The Effect of Caffeine Exposure on Sleep Patterns in Zebrafish Larvae and Its Underlying Mechanism
Abstract
1. Introduction
2. Results
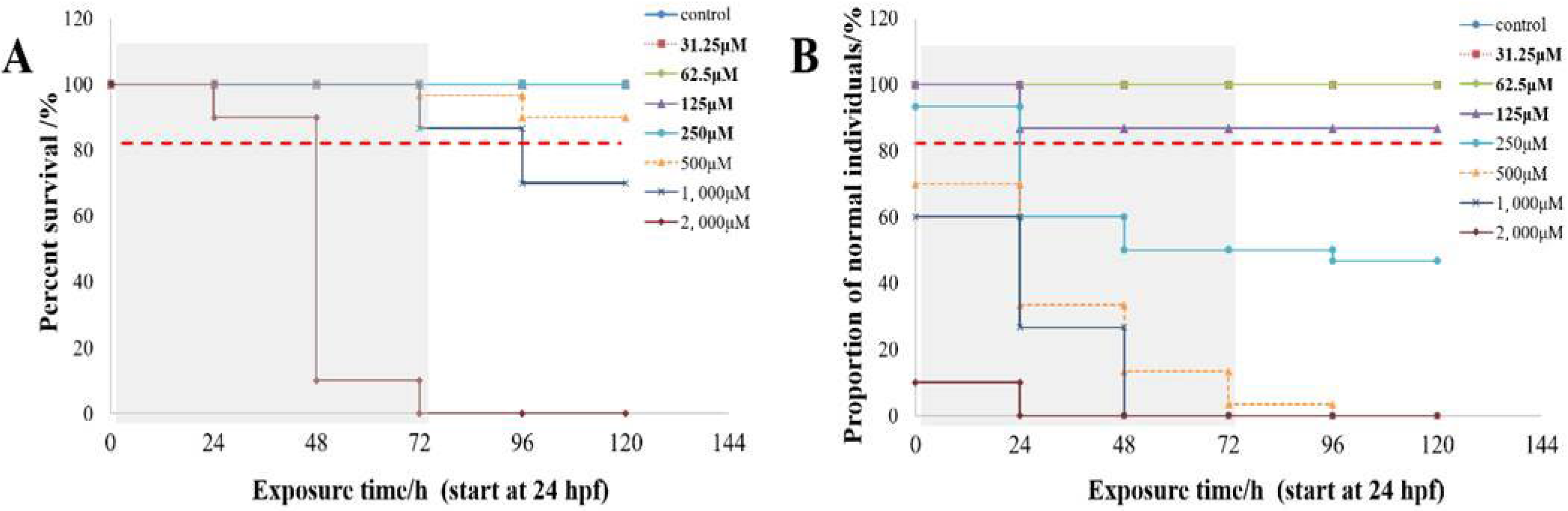
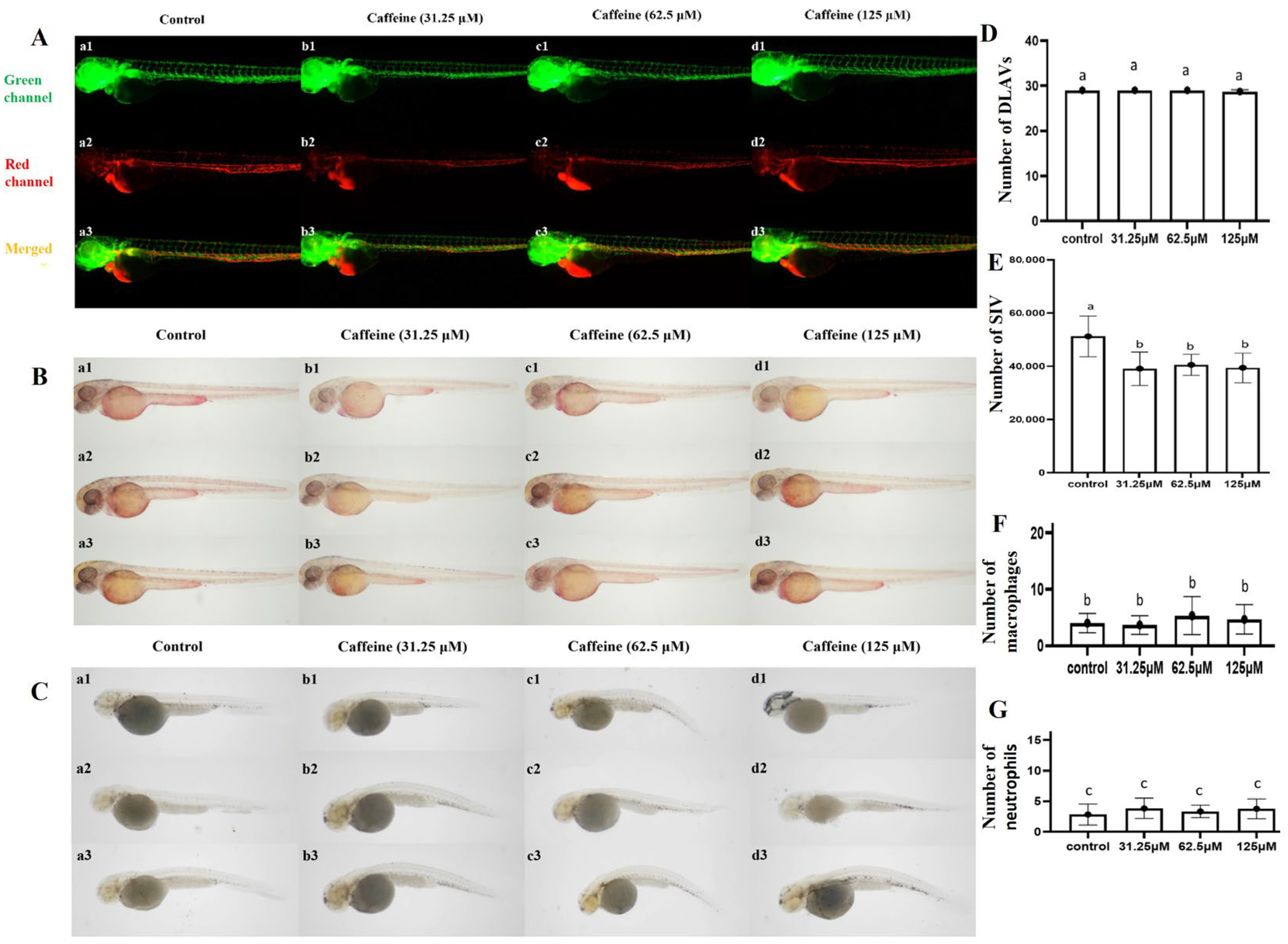
2.1. Toxicity of Caffeine on Developing
2.2. Sleep/Wake Behavioral Changes Induced by Caffeine in Zebrafish Larvae
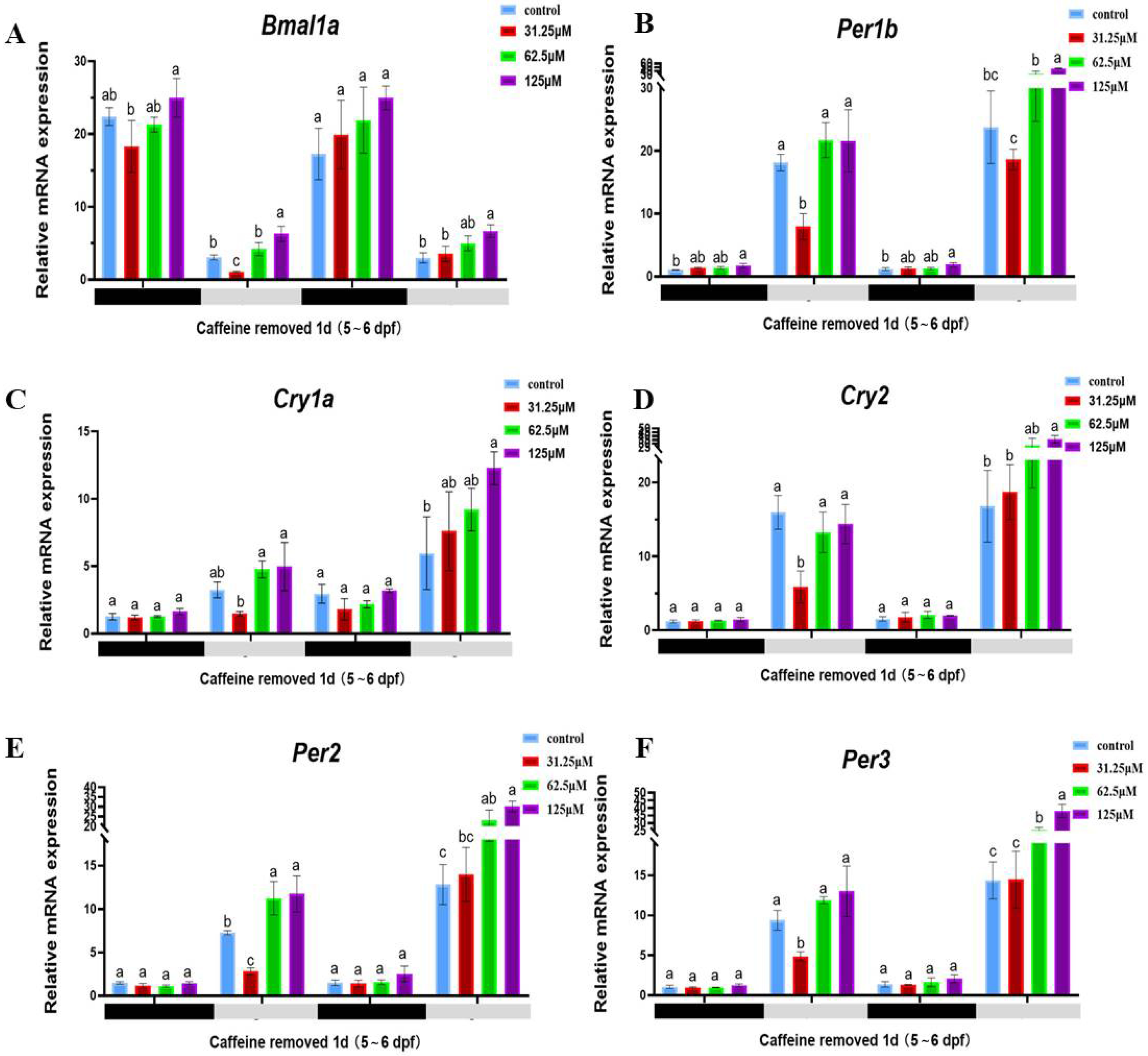
2.3. Caffeine Stimulates the Upregulation of Circadian Rhythm-Related Genes
2.4. Caffeine Can Induce the Up-Regulation of ADA Enzyme Activity and Adenosine Receptor Gene Expression
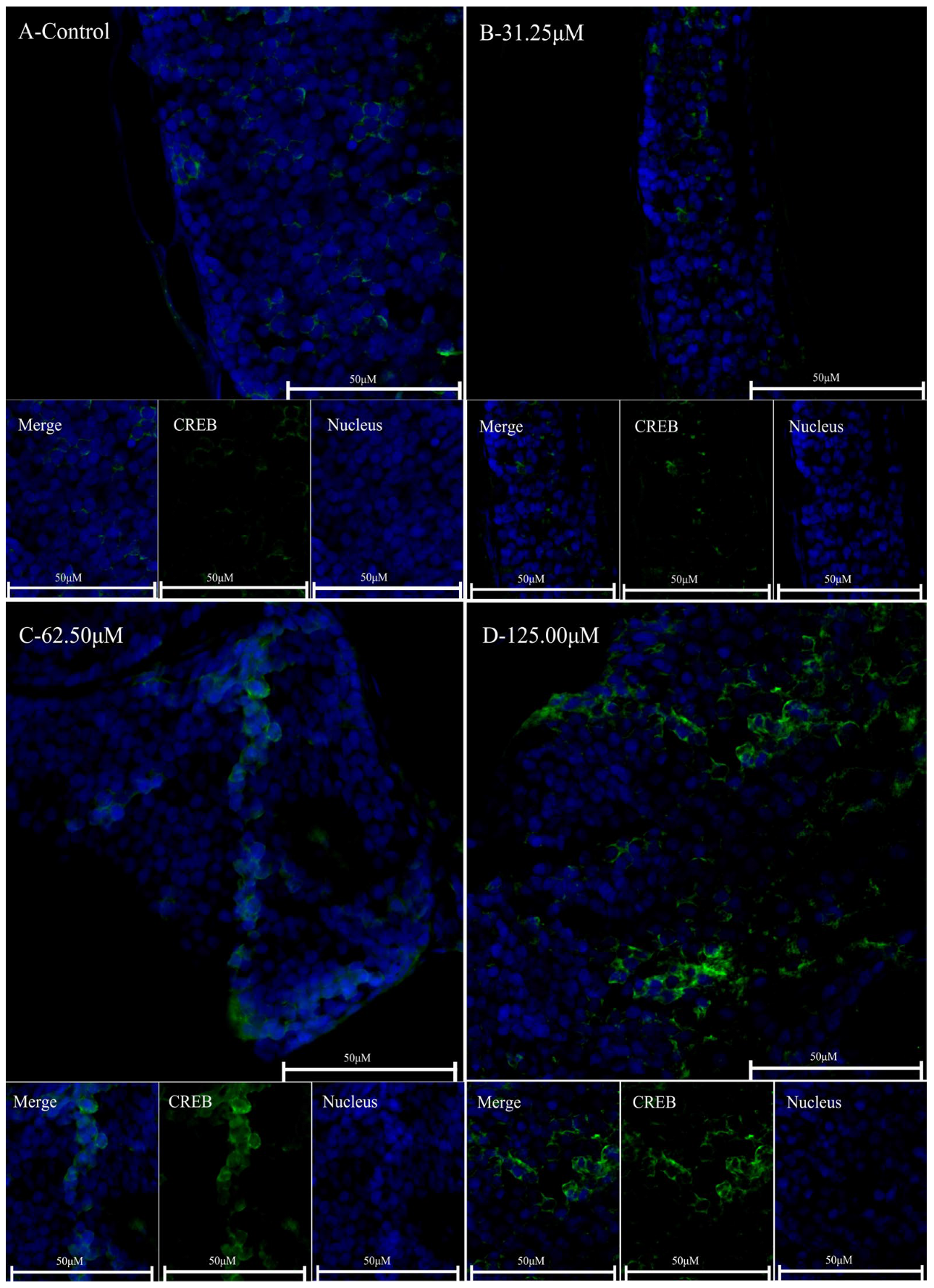
2.5. Caffeine May Regulate Circadian Clock Genes Through the ADA/ADORA/CREB Pathway
3. Discussion
4. Materials and Methods
4.1. Fish Lines and Maintenance
4.2. Determine the Optimal Dose and Duration of Caffeine Exposure
4.3. Sleep/Wake Behavior Test
4.4. Quantitative PCR
4.5. Adenosine Deaminase Assays
4.6. Immunohistochemical Analysis
4.7. Statistical Analysis
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Zada, D.; Bronshtein, I.; Lerer-Goldshtein, T.; Garini, Y.; Appelbaum, L. Sleep increases chromosome dynamics to enable reduction of accumulating DNA damage in single neurons. Nat. Commun. 2019, 10, 895. [Google Scholar] [CrossRef] [PubMed]
- Joiner, W.J. Unraveling the Evolutionary Determinants of Sleep. Curr. Biol. 2016, 26, R1073–R1087. [Google Scholar] [CrossRef] [PubMed]
- Barlow, I.L.; Rihel, J. Zebrafish sleep: From geneZZZ to neuronZZZ. Curr. Opin. Neurobiol. 2017, 44, 65–71. [Google Scholar] [CrossRef] [PubMed]
