analysis, complementing the visual representation in Figure 4.

Table 2.
Summary of Topic Trends and Statistical Validation.
The more detailed analysis of Table 2 is presented in Section 4. This system provides a relevance score for article and paper titles based on a selected topic. An example of a topic search and the resulting scores are shown in Figure 5. Based on these recommended articles, titles were located within the dataset. Subsequently, the DOI or a direct link to the reference source can be retrieved to facilitate the downloading of the papers intended for Systematic Literature Review (SLR).
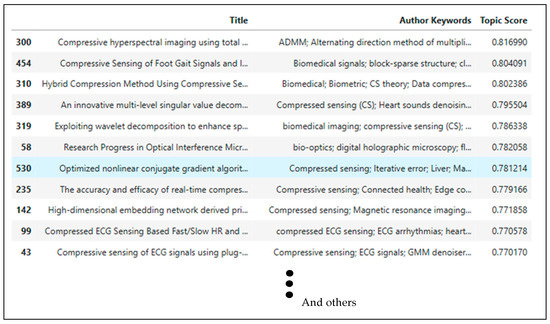
Figure 5.
Article or paper titles recommendation based on a selected topic.
The systematic approach in the algorithm identifies prominent research themes within a corpus of articles, specifically by analyzing author-provided keywords. The top five article recommendations based on author keywords and topic score calculations can be found in reference number [37,38,39,40,41]. The initial phase involves meticulous extraction of author keywords from the dataset, ensuring the exclusion of null entries. This raw textual input then underwent a series of preprocessing steps: each keyword string was uniformly converted to lowercase, all numerical characters were removed, and punctuation was eliminated. The cleaned strings were subsequently tokenized into their constituent words, which were then reassembled into a single string.
Following this preprocessing, the refined text is transformed into a document-term matrix utilizing Term Frequency-Inverse Document Frequency (TF-IDF) vectorization. This transformation assigns a numerical weight to each term, reflecting its significance in a broader dataset. For optimal performance, a maximum of 1000 unique features were retained, and common English stop words were filtered out to enhance the specificity of the extracted topics.
An LDA model was instantiated to uncover 10 distinct latent topics from the generated TF-IDF matrix. The model was then iteratively trained using vectorized textual data. To facilitate the interpretation of these discovered topics, a dedicated function extracts the top 10 most influential keywords associated with each identified topic. These keywords are precisely determined by ranking the feature names based on their respective weights within each topic’s component distribution. Ultimately, the derived topics and their corresponding salient keywords were structured into a Pandas DataFrame, providing a clear and accessible format for subsequent visualization and in-depth analysis. This method offers a robust mechanism for inferring the primary research areas directly from author-contributed metadata.
3.2. The SLR Result of RQ1: Issue, Method, and Performance Metrics of CS
In a Systematic Literature Review (SLR), the formulation of research questions is critical for guiding the analysis and synthesis of relevant studies. The first research question focuses on three key elements: issues, methods, and performance metrics to explore emerging trends in a research topic, specifically related to the provided dataset on CS and biomedical signal fusion. Table 3 addresses the key elements by analyzing the dataset and refining the research questions.

Table 3.
Issues and Methods Related to Topic.
Each methodology (in Table 3) implemented to address a specific issue is subject to performance measurement. Owing to the diverse objectives inherent to each issue, the criteria for evaluating their performance also differ. Table 4 provides a comprehensive breakdown of the performance metrics applied to each issue group, as shown in Table 3.

Table 4.
Performance Metrics Related to Method and Issue Group.
The analysis of the identified performance metrics will be elaborated upon in Section 4.
3.3. The SLR Result of RQ2: The Limitations and Potential Research of CS
The second research question focused on the limitations of related topics and how to identify their potential research. The results show in Table 5 corresponding to the issue group from RQ1.

Table 5.
Limitations and Potential Research.
This detailed analysis of the limitations and potential research directions provides a clear roadmap for future advancement in these critical areas. Addressing these identified challenges will not only refine existing methodologies but also open new frontiers for innovation in various engineering and medical applications. More analysis regarding the identified limitations and potential research of this topic will be elaborated upon in Section 4.
3.4. The SLR Result of RQ3: Methods, Limitations, and Potential Gaps in Biomedical Signal Fusion
The third research question focuses on the methods, limitations, and potential gaps in biomedical signal fusion. It is important to acknowledge its inherent limitations and to propose avenues for further exploration. These are detailed in Table 6.

Table 6.
Methods, Limitations, and Potential Gaps in Biomedical Signal Fusion.
