Improving Foraminifera Classification Using Convolutional Neural Networks with Ensemble Learning
Abstract
1. Introduction
Related Work
2. Materials and Methods
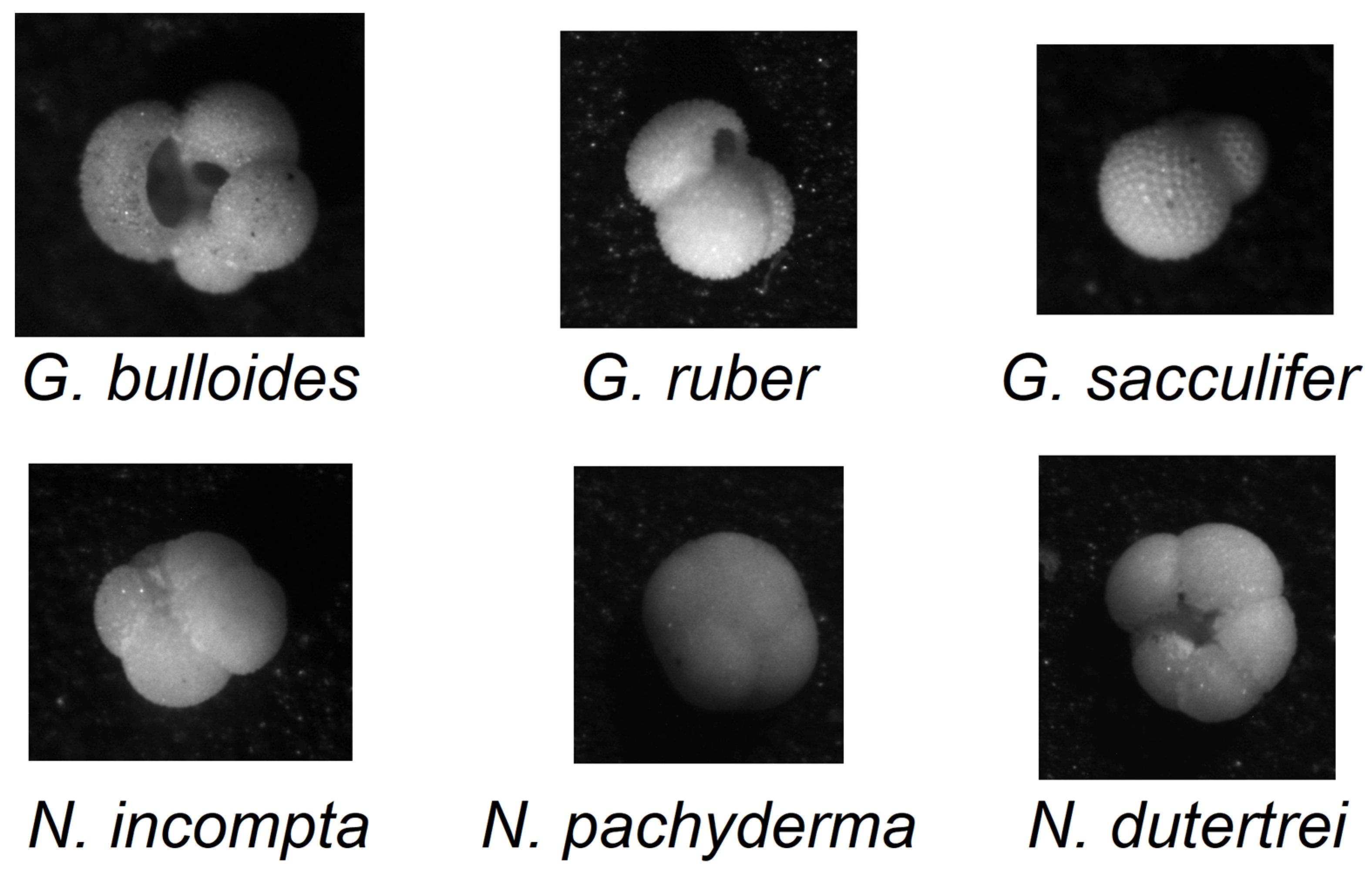
- 178 images are G. bulloides;
- 182 images are G. ruber;
- 150 images are G. sacculifer;
- 174 images are N. incompta;
- 152 images are N. pachyderma;
- 151 images are N. dutertrei;
- 450 images are “rest of the world”, i.e., they belong to other species of planktic foraminifera.
2.1. CNN Ensemble Learning
- Data level: by splitting the dataset into different subsets;
- Feature level: by pre-processing the dataset with different methods;
- Classifier level: by training different classifiers on the same dataset;
- Decision level: by combining the decisions of multiple models.
