ChronobioticsDB: The Database of Drugs and Compounds Modulating Circadian Rhythms
Abstract
1. Introduction
2. Results
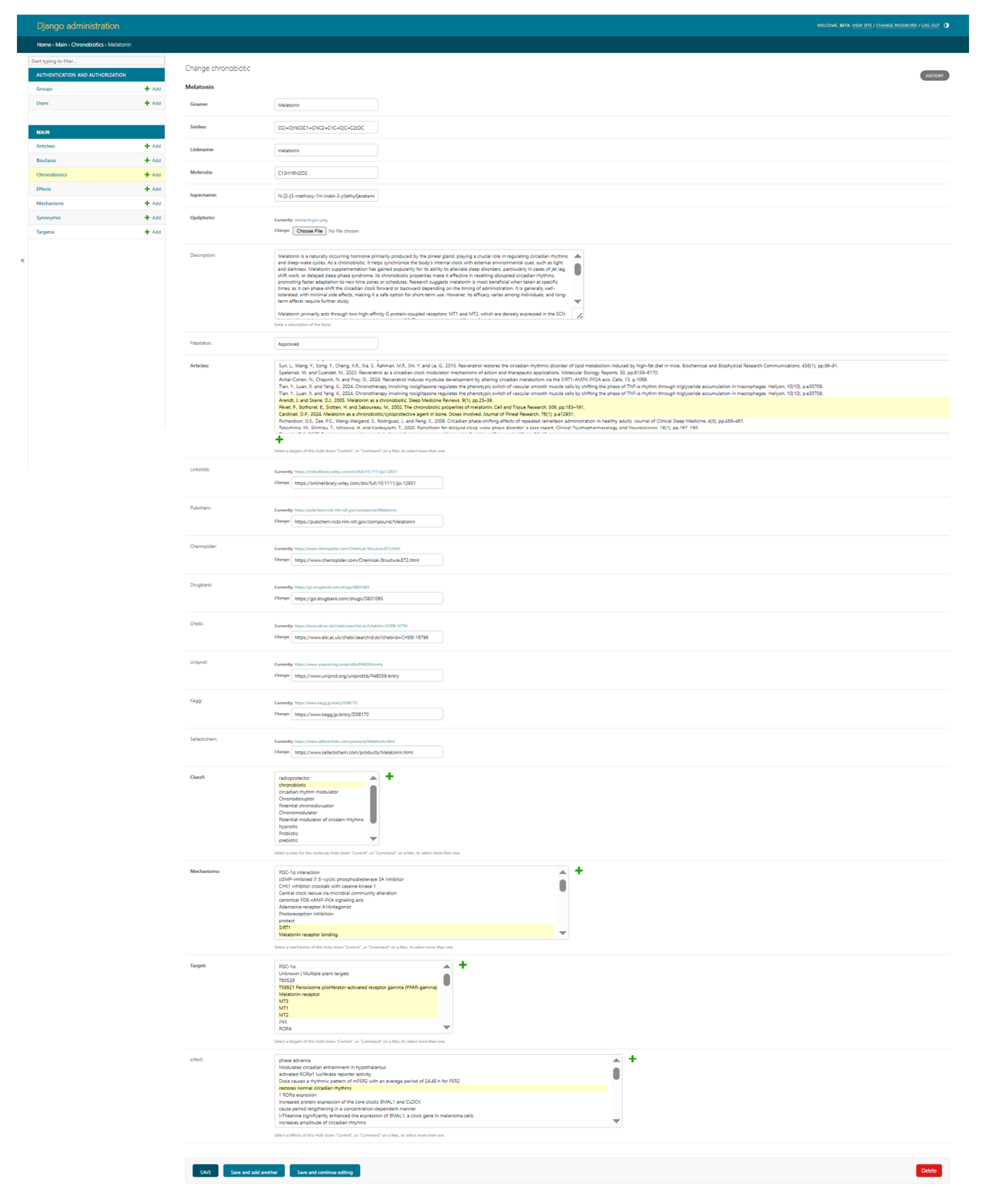
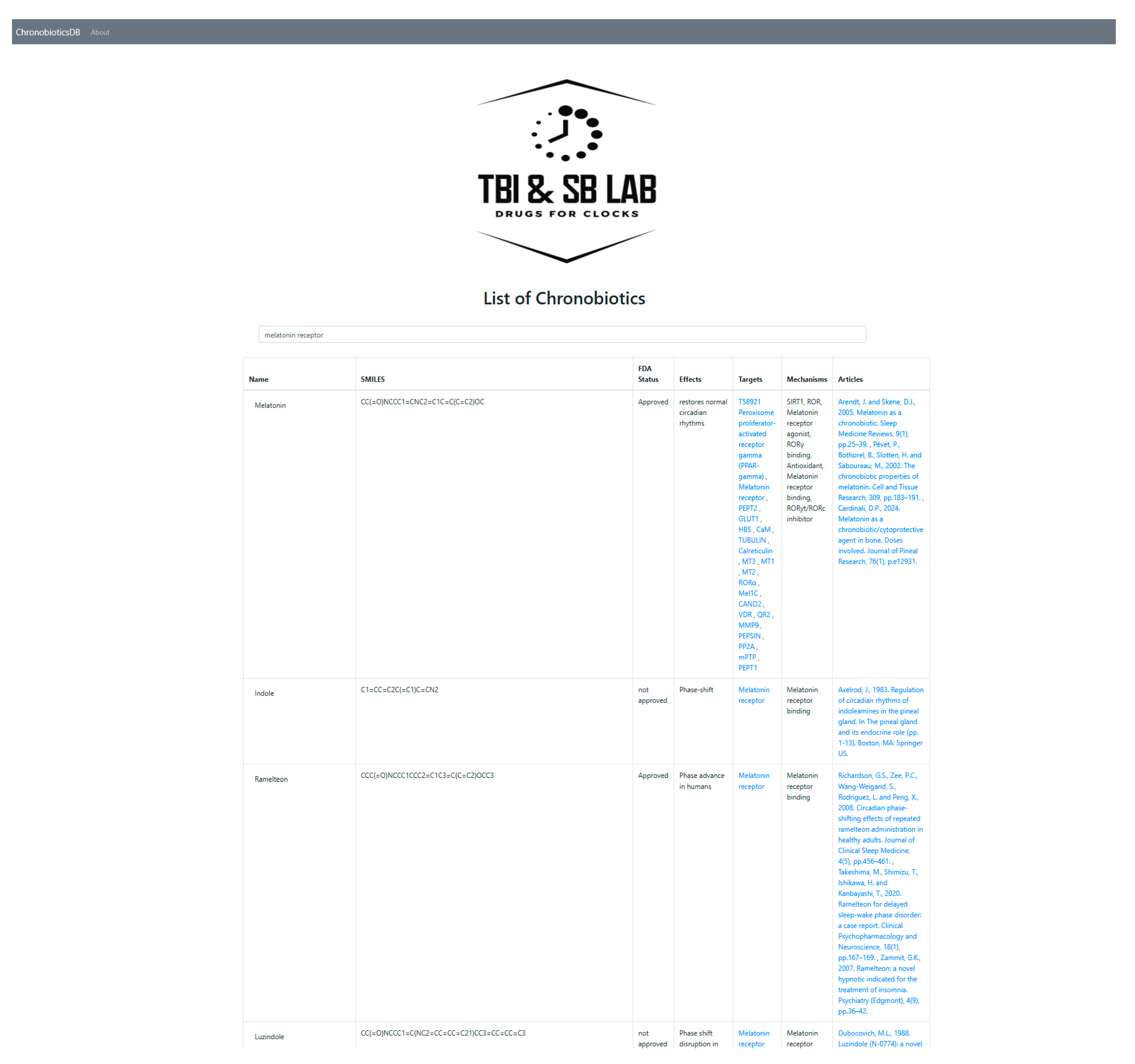