- Toth, L.A.; Bhargava, P. Animal models of sleep disorders. Comp. Med. 2013, 63, 91–104. [Google Scholar] [PubMed]
- Zhdanova, I.V. Sleep and its regulation in zebrafish. Rev. Neurosci. 2011, 22, 27–36. [Google Scholar] [CrossRef]
- Kazimi, N.; Cahill, G.M. Development of a circadian melatonin rhythm in embryonic zebrafish. Brain Res. Dev. Brain Res. 1999, 117, 47–52. [Google Scholar] [CrossRef]
- Oikonomou, G.; Prober, D.A. Attacking sleep from a new angle: Contributions from zebrafish. Curr. Opin. Neurobiol. 2017, 44, 80–88. [Google Scholar] [CrossRef]
- Zhdanova, I.V.; Wang, S.Y.; Leclair, O.U.; Danilova, N.P. Melatonin promotes sleep-like state in zebrafish. Brain Res. 2001, 903, 263–268. [Google Scholar] [CrossRef]
- Prober, D.A.; Rihel, J.; Onah, A.A.; Sung, R.J.; Schier, A.F. Hypocretin/orexin overexpression induces an insomnia-like phenotype in zebrafish. J. Neurosci. 2006, 26, 13400–13410. [Google Scholar] [CrossRef]
- Howe, K.; Clark, M.D.; Torroja, C.F.; Torrance, J.; Berthelot, C.; Muffato, M.; Collins, J.E.; Humphray, S.; McLaren, K.; Matthews, L.; et al. The zebrafish reference genome sequence and its relationship to the human genome. Nature 2013, 496, 498–503. [Google Scholar] [CrossRef]
- Rihel, J.; Schier, A.F. Behavioral screening for neuroactive drugs in zebrafish. Dev. Neurobiol. 2012, 72, 373–385. [Google Scholar] [CrossRef]
- Wang, D.; Hu, G.; Wang, J.; Yan, D.; Wang, M.; Yang, L.; Serikuly, N.; Alpyshov, E.; Demin, K.A.; Galstyan, D.S.; et al. Studying CNS effects of Traditional Chinese Medicine using zebrafish models. J. Ethnopharmacol. 2021, 267, 113383. [Google Scholar] [CrossRef] [PubMed]