This comprehensive analysis of the current methods, their inherent limitations, and the identified research gaps in biomedical signal fusion highlight critical areas for future investigation. Addressing these challenges is paramount for advancing the field, paving the way for more robust, user-friendly, and clinically impactful smart healthcare systems. A comprehensive comparative analysis, presented in Table 7, offers a more granular understanding of the current methods’ capabilities and limitations in addressing these critical areas. This table meticulously outlines the characteristics, advantages, and disadvantages of key Compressive Sensing techniques, specifically evaluating their performance and design considerations regarding data storage, computational complexity, and transmission efficiency.

Table 7.
Compressive Sensing Focus Areas for Optimization of Data Storage, Computation, and Transmission.
Understanding strategies for effectively integrating diverse data modalities is equally crucial, following the analysis of Compressive Sensing methods’ capabilities in optimizing individual biomedical signal streams for efficient storage, computation, and transmission (Table 7). Multimodal fusion strategies determine how information from disparate sources combines, leading to richer insights or more robust decisions. A comprehensive understanding of these critical integration paradigms is presented. Table 8 offers a detailed comparison of the primary multimodal fusion strategies for biomedical signals.

Table 8.
Comparison of Multimodal Fusion Strategies for Biomedical Signals.
This systematic comparison highlights that while significant advancements have been made in each area, trade-offs frequently exist between high compression ratios, computational efficiency, and real-time transmission capabilities. Understanding these specific characteristics, advantages, and disadvantages for each method is crucial for identifying optimal CS solutions for varied biomedical signal applications and for guiding future research towards more holistic optimization strategies.
4. Discussion
The Systematic Mapping Study (SMS) and Systematic Literature Review (SLR) conducted in this research emphasize the escalating importance of Compressive Sensing (CS) within biomedical signal processing, particularly concerning Wireless Body Sensor Networks (WBSN). A bibliometric analysis, performed using VOSviewer, revealed prominent research clusters formed around keywords such as “compressive sensing,” “deep learning,” and “ECG.” Notably, “compressed sensing” exhibited the highest occurrence (168) and a total link strength of 195, signifying a strong research emphasis on optimizing CS techniques for physiological signals, including ECG and EEG. Furthermore, trend analysis, visually represented through Structural Topic Modelling (STM) with Latent Dirichlet Allocation (LDA), showed a positive slope in topic prevalence. This indicates a consistent surge of interest in CS applications for biomedical signals throughout the 2014 to 2025 period. This observed trend highlights CS’s substantial potential to mitigate critical challenges in data compression, storage, and transmission. These are the principal factors for the development of energy-efficient and real-time healthcare monitoring systems. Figure 4 describes the STM with LDA result for topic 4 as the highest proportion of articles (approximately 35%) with author keyword combinations: compressed, sensing, learning, ECG, signal, dictionary, body, EEG, reconstruction, and compressive. Topic 9 showed the lowest proportion of articles whit stagnant trends from 2014 till 2025. Topic 3 showed the highest trend, whit a slope of 0.00287, but its proportion is still low. The negative slopes indicate that the proportion available for research or publication consequently decreases, these are topics 2, 4, 5, 7, and 8. Confidence interval estimates of the model’s confidence in the predicted trend, and topic 3 had a good confidence interval for this study. This means that the number of articles (with their author keywords) has consistently increased each year.
A significant challenge identified in the SLR is the inherent trade-off between compression ratio and reconstruction accuracy, which is particularly critical for resource-constrained devices in Internet of Medical Things (IoMT) applications. For example, while approaches, such as dynamic CS for multi-lead ECG have demonstrated compression ratios of up to 16 without degrading signal metrics, their high computational complexity poses a bottleneck for integration into wearable devices. Furthermore, the incorporation of adaptive CS techniques, such as the Compressive Adaptive Sense and Search (CASS) algorithm, can achieve near-optimal performance at lower signal-to-noise ratios (SNR).