2.1. Database Web Interface
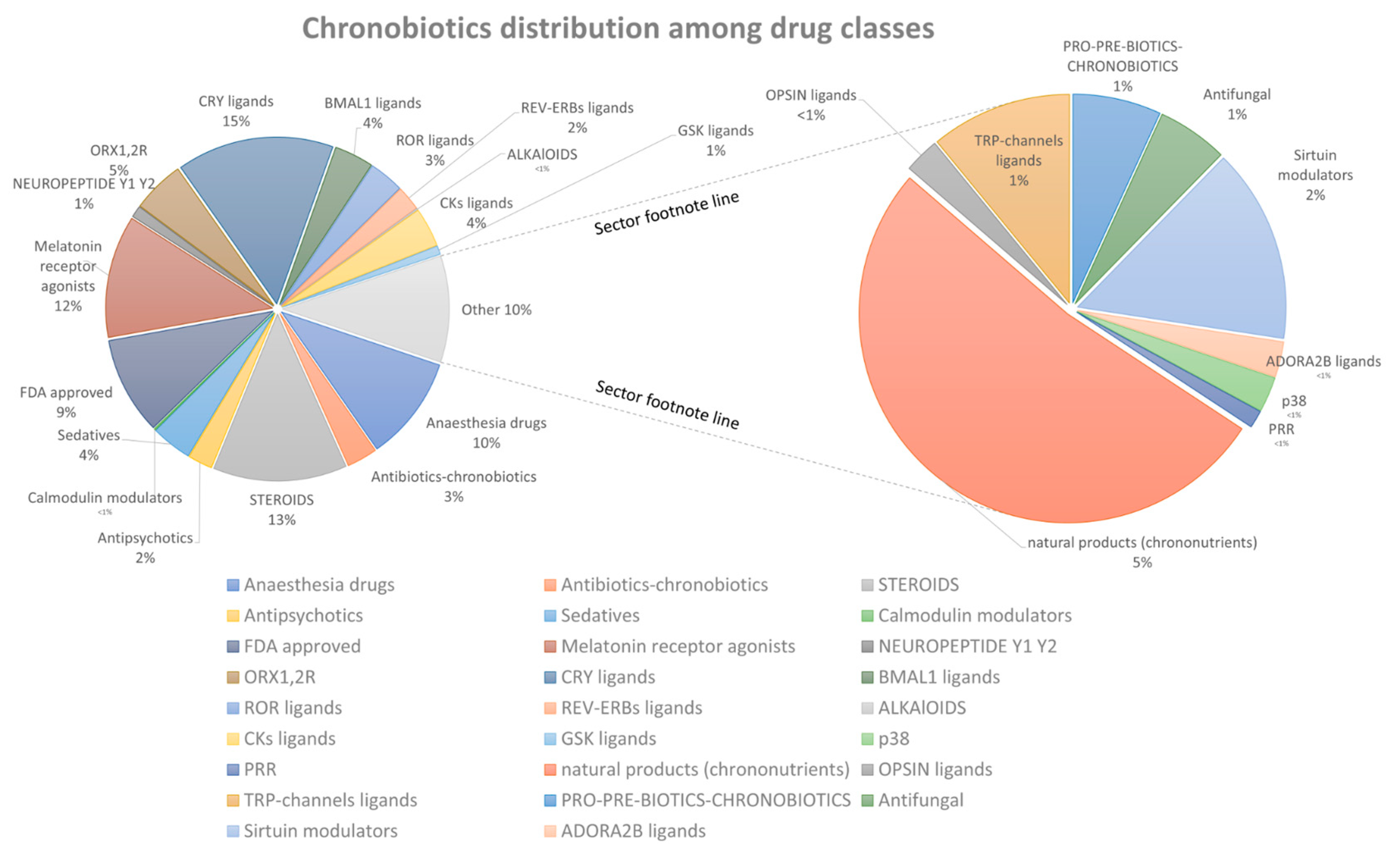
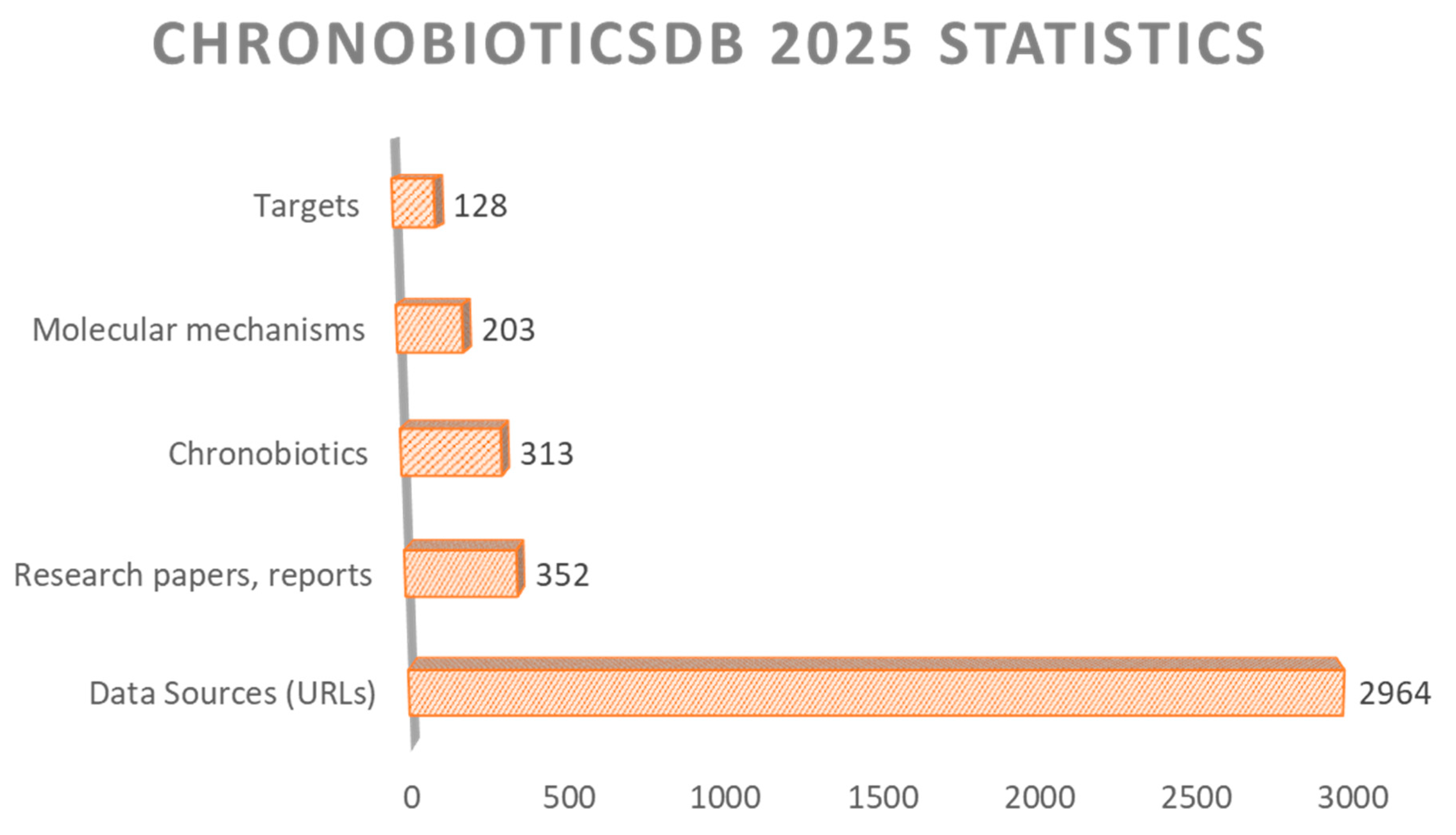
2.2. Database Statistics and Analysis
2.3. The Classification of Molecules According to the Major Chemical Groups
- Terpenes/Steroids;
- Alkaloids;
- Sulfonamides/Sulfonates;
- Aromatic compounds (Phenols and Flavonoids);
- Organofluorines/Organochlorines;
- Quaternary ammonium compounds;
- Amides/Peptides;
- Esters/Lipids;
- Heterocycles (Pyridine and Thiophene);
- Nitro compounds;
- Phosphates/Nucleotides;
- Organometallics/Salts;
- Macrocyclic compounds;
- Disulfides;
- Alkyne derivatives;
- Barbiturates;
- Metal salts.