- Burke, T.M.; Markwald, R.R.; McHill, A.W.; Chinoy, E.D.; Snider, J.A.; Bessman, S.C.; Jung, C.M.; O’Neill, J.S.; Wright, K.P., Jr. Effects of caffeine on the human circadian clock in vivo and in vitro. Sci. Transl. Med. 2015, 7, 305ra146. [Google Scholar] [CrossRef] [PubMed]
- Narishige, S.; Kuwahara, M.; Shinozaki, A.; Okada, S.; Ikeda, Y.; Kamagata, M.; Tahara, Y.; Shibata, S. Effects of caffeine on circadian phase, amplitude and period evaluated in cells in vitro and peripheral organs in vivo in PER 2:: LUCIFERASE mice. Br. J. Pharmacol. 2014, 171, 5858–5869. [Google Scholar] [CrossRef]
- Ruiz-Oliveira, J.; Silva, P.F.; Luchiari, A.C. Coffee time: Low caffeine dose promotes attention and focus in zebrafish. Learn. Behav. 2019, 47, 227–233. [Google Scholar] [CrossRef]
- Chaudhary, N.S.; Grandner, M.A.; Jackson, N.J.; Chakravorty, S. Caffeine consumption, insomnia, and sleep duration: Results from a nationally representative sample. Nutrition 2016, 32, 1193–1199. [Google Scholar] [CrossRef]
- Khor, Y.M.; Soga, T.; Parhar, I.S. Caffeine neuroprotects against dexamethasone-induced anxiety-like behaviour in the Zebrafish (Danio rerio). Gen. Comp. Endocrinol. 2013, 181, 310–315. [Google Scholar] [CrossRef]
- Santos, L.C.; Ruiz-Oliveira, J.; Silva, P.F.; Luchiari, A.C. Caffeine dose–response relationship and behavioral screening in zebrafish. Quest. Caffeine 2017, 21, 87–105. [Google Scholar]
- Collier, A.D. Anxiety-Like Behaviors and c-Fos Expression in Adult Zebrafish: Effects of Housing Conditions, Alcohol, and Caffeine; The University of Southern Mississippi: Hattiesburg, MS, USA, 2017. [Google Scholar]