However, the dependence on costly adaptive sensing hardware restricts scalability. This “adaptive sensing hardware” typically refers to specialized analog-to-digital converters (ADCs) or front-end circuitry that can dynamically adjust measurement parameters (e.g., sampling rate, measurement matrix configuration) based on real-time signal characteristics or changing environmental conditions. Such dynamic configurability, while offering performance benefits, often leads to increased design complexity, power consumption, and manufacturing costs compared to fixed-rate, non-adaptive hardware. Consequently, future research should prioritize the development of low-complexity algorithms that effectively balance reconstruction accuracy with computational efficiency. This can potentially be achieved by leveraging hardware-accelerated implementations to minimize power consumption in WBSN deployments. The SLR results also emphasize the potential of multimodal signal fusion for improving diagnostic accuracy in healthcare applications. For instance, techniques such as spatiotemporal ECG and PPG feature fusion, augmented by the Choquet integral, have demonstrated high classification accuracy (e.g., 99.49% in Zone A + B for blood glucose monitoring), consistently outperforming approaches based on single-modality data. The practical implementation of such systems is challenged by factors including the inherent complexity of EEG equipment and its sensitivity to non-physiological factors, such as motion artifacts. Addressing these limitations necessitates the development of user-friendly, low-power EEG sensors and the creation of robust feature engineering techniques capable of mitigating non-physiological noise. These advancements have significantly enhanced the scalability of multimodal fusion systems. Furthermore, exploring edge-computing frameworks to manage data synchronization and heterogeneity within IoMT environments could further optimize real-time performance, thereby overcoming the limitations of current fusion methodologies.
The integration of deep learning with CS, exemplified by methods such as deep CS with multiscale feature fusion and rU-Net architectures, holds significant promise for improving the reconstruction performance even at low sampling rates. However, substantial barriers persist, notably high computational demands and limited generalizability across diverse data sets. These issues can be mitigated through the application of transfer and federated learning, as suggested in Table 6. Such approaches would reduce computational overhead and enable model adaptation to various physiological signal types. Furthermore, automating parameter optimization using techniques such as Deep Reinforcement Learning (DRL) can enhance the adaptability of CS frameworks, making them more suitable for dynamic Internet of Medical Things (IoMT) scenarios. These advancements will facilitate the deployment of CS-based systems in resource-constrained environments, thereby paving the way for next-generation healthcare monitoring solutions.
Building upon the identified limitations and research gaps, several critical open questions emerge, guiding future investigations in Compressive Sensing (CS) for multimodal biomedical signal processing within WBSN and IoMT contexts. These questions aim to advance the field towards more robust, efficient, and user-centric healthcare monitoring systems.
For CS optimization in resource-constrained devices, future research must develop adaptive compression algorithms that optimally balance compression ratio and reconstruction accuracy for diverse 1D biomedical signals from wearables. This includes designing novels, low-complexity, hardware-accelerated CS reconstruction algorithms to minimize computational overhead and power consumption in real-time WBSN deployments and minimizing signal distortion in lossy CS compression while maintaining high ratios for IoMT. Additionally, exploring alternative validation techniques beyond traditional neural networks is crucial to assessing the clinical utility of CS-reconstructed biomedical signals across diverse patient populations.
Regarding adaptive sensing hardware and noise robustness, a key challenge lies in designing cost-effective adaptive sensing hardware architectures that offer dynamic measurement schemes without significantly increasing complexity or power draw for continuous monitoring. Validating adaptive CS algorithms for robust performance under diverse and realistic noise models (e.g., motion artifacts, environmental noise) in real-world acquisition is essential. Furthermore, simplifying mathematical formulations for adaptive CS is needed to bridge theoretical performance gains with practical benefits in constrained clinical settings.
In the domain of multimodal signal fusion in IoMT, developing user-friendly, low-power, and scalable EEG sensors is necessary to overcome the complexity and discomfort of conventional high-electrode systems for large-scale data acquisition. Research should also focus on advanced feature engineering and model architectures to enhance the robustness of fusion models to non-physiological factors and ensure generalizability. Integrating explainable AI (XAI) and interpretable machine learning approaches into multimodal fusion systems is vital to enhance reliability and trustworthiness of diagnostic predictions. Finally, identifying effective edge-computing frameworks and strategies is crucial for addressing data synchronization, heterogeneity, and transmission constraints for real-time fusion within distributed IoMT environments.
Pertaining to deep learning integration with CS, future work should leverage transfer and federated learning to reduce high computational, and memory demands of deep CS frameworks, enabling deployment on resource-constrained WBSN nodes and improving generalizability across varied signal types. Developing novel deep learning frameworks that automate parameter optimization in CS algorithms will enhance their adaptability to dynamic IoMT scenarios. Lastly, improving interpretability in deep learning models integrated with CS for biomedical signal analysis is critical for clinical acceptance and trust. Addressing these questions will be pivotal in unlocking the full transformative potential of CS combined with multimodal fusion and deep learning, paving the way for the next-generation, ubiquitous, and clinically impactful healthcare monitoring solutions.
5. Conclusions
The findings from the SMS and SLR collectively emphasize the transformative potential of CS in biomedical signal processing, especially when integrated with deep learning and multimodal fusion techniques. Despite these advancements, significant research gaps remain in literature. These include the necessity for cost-effective adaptive sensing hardware, the development of simplified algorithms suitable for resource-constrained environments, and the crucial need for robust validation across diverse IoMT scenarios. Addressing these identified gaps by combining expertise in signal processing, machine learning, and hardware optimization will be critical for fully realizing CS’s potential in revolutionizing WBSN-based healthcare systems.