2.2. Image Pre-Processing
- A “Gaussian” image processing method that encodes each color channel based on the normal distribution of the grayscale intensities of the sixteen images;
- Two “mean-based” methods focused on utilizing an average or mean of the sixteen images to reconstruct the R, G, and B values;
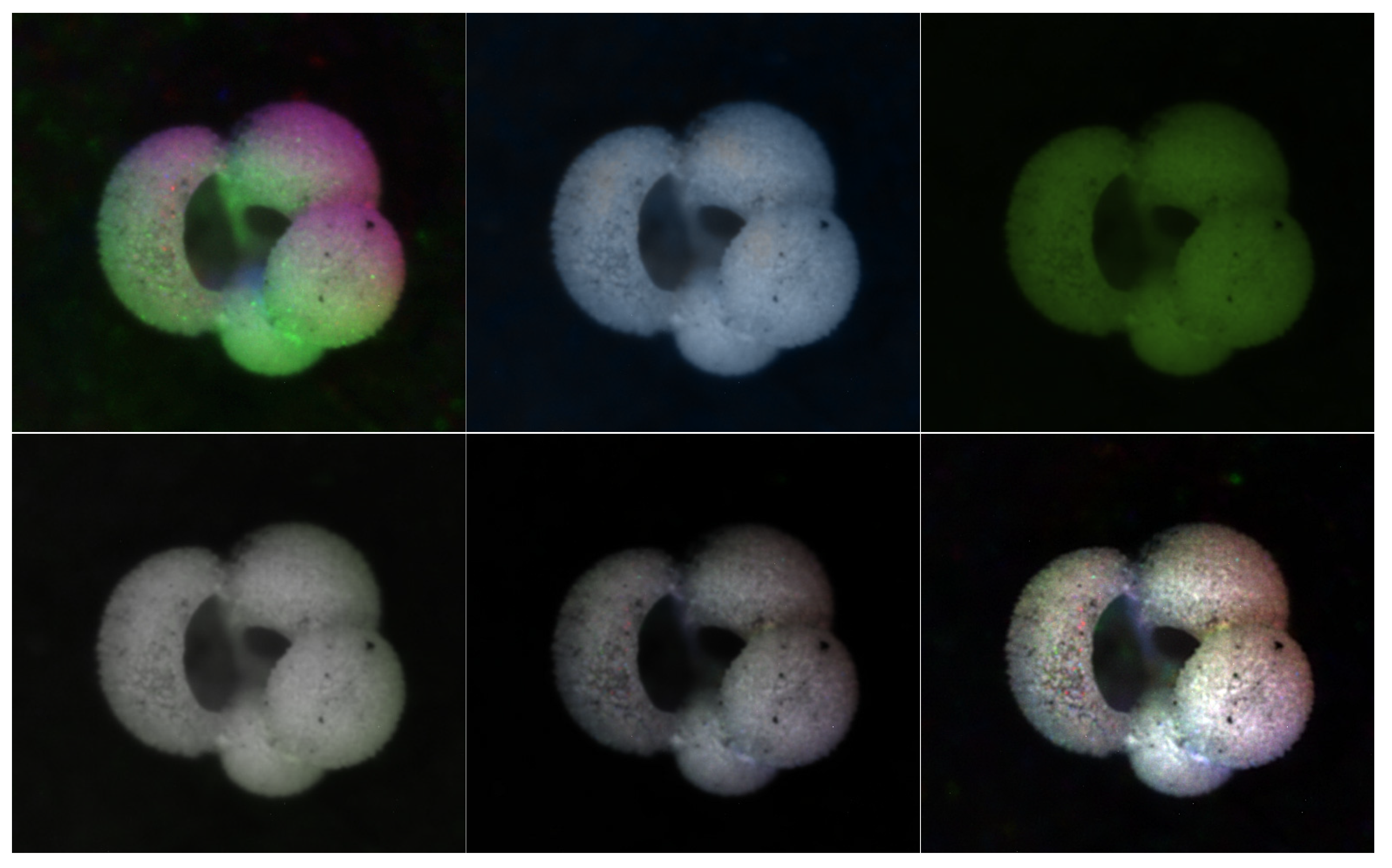
- The “HSVPP” method, that utilizes a different color space composed of hue, saturation, and value of brightness information, see Figure 4;
- The “GraySet” method, which takes each of the 16 grayscale images for every pattern, and creates 16 RGB images by copying grayscale values in every color channel.