- Alia, A.O.; Petrunich-Rutherford, M.L.J.P. Anxiety-like behavior and whole-body cortisol responses to components of energy drinks in zebrafish (Danio rerio). Peerj 2019, 7, e7546. [Google Scholar] [CrossRef]
- Steele, W.B.; Mole, R.A.; Brooks, B.W. Experimental Protocol for Examining Behavioral Response Profiles in Larval Fish: Application to the Neuro-stimulant Caffeine. J. Vis. Exp. 2018, 137, e57938. [Google Scholar] [CrossRef]
- Bailey, J.; Oliveri, A.; Levin, E.D. Zebrafish model systems for developmental neurobehavioral toxicology. Birth Defects Res. C Embryo Today 2013, 99, 14–23. [Google Scholar] [CrossRef] [PubMed]
- Padilla, S.; MacPhail, R. Using zebrafish to assess developmental neurotoxicity. In Reproductive and Developmental Toxicology; Elsevier: Amsterdam, The Netherlands, 2011; pp. 179–191. [Google Scholar]
- Reichert, S.; Arocas, O.P.; Rihel, J.J.N. The neuropeptide galanin is required for homeostatic rebound sleep following increased neuronal activity. Neuron 2019, 104, 370–384.e375. [Google Scholar] [CrossRef] [PubMed]
- Grone, B.P.; Qu, T.; Baraban, S.C. Behavioral Comorbidities and Drug Treatments in a Zebrafish scn1lab Model of Dravet Syndrome. eNeuro 2017, 4, ENEURO.0066-17.2017. [Google Scholar] [CrossRef] [PubMed]
- Yeh, C.H.; Liao, Y.F.; Chang, C.Y.; Tsai, J.N.; Wang, Y.H.; Cheng, C.C.; Wen, C.C.; Chen, Y.H. Caffeine treatment disturbs the angiogenesis of zebrafish embryos. Drug Chem. Toxicol. 2012, 35, 361–365. [Google Scholar] [CrossRef]