Author Contributions
Conceptualization: A.L.P. and B.E.; methodology: A.L.P. and B.E.; validation: A.L.P.; formal analysis: B.E.; investigation: A.R.; data curation: A.L.P.; writing—original draft preparation: A.L.P.; writing—review and editing: B.E.; visualization: S.Z.; supervision: A.R.; project administration: S.Z.; funding acquisition: A.R. All authors have read and agreed to the published version of the manuscript.
Funding
This research received funding from Telkom Foundation, which was channeled through Telkom University.
Data Availability Statement
Supporting data related to this work are available upon request. Please contact Anggunmeka (anggunmeka@telkomuniversity.ac.id) for access.
Acknowledgments
This research was generously supported by Telkom Foundation and Telkom University. The authors extend their sincere gratitude for the institutional backing that made this work possible. It is important to note that the opinions, findings, conclusions, and recommendations presented in this material are solely those of the authors and do not necessarily reflect the official views or policies of the supporting institution.
Conflicts of Interest
The authors declare that they have no competing interests.
Abbreviations
The following abbreviations are used in this manuscript:
| CS | Compressive Sensing or Compressed Sensing |
| SMS | Systematic Mapping Study |
| SLR | Systematic Literature Review |
| DLR | Deep Reinforcement Learning |
| EEG | Electroencephalogram |
| ECG | Electrocardiogram |
| PPG | Photoplethysmogram |
| SCG | Seismocardiogram |
| SNR | Signal-to-noise-ratios |
| CASS | Compressive Adaptive Sense and Search |
| KNN | K-Nearest Neighbors |
| DT | Decision Tree |
| SVM | Support Vector Machine |
| WVSN | Wireless Vehicle Sensor Network |
| WBSN | Wireless Body Sensor Network |
| IoMT | Internet of Medical Things |
| BSBL | Block Sparse Bayesian Learning |
| OMP | Orthogonal Matching Pursuit |
| SOMP | Simultaneous Orthogonal Matching Pursuit |
References
- Gravina, R.; Alinia, P.; Ghasemzadeh, H.; Fortino, G. Multi-Sensor Fusion in Body Sensor Networks: State-of-the-Art and Research Challenges. Inf. Fusion 2017, 35, 68–80. [Google Scholar] [CrossRef]
- Li, J.; Wang, Q. Multi-Modal Bioelectrical Signal Fusion Analysis Based on Different Acquisition Devices and Scene Settings: Overview, Challenges, and Novel Orientation. Inf. Fusion 2022, 79, 229–247. [Google Scholar] [CrossRef]
- Muhammad, G.; Alshehri, F.; Karray, F.; El Saddik, A.; Alsulaiman, M.; Falk, T.H. A Comprehensive Survey on Multimodal Medical Signals Fusion for Smart Healthcare Systems. Inf. Fusion 2021, 76, 355–375. [Google Scholar] [CrossRef]
- Gurve, D.; Delisle-Rodriguez, D.; Bastos-Filho, T.; Krishnan, S. Trends in Compressive Sensing for EEG Signal Processing Applications. Sensors 2020, 20, 3703. [Google Scholar] [CrossRef] [PubMed]
- Izadi, V.; Shahri, P.K.; Ahani, H. A Compressed-Sensing-Based Compressor for ECG. Biomed. Eng. Lett. 2020, 10, 299–307. [Google Scholar] [CrossRef]
- Hassan, A.M.A.; Mohsen, S.; Abo-Zahhad, M.M. ECG Signals Compression Using Dynamic Compressive Sensing Technique toward IoT Applications. Multimed. Tools Appl. 2024, 83, 35709–35726. [Google Scholar] [CrossRef]
- Chen, C.; Pan, R.; Huang, H.; Zhang, Q.; Jiang, X.; Zhang, Y.; Zhao, J.; Li, Y. PSCS: A Physiological Sound Compression System Based on Compressive Sensing with Self-Adaptive Compression Ratio and Optimized DCT. In Proceedings of the 2024 IEEE International Symposium on Circuits and Systems (ISCAS), Singapore, 19–22 May 2024; pp. 1–5. [Google Scholar] [CrossRef]