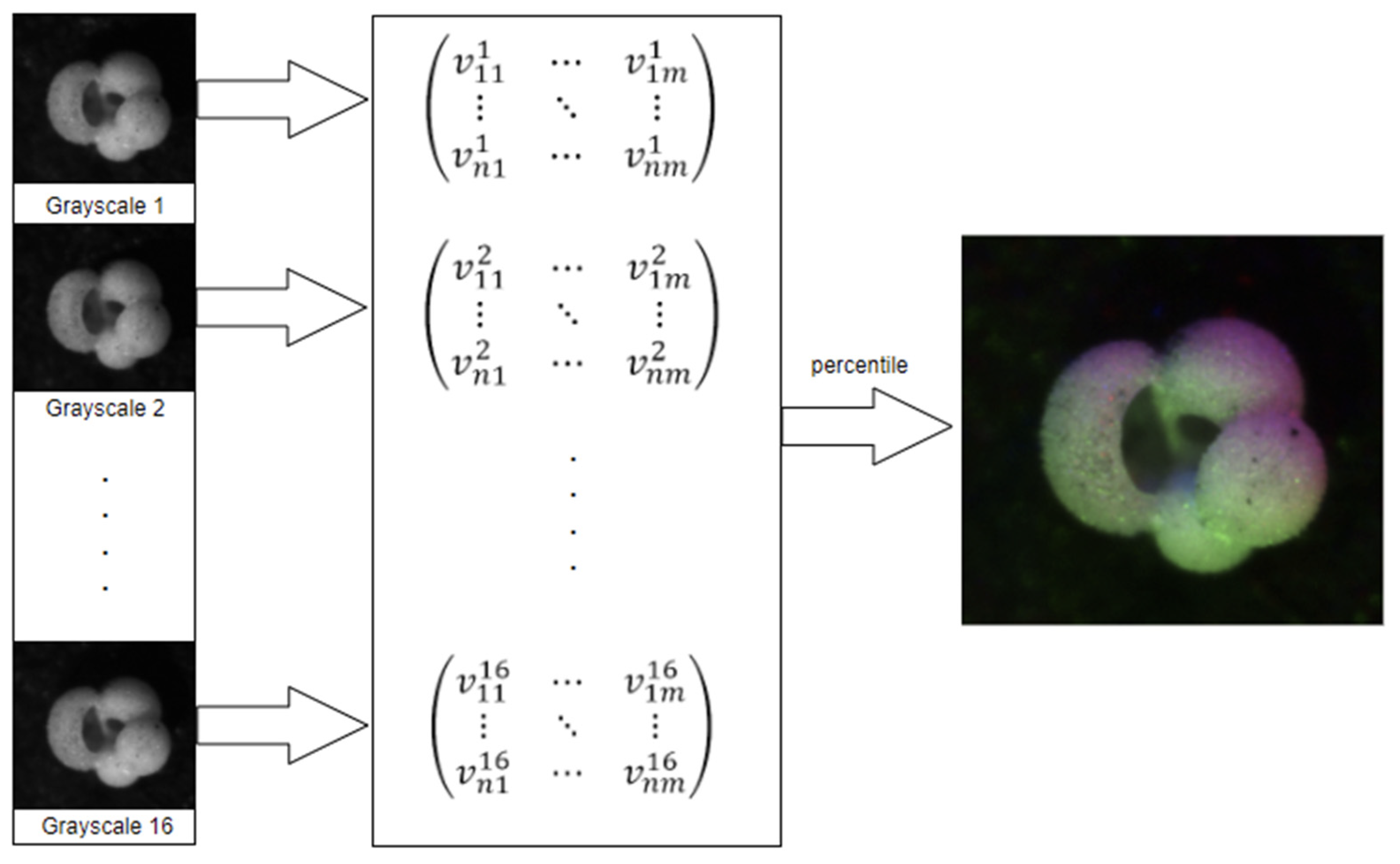
2.2.1. Percentile
- Read the sixteen images;
- Populate a matrix with the grayscale values;
- For each pixel, extract its sixteen grayscale values into a list;
- Sort the list;
- Use elements 2, 8, and 15 as RGB values for the new image.
2.2.2. Gaussian
2.2.3. Mean-Based
2.2.4. Luma Scaling
- ;
- ;
- .
2.2.5. Means Reconstruction
- ;
- ;
- .
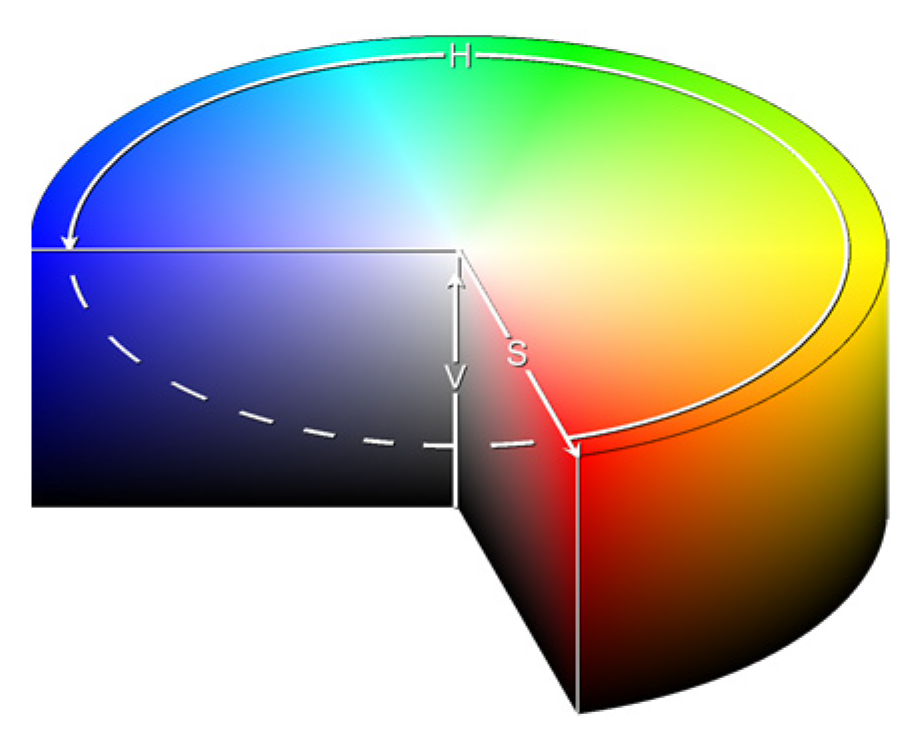
2.2.6. HSVPP: Hue, Saturation, Value of Brightness + Post-Processing
- H is assigned based on the index of the image, giving each a different color hue;
- S is set to 1 by default, for maximizing diversity between colors;
- V is set to the grayscale image’s original intensity, i.e., its brightness.