- Rodriguez, R.S.; Haugen, R.; Rueber, A.; Huang, C.C. Reversible neuronal and muscular toxicity of caffeine in developing vertebrates. Comp. Biochem. Physiol. C Toxicol. Pharmacol. 2014, 163, 47–54. [Google Scholar] [CrossRef]
- Zheng, X.; Dai, W.; Chen, X.; Wang, K.; Zhang, W.; Liu, L.; Hou, J. Caffeine reduces hepatic lipid accumulation through regulation of lipogenesis and ER stress in zebrafish larvae. J. Biomed. Sci. 2015, 22, 105. [Google Scholar] [CrossRef]
- Ribatti, D.; Crivellato, E. Immune cells and angiogenesis. J. Cell Mol. Med. 2009, 13, 2822–2833. [Google Scholar] [CrossRef]
- Irwin, M.R. Sleep and inflammation: Partners in sickness and in health. Nat. Rev. Immunol. 2019, 19, 702–715. [Google Scholar] [CrossRef]
- Capiotti, K.M.; Menezes, F.P.; Nazario, L.R.; Pohlmann, J.B.; de Oliveira, G.M.; Fazenda, L.; Bogo, M.R.; Bonan, C.D.; Da Silva, R.S. Early exposure to caffeine affects gene expression of adenosine receptors, DARPP-32 and BDNF without affecting sensibility and morphology of developing zebrafish (Danio rerio). Neurotoxicol. Teratol. 2011, 33, 680–685. [Google Scholar] [CrossRef]
- Abdelkader, T.S.; Chang, S.-N.; Lee, J.-M.; Song, J.; Oh, H.; Park, J.-H.J.L. Effect of caffeine on Alzheimer’s molecular factors in correlation with involved cell communication systems in developing zebrafish Danio rerio. Lab. J. Lab. Med. 2015, 39, 165–174. [Google Scholar] [CrossRef]
- Lantz-McPeak, S.; Guo, X.; Cuevas, E.; Dumas, M.; Newport, G.D.; Ali, S.F.; Paule, M.G.; Kanungo, J. Developmental toxicity assay using high content screening of zebrafish embryos. J. Appl. Toxicol. 2015, 35, 261–272. [Google Scholar] [CrossRef] [PubMed]