- Hua, J.; Zou, J.; Rao, J.; Yin, H.; Chen, J. ECG Signals Deep Compressive Sensing Framework Based on Multiscale Feature Fusion and SE Block. IEEE Access 2023, 11, 104359–104372. [Google Scholar] [CrossRef]
- Hua, J.; Rao, J.; Peng, Y.; Liu, J.; Tang, J. Deep Compressive Sensing on ECG Signals with Modified Inception Block and LSTM. Entropy 2022, 24, 1024. [Google Scholar] [CrossRef] [PubMed]
- Zhu, Y.; Liu, W.; Shen, Q. Adaptive Algorithm on Block-Compressive Sensing and Noisy Data Estimation. Electronics 2019, 8, 753. [Google Scholar] [CrossRef]
- Wang, Y.; Du, L.; Tang, G.; Ling, S. A Biometric Identification for Multi-Modal Biomedical Signals in Geriatric Care. Sensors 2024, 24, 6558. [Google Scholar] [CrossRef]
- Wang, S.; Celebi, M.E.; Zhang, Y.-D.; Yu, X.; Lu, S.; Yao, X.; Zhou, Q.; Martinez-Garcia, M.; Tian, Y.; Gorriz, J.M.; et al. Advances in Data Preprocessing for Biomedical Data Fusion: An Overview of the Methods, Challenges, and Prospects. Inf. Fusion 2021, 76, 376–421. [Google Scholar] [CrossRef]
- Duan, J.; Xiong, J.; Li, Y.; Ding, W. Deep Learning Based Multimodal Biomedical Data Fusion: An Overview and Comparative Review. Inf. Fusion 2024, 112, 102536. [Google Scholar] [CrossRef]
- Azam, K.S.F.; Ryabchykov, O.; Bocklitz, T. A Review on Data Fusion of Multidimensional Medical and Biomedical Data. Molecules 2022, 27, 7448. [Google Scholar] [CrossRef]
- Siirtola, P.; Tamminen, S.; Chandra, G.; Ihalapathirana, A.; Röning, J. Predicting Emotion with Biosignals: A Comparison of Classification and Regression Models for Estimating Valence and Arousal Level. Sensors 2022, 22, 1598. [Google Scholar] [CrossRef]
- Wang, Y.; Song, W.; Tao, W.; Liotta, A.; Yang, D.; Li, X.; Gao, S.; Sun, Y.; Ge, W.; Zhang, W.; et al. A Systematic Review on Affective Computing: Emotion Models, Databases, and Recent Advances. Inf. Fusion 2022, 83–84, 19–52. [Google Scholar] [CrossRef]
- Khan, H.; Sharma, T.; Soni, A.; Choudhary, M.; Islam, S. Analysis of Mental Health Based on the Dreams by Analyzing the Biomedical Signals of the Body. NeuroQuantology 2022, 20, 4204. [Google Scholar]
- Moon, K.S.; Lee, S.Q. A Wearable Multimodal Wireless Sensing System for Respiratory Monitoring and Analysis. Sensors 2023, 23, 6790. [Google Scholar] [CrossRef] [PubMed]
- Antaki, B.; Dalloul, A.H.; Miramirkhani, F. Intelligent Health Monitoring in 6G Networks: Machine Learning-Enhanced VLC-Based Medical Body Sensor Networks. Sensors 2025, 25, 3280. [Google Scholar] [CrossRef]
- Ancans, A.; Greitans, M.; Kagis, S. An Efficient Communication Protocol for Real-Time Body Sensor Data Acquisition and Feedback in Interactive Wearable Systems. J. Sens. Actuator Netw. 2025, 14, 4. [Google Scholar] [CrossRef]
- Matsuo, Y.; LeCun, Y.; Sahani, M.; Precup, D.; Silver, D.; Sugiyama, M.; Uchibe, E.; Morimoto, J. Deep Learning, Reinforcement Learning, and World Models. Neural Netw. 2022, 152, 267–275. [Google Scholar] [CrossRef]
- Chen, J.; Jia, J.; Deng, Y.; Wang, X.; Aghvami, A.A. Adaptive Compressive Sensing and Data Recovery for Periodical Monitoring Wireless Sensor Networks. Sensors 2018, 18, 3369. [Google Scholar] [CrossRef]
- Zeng, H.; Yu, Y.; Liu, G.; Wu, Y. A Robust Method Based on Deep Learning for Compressive Spectrum Sensing. Sensors 2025, 25, 2187. [Google Scholar] [CrossRef]
- Kumar, S.S.; Ramachandran, P. Review on Compressive Sensing Algorithms for ECG Signal for IoT Based Deep Learning Framework. Appl. Sci. 2022, 12, 8368. [Google Scholar] [CrossRef]