2.2.7. GraySet
- for the first RGB image;
- for the second one;
- And so on, up to for the last image in the pattern.
2.3. Training
- Mini Batch Size: 30;
- Max Epochs: 20;
- Learning Rate: 10−3.
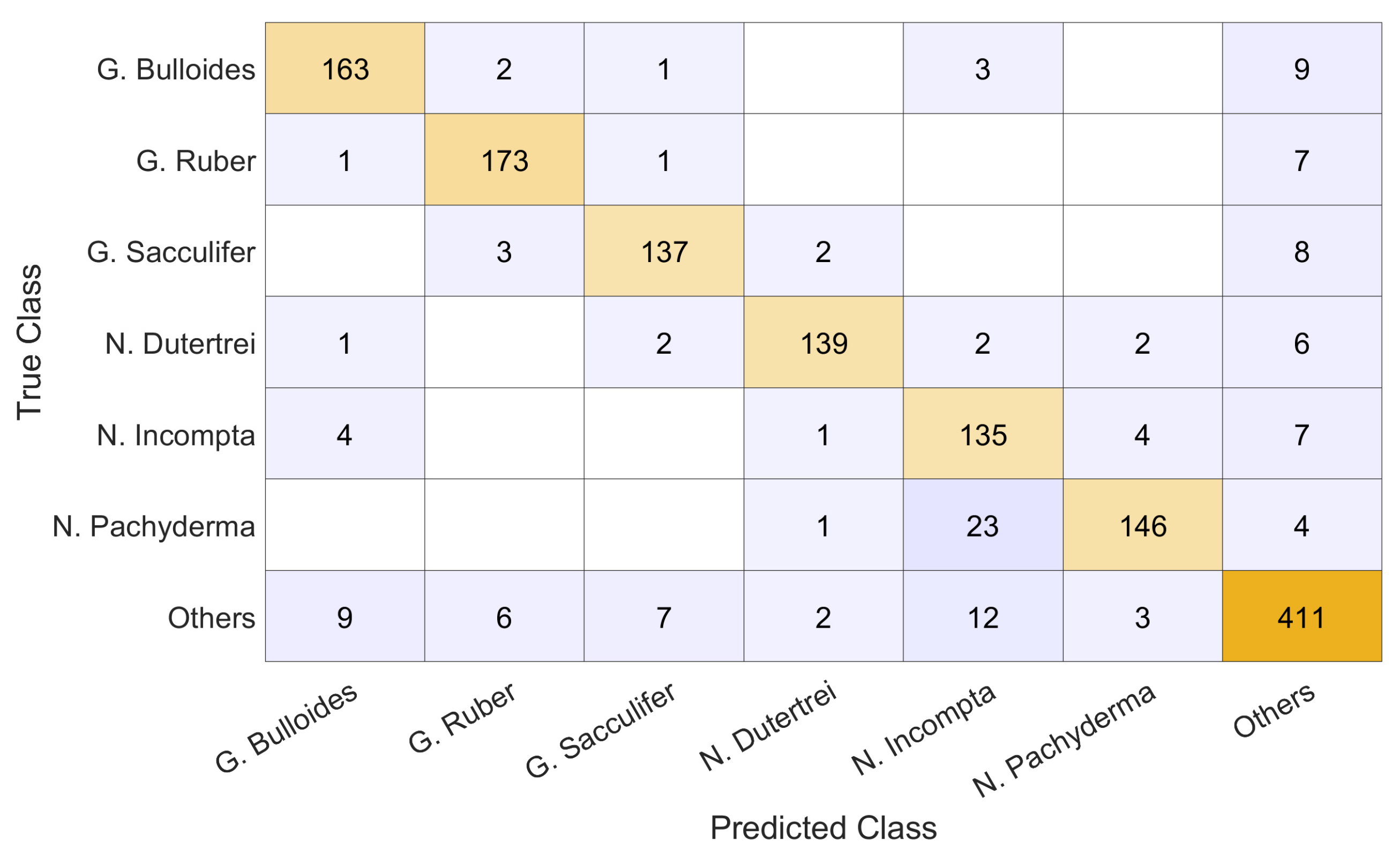
3. Results
- The best-performing ensemble produces results that significantly improve those obtained by the method presented in [3] (percentile), whose F1 score was reported as 85%;
- Among stand-alone approaches the best performance is obtained by GraySet;
- It appears that, in general, increasing the diversity of the ensemble yields better results. The approaches combining multiple pre-processed images sets consistently rank higher in F1 scores than any individual method, iterated ten times. Combining fewer iterations of all the approaches yielded the best results overall. Similar conclusions are obtained with the different topologies;
- The ensemble based on ResNet50_DA obtains performance similar to the one based on ResNet50, but clearly data augmentation improves the stand-alone approaches.