2.4. Key Observations on the Proposed Classes in ChronobioticsDB
3. Discussion
3.1. Ethical Considerations
3.2. Future Updates and Community Engagement
3.3. Limitations, International Validation
4. Materials and Methods
4.1. Data Acquisition
4.1.1. Data Sources and Extraction
4.1.2. Data Quality Assessment and Validation
- (a)
- Peer-reviewed articles published in the past five decades.
- (b)
- Research demonstrating both acute and chronic effects on circadian rhythms.
- (c)
- Compounds studied through both in vitro and in vivo methodologies.
4.1.3. Data Integration and Standardization
- (a)
- Compound names were normalized following the IUPAC nomenclature (if available).
- (b)
- Controlled vocabularies were established for biological terms relating to chronobiotic effects to ensure uniformity across entries.
4.1.4. Database Implementation
4.2. Database Organization (Primary and Secondary Data)
4.3. Database Architecture
4.4. Core Components of the Architecture
4.4.1. Central Table: Chronobiotic
4.4.2. Auxiliary Tables
- One-to-Many Relationships: The synonyms table is connected to Chronobiotic via the foreign key originalbiotic.
- Many-to-Many Relationships: The target, mechanism, effects, article, and class tables are linked to Chronobiotic through intermediary tables, which are automatically generated by Django.
4.4.3. Schema of Table Relationships
- Chronobiotic → synonyms: A single compound may have multiple synonyms.
- Chronobiotic → target: A single compound may interact with multiple targets, and a single target may be associated with multiple compounds.
- Chronobiotic → mechanism: A single compound may exhibit multiple mechanisms of action, and a single mechanism may be associated with multiple compounds.
- Chronobiotic → class: A single compound may belong to multiple classes, and a single class may encompass multiple compounds.
- Chronobiotic → effects. A single compound may exhibit multiple effects on circadian rhythms.
- Chronobiotic → article. A single compound may exhibit multiple literature sources where it is described, and one source also may contain many compounds.
4.4.4. Technologies and Tools
- DBMS: PostgreSQL, a robust and reliable relational database, provides high performance and supports complex queries [15].
- ORM: Django ORM (Django 5.1.2) is employed for database interactions at the Python (v3.11) code level. This eliminates the need for manual SQL query writing and facilitates efficient data management [16].
- Indexes: Indexes have been created on frequently queried fields, such as gname, smiles, and targetsname, to optimize search performance.
- Migrations: Django’s built-in migration system allows for seamless modifications to the database structure without data loss.
4.4.5. Ensuring Data Integrity and Security
- Foreign Keys: All inter-table relationships are implemented through foreign keys, ensuring data integrity.
- Unique Constraints: Unique fields (gname, smiles, molecula, and iupacname) prevent record duplication.
- Role-Based Access Control: Database access is restricted at the user and role levels, ensuring data security.
- Encryption: Confidential data is stored in an encrypted format.