- Tedja, B.; Al Musadieq, M.; Kusumawati, A. Systematic Literature Review Using PRISMA: Exploring the Influence of Service Quality and Perceived Value on Satisfaction and Intention to Continue Relationship. Future Bus. J. 2024, 10, 39. [Google Scholar] [CrossRef]
- Hapsari, G.I.; Munadi, R.; Erfianto, B.; Irawati, I.D. Future Research and Trends in Ultra-Wideband Indoor Tag Localization. IEEE Access 2025, 13, 21827–21836. [Google Scholar] [CrossRef]
- Wang, J.; Zakaria, S.A. Design Application and Evolution of 3D Visualization Technology in Architectural Heritage Conservation: A CiteSpace-Based Knowledge Mapping and Systematic Review (2005–2024). Buildings 2025, 15, 1854. [Google Scholar] [CrossRef]
- Salama, M.; Bahsoon, R.; Bencomo, N. Managing Trade-offs in Self-Adaptive Software Architectures: A Systematic Mapping Study. In Managing Trade-Offs in Adaptable Software Architectures; Mistrik, I., Ali, N., Kazman, R., Grundy, J., Schmerl, B., Eds.; Morgan Kaufmann: Cambridge, MA, USA, 2017; pp. 249–297. [Google Scholar] [CrossRef]
- Werneck, H.; Silva, N.; Viana, M.; Pereira, A.C.M.; Mourão, F.; Rocha, L. Points of Interest Recommendations: Methods, Evaluation, and Future Directions. Inf. Syst. 2021, 101, 101789. [Google Scholar] [CrossRef]
- Hosseini, M.; Shahri, A.; Phalp, K.; Taylor, J.; Ali, R. Crowdsourcing: A Taxonomy and Systematic Mapping Study. Comput. Sci. Rev. 2015, 17, 43–69. [Google Scholar] [CrossRef]
- Carrera-Rivera, A.; Ochoa, W.; Larrinaga, F.; Lasa, G. How-to Conduct a Systematic Literature Review: A Quick Guide for Computer Science Research. MethodsX 2022, 9, 101895. [Google Scholar] [CrossRef] [PubMed]
- Machchhar, R.J.; Toller, C.N.K.; Bertoni, A.; Bertoni, M. Data-Driven Value Creation in Smart Product-Service System Design: State-of-the-Art and Research Directions. Comput. Ind. 2022, 137, 103606. [Google Scholar] [CrossRef]
- van Eck, N.J.; Waltman, L. Software Survey: VOSviewer, a Computer Program for Bibliometric Mapping. Scientometrics 2010, 84, 523–538. [Google Scholar] [CrossRef]
- Donthu, N.; Kumar, S.; Mukherjee, D.; Pandey, N.; Lim, W.M. How to Conduct a Bibliometric Analysis: An Overview and Guidelines. J. Bus. Res. 2021, 133, 285–296. [Google Scholar] [CrossRef]
- He, L.; Han, D.; Zhou, X.; Qu, Z. The Voice of Drug Consumers: Online Textual Review Analysis Using Structural Topic Model. Int. J. Environ. Res. Public Health 2020, 17, 3648. [Google Scholar] [CrossRef]
- Yin, R.; Tian, R.; Wu, J.; Gan, F. Exploring the Factors Associated with Mental Health Attitude in China: A Structural Topic Modeling Approach. Int. J. Environ. Res. Public Health 2022, 19, 12579. [Google Scholar] [CrossRef]
- Lee, D.J.; Shields, E.A. Compressive Hyperspectral Imaging Using Total Variation Minimization. In Proceedings of the SPIE International Society for Optical Engineering, San Diego, CA, USA, 19–23 August 2018; Volume 10768, p. 1076804. [Google Scholar] [CrossRef]
- Pant, J.K.; Krishnan, S. Compressive Sensing of Foot Gait Signals and Its Application for the Estimation of Clinically Relevant Time Series. IEEE Trans. Biomed. Eng. 2016, 63, 1401–1415. [Google Scholar] [CrossRef]
- Thanki, R.; Dwivedi, V.; Borisagar, K. Hybrid Compression Method Using Compressive Sensing (CS) Theory for Various Biometric Data and Biomedical Data. Adv. Intell. Syst. Comput. 2018, 671, 1–13. [Google Scholar] [CrossRef]
- Zheng, Y.; Guo, X.; Jiang, H.; Zhou, B. An Innovative Multi-Level Singular Value Decomposition and Compressed Sensing Based Framework for Noise Removal from Heart Sounds. Biomed. Signal Process. Control 2017, 38, 34–43. [Google Scholar] [CrossRef]