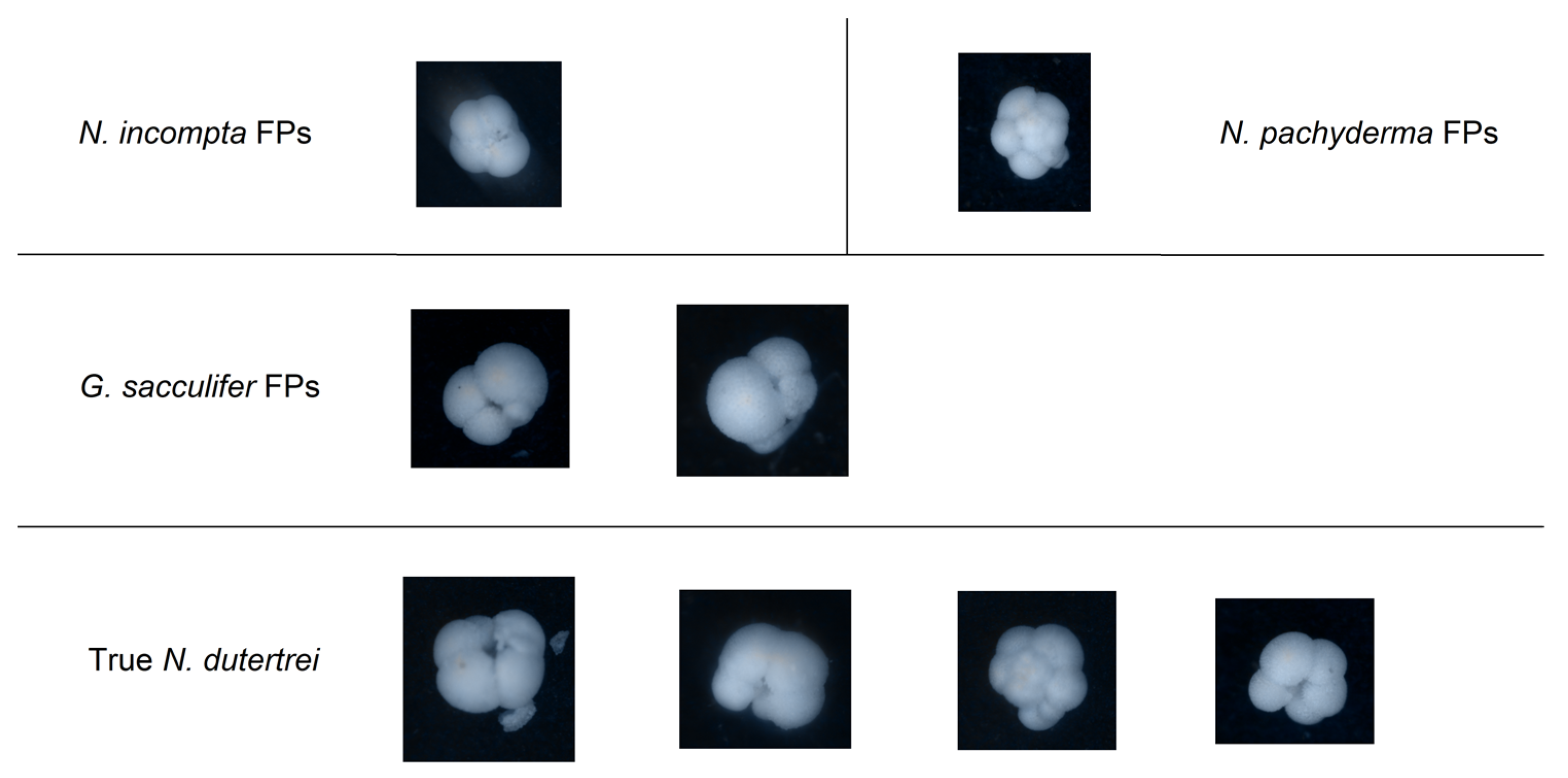
4. Discussion
5. Conclusions
Author Contributions
Funding
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Li, Z.; Liu, F.; Yang, W.; Peng, S.; Zhou, J. A Survey of Convolutional Neural Networks: Analysis, Applications, and Prospects. IEEE Trans. Neural Netw. Learn. Syst. 2022, 33, 6999–7019. [Google Scholar] [CrossRef]
- Menart, C. Evaluating the variance in convolutional neural network behavior stemming from randomness. Proc. SPIE 11394 Autom. Target Recognit. XXX 2020, 11394, 1139410. [Google Scholar] [CrossRef]
- Mitra, R.; Marchitto, T.M.; Ge, Q.; Zhong, B.; Kanakiya, B.; Cook, M.S.; Fehrenbacher, J.D.; Ortiz, J.D.; Tripati, A.; Lobaton, E. Automated species-level identification of planktic foraminifera using convolutional neural networks, with comparison to human performance. Mar. Micropaleontol. 2019, 147, 16–24. [Google Scholar] [CrossRef]
- Edwards, R.; Wright, A. Foraminifera. In Handbook of Sea-Level Research; Shennan, I., Long, A.J., Horton, B.P., Eds.; Wiley: Hoboken, NJ, USA, 2015. [Google Scholar] [CrossRef]
- Liu, S.; Thonnat, M.; Berthod, M. Automatic classification of planktonic foraminifera by a knowledge-based system. In Proceedings of the Tenth Conference on Artificial Intelligence for Applications, San Antonio, TX, USA, 1–4 March 1994; pp. 358–364. [Google Scholar] [CrossRef]
- Beaufort, L.; Dollfus, D. Automatic recognition of coccoliths by dynamical neural networks. Mar. Micropaleontol. 2004, 51, 57–73. [Google Scholar] [CrossRef]
- Pedraza, L.F.; Hernández, C.A.; López, D.A. A Model to Determine the Propagation Losses Based on the Integration of Hata-Okumura and Wavelet Neural Models. Int. J. Antennas Propag. 2017, 2017, 1034673. [Google Scholar] [CrossRef]
- Zhuang, F.; Qi, Z.; Duan, K.; Xi, D.; Zhu, Y.; Zhu, H.; Xiong, H.; He, Q. A Comprehensive Survey on Transfer Learning. Proc. IEEE 2021, 109, 43–76. [Google Scholar] [CrossRef]