4.5. Use of Artificial Intelligence
5. Conclusions and Future Perspectives
5.1. Content Expansion and Data Curation
5.2. Integration of AI-Driven Search and Assisting Tools
5.3. Computational Chronobiotics Discovery
5.4. Gerontological Applications and Predictive Modeling
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Chen, Z.; Yoo, S.H.; Takahashi, J.S. Development and therapeutic potential of small-molecule modulators of circadian systems. Annu. Rev. Pharmacol. Toxicol. 2018, 58, 231–252. [Google Scholar] [CrossRef] [PubMed]
- Solovev, I.A.; Golubev, D.A. Chronobiotics: Classifications of existing circadian clock modulators, future perspectives. Biomeditsinskaya Khimiya 2024, 70, 381–393. [Google Scholar] [CrossRef] [PubMed]
- Cha, H.K.; Chung, S.; Lim, H.Y.; Jung, J.W.; Son, G.H. Small molecule modulators of the circadian molecular clock with implications for neuropsychiatric diseases. Front. Mol. Neurosci. 2019, 11, 496. [Google Scholar] [CrossRef] [PubMed]
- Robles-Piedras, A.L.; Bautista-Sánchez, U.; Olvera-Hernández, E.G.; Chehue-Romero, A. Chronopharmacokinetics: A Brief Analysis of the Influence of Circadian Rhythm on the Absorption, Distribution, Metabolism, and Elimination of Drugs. Biomed. Pharmacol. J. 2024, 17, 2011–2017. [Google Scholar] [CrossRef]
- Tamai, T.K.; Nakane, Y.; Ota, W.; Kobayashi, A.; Ishiguro, M.; Kadofusa, N.; Ikegami, K.; Yagita, K.; Shigeyoshi, Y.; Sudo, M.; et al. Identification of circadian clock modulators from existing drugs. EMBO Mol. Med. 2018, 10, e8724. [Google Scholar] [CrossRef] [PubMed]
- Imming, P. Medicinal chemistry: Definitions and objectives, drug activity phases, drug classification systems. In The Practice of Medicinal Chemistry; Academic Press: Cambridge, MA, USA, 2015; pp. 3–13. [Google Scholar]
- Pizarro, A.; Hayer, K.; Lahens, N.F.; Hogenesch, J.B. CircaDB: A database of mammalian circadian gene expression profiles. Nucleic Acids Res. 2012, 41, D1009–D1013. [Google Scholar] [CrossRef] [PubMed]
- Zhu, X.; Han, X.; Li, Z.; Zhou, X.; Yoo, S.H.; Chen, Z.; Ji, Z. CircaKB: A comprehensive knowledgebase of circadian genes across multiple species. Nucleic Acids Res. 2025, 53, D67–D78. [Google Scholar] [CrossRef] [PubMed]
- Kim, S.; Bolton, E.E. PubChem: A Large-Scale Public Chemical Database for Drug Discovery. In Open Access Databases and Datasets for Drug Discovery; Wiley: Hoboken, NJ, USA, 2024; pp. 39–66. [Google Scholar]
- Knox, C.; Wilson, M.; Klinger, C.M.; Franklin, M.; Oler, E.; Wilson, A.; Pon, A.; Cox, J.; Chin, N.E.; Strawbridge, S.A.; et al. DrugBank 6.0: The DrugBank knowledgebase for 2024. Nucleic Acids Res. 2024, 52, D1265–D1275. [Google Scholar] [CrossRef] [PubMed]
- Pence, H.E.; Williams, A. ChemSpider: An online chemical information resource. J. Chem. Educ. 2010, 87, 1123–1124. [Google Scholar] [CrossRef]
- Califf, R.M.; Cutler, T.L.; Marston, H.D.; Meeker-O’Connell, A. The importance of ClinicalTrials. gov in informing trial design, conduct, and results. J. Clin. Transl. Sci. 2025, 9, e42. [Google Scholar] [CrossRef] [PubMed]
- Odell, S.G.; Lazo, G.R.; Woodhouse, M.R.; Hane, D.L.; Sen, T.Z. The art of curation at a biological database: Principles and application. Curr. Plant Biol. 2017, 11, 2–11. [Google Scholar] [CrossRef]
- Bittrich, S.; Segura, J.; Duarte, J.M.; Burley, S.K.; Rose, Y. RCSB protein Data Bank: Exploring protein 3D similarities via comprehensive structural alignments. Bioinformatics 2024, 40, btae370. [Google Scholar] [CrossRef] [PubMed]
- Simkovics, S.; Petersgasse, P. Enhancement of the ANSI SQL Implementation of PostgreSQL. 1998. Available online: https://www.ic.unicamp.br/~celio/livrobd/postgres/ansi_sql_implementation_postgresql.pdf (accessed on 14 May 2025).