- Ambrosanio, M.; Kosmas, P.; Pascazio, V. Exploiting Wavelet Decomposition to Enhance Sparse Recovery in Microwave Imaging. In Proceedings of the 2017 11th European Conference on Antennas and Propagation (EUCAP), Paris, France, 19–24 March 2017; pp. 1607–1610. [Google Scholar] [CrossRef]
- Chen, Y.; Chen, J.; Sun, S.; Su, J.; Li, Q.; Lyu, Z. Atrial fibrillation detection from compressed ecg measurements for wireless body sensor network. ACM Trans. Internet Technol. 2024. [Google Scholar] [CrossRef]
- Craven, D.; McGinley, B.; Kilmartin, L.; Glavin, M.; Jones, E. Compressed sensing for bioelectric signals: A review. IEEE J. Biomed. Health Inform. 2015, 19, 529–540. [Google Scholar] [CrossRef] [PubMed]
- Emara, H.M.; El-Shafai, W.; Algarni, A.D.; Soliman, N.F.; Abd El-Samie, F.E. A hybrid compressive sensing and classification approach for dynamic storage management of vital biomedical signals. IEEE Access 2023, 11, 10823–10836. [Google Scholar] [CrossRef]
- Chen, B.; Zhang, X.; Liu, S.; Zhang, Y.; Zhang, J. Self-supervised scalable deep compressed sensing. Int. J. Comput. Vis. 2025, 133, 688–723. [Google Scholar] [CrossRef]
- Polanía, L.F.; Plaza, R.I. Compressed sensing ECG using restricted Boltzmann machines. Biomed. Signal Process. Control 2018, 45, 237–245. [Google Scholar] [CrossRef]
- Bertsimas, D.; Johnson, N.A.G. Compressed sensing: A discrete optimization approach. Mach. Learn. 2024, 113, 6725–6764. [Google Scholar] [CrossRef]
- Ndaoud, M.; Tsybakov, A.B. Optimal variable selection and adaptive noisy compressed sensing. IEEE Trans. Inf. Theory 2020, 66, 2517–2532. [Google Scholar] [CrossRef]
- Nakos, V.; Shi, X.; Woodruff, D.P.; Zhang, H. Improved Algorithms for Adaptive Compressed Sensing. arXiv 2018, arXiv:1804.09673. [Google Scholar] [CrossRef]
- Hadi, M.A.; Alshebeili, S.; Jamil, K.; El-Samie, F.E.A. Compressive sensing applied to radar systems: An overview. Signal Image Video Process. 2015, 9, 25–39. [Google Scholar] [CrossRef]
- Hao, J.; Zhang, B.; Jiao, Z.; Mao, S. Adaptive compressive sensing based sample scheduling mechanism for wireless sensor networks. Pervasive Mob. Comput. 2015, 22, 113–125. [Google Scholar] [CrossRef]
- Wang, J.; Wang, W.; Chen, J. Adaptive rate block compressive sensing based on statistical characteristics estimation. IEEE Trans. Image Process. 2022, 31, 734–747. [Google Scholar] [CrossRef]
- Fayed, S.; Youssef, S.M.; El-Helw, A.; Patwary, M.; Moniri, M. Adaptive compressive sensing for target tracking within wireless visual sensor networks-based surveillance applications. Multimedia Tools Appl. 2016, 75, 6347–6371. [Google Scholar] [CrossRef]
- Davenport, M.A.; Massimino, A.K.; Needell, D.; Woolf, T. Constrained adaptive sensing. IEEE Trans. Signal Process. 2016, 64, 5437–5449. [Google Scholar] [CrossRef]
- Wang, Y.; Li, X.; Xu, K.; Ren, F.; Yu, H. Data-driven sampling matrix Boolean optimization for energy-efficient biomedical signal acquisition by compressive sensing. IEEE Trans. Biomed. Circuits Syst. 2017, 11, 255–265. [Google Scholar] [CrossRef]
- Cai, H.; Qu, Z.; Li, Z.; Zhang, Y.; Hu, X.; Hu, B. Feature-level fusion approaches based on multimodal EEG data for depression recognition. Inf. Fusion 2020, 59, 127–138. [Google Scholar] [CrossRef]
- Li, J.; Ma, J.; Omisore, O.M.; Liu, Y.; Tang, H.; Ao, P.; Yan, Y.; Wang, L.; Nie, Z. Noninvasive blood glucose monitoring using spatiotemporal ECG and PPG feature fusion and weight-based Choquet integral multimodel approach. IEEE Trans. Neural Netw. Learn. Syst. 2024, 35, 3521–3535. [Google Scholar] [CrossRef]