- Huang, B.; Yang, F.; Yin, M.; Mo, X.; Zhong, C. A Review of Multimodal Medical Image Fusion Techniques. Comput. Math. Methods Med. 2020, 2020, 8279342. [Google Scholar] [CrossRef] [PubMed]
- Sellami, A.; Abbes, A.B.; Barra, V.; Farah, I.R. Fused 3-D spectral-spatial deep neural networks and spectral clustering for hyperspectral image classification. Pattern Recognit. Lett. 2020, 138, 594–600. [Google Scholar] [CrossRef]
- Zhang, X.; Ye, P.; Xiao, G. VIFB: A Visible and Infrared Image Fusion Benchmark. In Proceedings of the IEEE/CVF Conference on Computer Vision and Pattern Recognition (CVPR) Workshops, Seattle, WA, USA, 14–19 June 2020; pp. 104–105. [Google Scholar]
- James, A.P.; Dasarathy, B.V. Medical image fusion: A survey of the state of the art. Inf. Fusion 2014, 19, 4–19. [Google Scholar] [CrossRef]
- Hermessi, H.; Mourali, O.; Zagrouba, E. Multimodal medical image fusion review: Theoretical background and recent advances. Signal Process. 2021, 183, 108036. [Google Scholar] [CrossRef]
- Li, X.; Jing, D.; Li, Y.; Guo, L.; Han, L.; Xu, Q.; Xing, M.; Hu, Y. Multi-Band and Polarization SAR Images Colorization Fusion. Remote Sens. 2022, 14, 4022. [Google Scholar] [CrossRef]
- Moon, W.K.; Lee, Y.W.; Ke, H.H.; Lee, S.H.; Huang, C.S.; Chang, R.F. Computer-aided diagnosis of breast ultrasound images using ensemble learning from convolutional neural networks. Comput. Methods Programs Biomed. 2020, 190, 105361. [Google Scholar] [CrossRef] [PubMed]
- Maqsood, S.; Javed, U. Multi-modal Medical Image Fusion based on Two-scale Image Decomposition and Sparse Representation. Biomed. Signal Process. Control 2020, 57, 101810. [Google Scholar] [CrossRef]