- Holovaty, A.; Willison, S. Django Web Framework. 2003. Available online: http://www.djangoproject.com (accessed on 14 May 2025).
- Rajan, K.; Brinkhaus, H.O.; Agea, M.I.; Zielesny, A.; Steinbeck, C. DECIMER.ai: An open platform for automated optical chemical structure identification, segmentation and recognition in scientific publications. Nat. Commun. 2023, 14, 5045. [Google Scholar] [CrossRef] [PubMed]

| Trait of Research | CRITERIA | |
|---|---|---|
| Research Inclusion | Research Exclusion | |
| Date of Publication | Present | Absent |
| Age of an article, years | >0 | >50 |
| The source/subject is relevant to the research question | Yes | No |
| Appropriate academic and technical level | Yes | No |
| Source authority and authorship | Peer-reviewed journal article, scientific report, governmental or academic website, description of authors, presence of affiliation, publisher information | Non-peer-reviewed sources, incomplete information about author, affiliations, publisher, journal or book, or online resource |
| Accuracy of information presentation in the source | There are no mistakes or unclear statements. Statements are supported by evidence. Information is reliable and is presented in reliable form. | Numerous mistakes, statements unsupported by evidence, unreliable information, and unclear presentation of it. |
| Purpose of the source publication | Academic or technical use | Entertainment, opinion, propaganda |
| Cited literature in the source | Bibliography, link in the text with a description of the source | Absence of any links and bibliographic records or inappropriate non-scholarly sources cited |
| Effect on circadian rhythm described in the article | Present | Absent |
| Sample size, objects, or patients treated | >30 | <30 |
| Reproducibility | Methods are reproducible (Clearly described source of compound or way of extraction/synthesis, doses, regimen, model organism strain or patients cohort described, the method of circadian rhythm measurements and statistics are represented properly) | Not reproducible, speculation (Not clearly described source of compound or way of extraction/synthesis, nonclear doses, regimen. Model organism strain or patient cohort is not appropriate for academic study, the methods of circadian rhythm measurements and statistics are not described or mentioned in general aspect without citation) |
| Interaction with target | Described | Not described |
| Model object | Having a circadian molecular clock and circadian rhythms of physiological and molecular processes | The circadian patterns in the object are not described and there is no molecular machinery of the oscillator |
| Ethical aspect | Ethically appropriate protocol of study, verified with an ethical committee if needed | Illegal or unethical protocol described |
| Presence of chemical compound or living organism (if probiotic) | Yes | No |
| Presence of the mechanism of activity | Yes | No |
| Presence of a chemical graphic formula | Yes | No |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Solovev, I.A.; Golubev, D.A.; Yagovkina, A.I.; Kotelina, N.O. ChronobioticsDB: The Database of Drugs and Compounds Modulating Circadian Rhythms. Clocks & Sleep 2025, 7, 30. https://doi.org/10.3390/clockssleep7030030
Solovev IA, Golubev DA, Yagovkina AI, Kotelina NO. ChronobioticsDB: The Database of Drugs and Compounds Modulating Circadian Rhythms. Clocks & Sleep. 2025; 7(3):30. https://doi.org/10.3390/clockssleep7030030
Chicago/Turabian StyleSolovev, Ilya A., Denis A. Golubev, Arina I. Yagovkina, and Nadezhda O. Kotelina. 2025. "ChronobioticsDB: The Database of Drugs and Compounds Modulating Circadian Rhythms" Clocks & Sleep 7, no. 3: 30. https://doi.org/10.3390/clockssleep7030030
APA StyleSolovev, I. A., Golubev, D. A., Yagovkina, A. I., & Kotelina, N. O. (2025). ChronobioticsDB: The Database of Drugs and Compounds Modulating Circadian Rhythms. Clocks & Sleep, 7(3), 30. https://doi.org/10.3390/clockssleep7030030