- Chen, J.; Zhou, X.; Feng, L.; Ling, B.W.-K.; Han, L.; Zhang, H. rU-Net, multi-scale feature fusion and transfer learning: Unlocking the potential of cuffless blood pressure monitoring with PPG and ECG. IEEE J. Biomed. Health Inform. 2025, 29, 346–357. [Google Scholar] [CrossRef]
- Ming, Y.; Wu, D.; Wang, Y.-K.; Shi, Y.; Lin, C.-T. EEG-based drowsiness estimation for driving safety using deep Q-learning. IEEE Trans. Emerg. Top. Comput. Intell. 2021, 5, 505–516. [Google Scholar]
- Machidon, A.L.; Pejović, V. Deep learning for compressive sensing: A ubiquitous systems perspective. Artif. Intell. Rev. 2023, 56, 3619–3658. [Google Scholar] [CrossRef]
- Yamaç, M.; Akpinar, U.; Sahin, E.; Kiranyaz, S.; Gabbouj, M. Generalized tensor summation compressive sensing network (GTSNET): An easy to learn compressive sensing operation. IEEE Trans. Image Process. 2023, 32, 5637–5652. [Google Scholar] [CrossRef] [PubMed]
- Marmenone, N.; Moya, P.; Ledesma, A.; Pérez, J.; Fornés, S.; Iribarren, O. Compressive Sensing-Based Brain Network Characterization from EEG Signals for Alzheimer’s Disease and Mild Cognitive Impairment. Sensors 2023, 23, 2185. [Google Scholar] [CrossRef]
- Moeabite, F.; Kachenoura, A.; Gougeon, M.; Attal, Y.; Naccache, L.; Naccache, P.; Pélégrini-Issac, M. Assessing the Compressibility of EEG Signals to Discriminate Brain States. Entropy 2021, 23, 1374. [Google Scholar] [CrossRef]
- Kalliannan, B.K.; Ganesan, N.; Arjunan, T. Low-power hardware implementation for multi-channel EEG acquisition using distributed compressive sensing. Biomed. Signal Process. Control 2023, 85, 104928. [Google Scholar] [CrossRef]
- Lee, H.; Lee, H.; Kim, Y. A Novel Automatic Sleep Stage Classification Based on Single-Channel EEG Using Compressive Sensing and Sparse Representation. Sensors 2021, 21, 5236. [Google Scholar] [CrossRef]
- Abahusain, K.O.; Mohammed, Y.; Almashary, M.; Al-Sharif, M.A.; Hamza, B.A. Optimized Compressive Sensing for Wireless Epilepsy Seizure Detection with Reduced Energy Consumption in Wearable EEG Devices. Sensors 2023, 23, 7306. [Google Scholar]
- Sheng, J.; Xiao, Z.; Zhang, X.; Yan, H. Optimized Measurement Matrix Design for Compressed Sensing-Based Wireless Body Area Networks. Sensors 2020, 20, 2397. [Google Scholar] [CrossRef]
- AlZubi, A.A.; Al-Qurran, Z.M.; Alqudah, A.M.; Althunibat, T.A.; Almobaideen, W.M. Machine Learning Techniques for Emotion Detection Using Physiological Signals. IEEE Trans. Biomed. Eng. 2018, 65, 2450–2463. [Google Scholar]
- Lee, G.; Kim, S.; Lee, K. Wearable Heart Rate Monitoring Using ECG and PPG Signals. Sensors 2019, 19, 2339. [Google Scholar] [CrossRef]
- Zhang, Y.; Ma, Y.; Liu, X.; Fan, X.; Ren, Y.; Yuan, T. Multimodal Feature Fusion for Sleep Apnea Detection Using EEG and ECG. J. Med. Syst. 2020, 44, 148. [Google Scholar]
- Wang, Z.; Deng, Y.; Gu, D.; Wang, Z. HOG-based feature extraction for medical image analysis. Med. Image Anal. 2017, 36, 12–22. [Google Scholar]
- Chen, J.; Li, S.; Wang, Y.; Li, Y.; Liu, Z. Late Fusion of EEG and EMG for Sleep Disorder Detection. In Proceedings of the 2019 IEEE International Conference on Bioinformatics and Biomedicine (BIBM), San Diego, CA, USA, 18–21 November 2019; pp. 1968–1973. [Google Scholar] [CrossRef]
- Penzel, T.; Kesper, K.; Schrading, L.; Jerrentrup, A.; Becker, H.F.; Glos, M. Multimodal biosignal analysis for respiratory monitoring. Physiol. Meas. 2020, 41, 05TR01. [Google Scholar]
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).