- Ding, I.J.; Zheng, N.W. CNN Deep Learning with Wavelet Image Fusion of CCD RGB-IR and Depth-Grayscale Sensor Data for Hand Gesture Intention Recognition. Sensors 2022, 22, 803. [Google Scholar] [CrossRef] [PubMed]
- Tasci, E.; Uluturk, C.; Ugur, A. A voting-based ensemble deep learning method focusing on image augmentation and preprocessing variations for tuberculosis detection. Neural Comput. Appl. 2021, 33, 15541–15555. [Google Scholar] [CrossRef] [PubMed]
- Mishra, P.; Biancolillo, A.; Roger, J.M.; Marini, F.; Rutledge, D.N. New data preprocessing trends based on ensemble of multiple preprocessing techniques. TrAC Trends Anal. Chem. 2020, 132, 116045. [Google Scholar] [CrossRef]
- Kuncheva, L.I. Combining Pattern Classifiers. Methods and Algorithms, 2nd ed.; Wiley: Hoboken, NJ, USA, 2014. [Google Scholar]
- LeCun, Y.; Boser, B.; Denker, J.S.; Henderson, D.; Howard, R.E.; Hubbard, W.; Jackel, L.D. Backpropagation Applied to Handwritten Zip Code Recognition. Neural Comput. 1989, 1, 541–551. [Google Scholar] [CrossRef]
- Bengio, Y.; LeCun, Y. Convolutional Networks for Images, Speech, and Time-Series. In The Handbook of Brain Theory and Neural Networks; MIT Press: Cambridge, MA, USA, 1998; pp. 255–258. [Google Scholar]
- LeCun, Y.; Bottou, L.; Bengio, Y.; Haffner, P. Gradient-based learning applied to document recognition. Proc. IEEE 1998, 86, 2278–2324. [Google Scholar] [CrossRef]
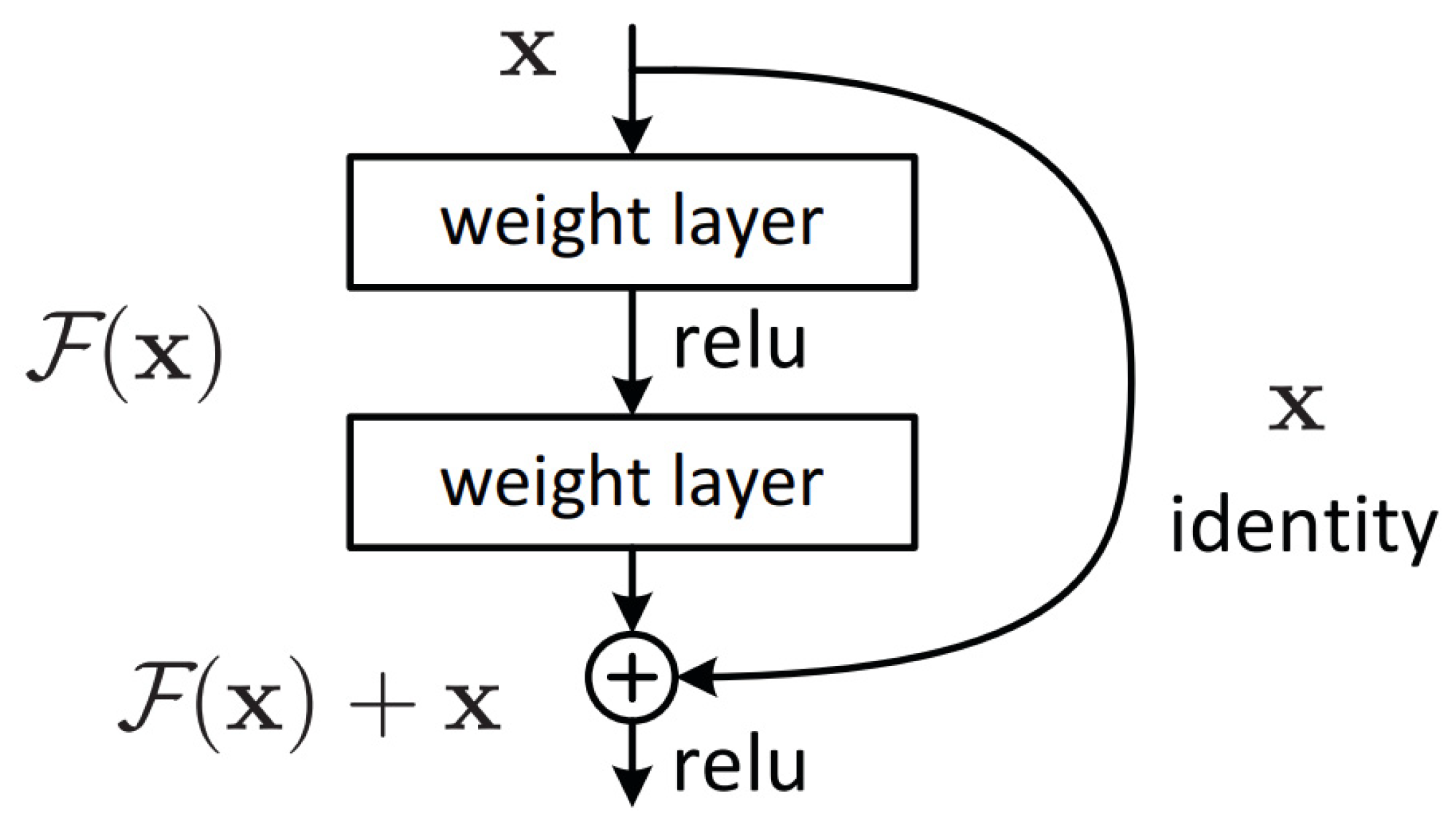
- He, K.; Zhang, X.; Ren, S.; Sun, J. Deep residual learning for image recognition. In Proceedings of the IEEE Conference on Computer Vision and Pattern Recognition, Las Vegas, NV, USA, 27–30 June 2016; pp. 770–778. [Google Scholar] [CrossRef]
- Vallazza, M.; Nanni, L. Convolutional Neural Network Ensemble for Foraminifera Classification. Bachelor’s Thesis, University of Padua, Padua, Italy, 2022. Available online: https://hdl.handle.net/20.500.12608/29285 (accessed on 13 May 2023).
- Hue Saturation Brightness, Wikipedia, Wikimedia Foundation. Available online: https://it.wikipedia.org/wiki/Hue_Saturation_Brightness (accessed on 13 May 2023).
- Szegedy, C.; Liu, W.; Jia, Y.; Sermanet, P.; Reed, S.; Anguelov, D.; Erhan, D.; Vanhoucke, V.; Rabinovich, A. Going deeper with convolutions. In Proceedings of the IEEE Conference on Computer Vision and Pattern Recognition (CVPR), Boston, MA, USA, 7–12 June 2015; pp. 1–9. [Google Scholar] [CrossRef]
- Sandler, M.; Howard, A.; Zhu, M.; Zhmoginov, A.; Chen, L.C. MobileNetV2: Inverted Residuals and Linear Bottlenecks. arXiv 2019, arXiv:1801.04381. [Google Scholar] [CrossRef]

| 4-Fold Cross-Validation | |||||
|---|---|---|---|---|---|
| ensemble [3] | 0.850 | ||||
| Vgg16 [3] | 0.810 | ||||
| ResNet50 | ResNet18 | GoogleNet | MobileNetV2 | ResNet50_DA | |
| Percentile(1) | 0.811 | 0.803 | 0.785 | 0.817 | 0.870 |
| Percentile(10) | 0.853 | 0.860 | 0.807 | 0.874 | --- |
| Luma Scaling(10) | 0.870 | 0.845 | 0.794 | 0.856 | --- |
| Means Reconstruction(10) | 0.874 | 0.859 | 0.810 | 0.879 | --- |
| Gaussian(10) | 0.873 | 0.850 | 0.805 | 0.867 | --- |
| HSVPP(10) | 0.843 | 0.833 | 0.789 | 0.841 | --- |
| GraySet(10) | 0.885 | 0.864 | 0.831 | 0.892 | --- |
| Percentile(3) + Luma Scaling(3) + Means Reconstruction(3) | 0.877 | 0.868 | 0.821 | 0.881 | 0.895 |
| Gaussian(3) + Luma Scaling(3) + Means Reconstruction(3) | 0.879 | 0.859 | 0.798 | 0.872 | 0.889 |
| Percentile(2) + Gaussian(2) + Luma Scaling(2) + Means Reconstruction(2)+HSVPP(2) | 0.885 | 0.880 | 0.840 | 0.888 | 0.903 |
| Percentile(1) + Gaussian(1) + Luma Scaling(1) + Means Reconstruction(1) + HSVPP(1) + GraySet(5) | 0.906 | 0.865 | 0.841 | 0.897 | 0.906 |
| Precision (%) | Recall (%) | F1 Score (%) | Accuracy (%) | |
|---|---|---|---|---|
| Novices (max) | 65 | 64 | 63 | 63 |
| Experts (max) | 83 | 83 | 83 | 83 |
| ResNet50 + Vgg16 [3] | 84 | 86 | 85 | 85 |
| Vgg16 [3] | 80 | 82 | 81 | 81 |
| Percentile(1) | 80.8 | 81.5 | 81.6 | 81.6 |
| Percentile(10) | 85.1 | 85.8 | 85.2 | 85.3 |
| GraySet(10) | 88.2 | 89.1 | 88.4 | 88.0 |
| Proposed Ensemble | 90.9 | 90.6 | 90.6 | 90.7 |
| Precision | Recall | F1 Score | |
|---|---|---|---|
| G. bulloides | 0.92 | 0.92 | 0.92 |
| G. ruber | 0.94 | 0.95 | 0.95 |
| G. sacculifer | 0.93 | 0.91 | 0.92 |
| N. dutertrei | 0.77 | 0.89 | 0.83 |
| N. incompta | 0.94 | 0.83 | 0.89 |
| N. pachyderma | 0.96 | 0.91 | 0.94 |
| Other | 0.91 | 0.91 | 0.91 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Nanni, L.; Faldani, G.; Brahnam, S.; Bravin, R.; Feltrin, E. Improving Foraminifera Classification Using Convolutional Neural Networks with Ensemble Learning. Signals 2023, 4, 524-538. https://doi.org/10.3390/signals4030028
Nanni L, Faldani G, Brahnam S, Bravin R, Feltrin E. Improving Foraminifera Classification Using Convolutional Neural Networks with Ensemble Learning. Signals. 2023; 4(3):524-538. https://doi.org/10.3390/signals4030028
Chicago/Turabian StyleNanni, Loris, Giovanni Faldani, Sheryl Brahnam, Riccardo Bravin, and Elia Feltrin. 2023. "Improving Foraminifera Classification Using Convolutional Neural Networks with Ensemble Learning" Signals 4, no. 3: 524-538. https://doi.org/10.3390/signals4030028
APA StyleNanni, L., Faldani, G., Brahnam, S., Bravin, R., & Feltrin, E. (2023). Improving Foraminifera Classification Using Convolutional Neural Networks with Ensemble Learning. Signals, 4(3), 524-538. https://doi.org/10.3390/signals4030028