- Pruvot, B.; Quiroz, Y.; Voncken, A.; Jeanray, N.; Piot, A.; Martial, J.A.; Muller, M. A panel of biological tests reveals developmental effects of pharmaceutical pollutants on late stage zebrafish embryos. Reprod. Toxicol. 2012, 34, 568–583. [Google Scholar] [CrossRef]
- Rah, Y.C.; Yoo, M.H.; Choi, J.; Park, S.; Park, H.C.; Oh, K.H.; Lee, S.H.; Kwon, S.Y. In vivo assessment of hair cell damage and developmental toxicity caused by gestational caffeine exposure using zebrafish (Danio rerio) models. Neurotoxicol. Teratol. 2017, 64, 1–7. [Google Scholar] [CrossRef] [PubMed]
- Maeda, H.; Hasumi, A.; Yoshida, K.-i. Caffeine-induced bradycardia, death, and anxiety-like behavior in zebrafish larvae. Forensic Toxicol. 2021, 39, 427–436. [Google Scholar] [CrossRef]
- Tran, S.; Fulcher, N.; Nowicki, M.; Desai, P.; Tsang, B.; Facciol, A.; Chow, H.; Gerlai, R. Time-dependent interacting effects of caffeine, diazepam, and ethanol on zebrafish behaviour. Prog. Neuropsychopharmacol. Biol. Psychiatry 2017, 75, 16–27. [Google Scholar] [CrossRef]
- Falcão, M.A.P.; de Souza, L.S.; Dolabella, S.S.; Guimarães, A.G.; Walker, C.I.B. Zebrafish as an alternative method for determining the embryo toxicity of plant products: A systematic review. Environ. Sci. Pollut. Res. 2018, 25, 35015–35026. [Google Scholar] [CrossRef]
- Atik, A.; De Matteo, R.; Boomgardt, M.; Rees, S.; Harding, R.; Cheong, J.; Rana, S.; Crossley, K.; Tolcos, M. Impact of High-Dose Caffeine on the Preterm Ovine Cerebrum and Cerebellum. Front. Physiol. 2019, 10, 990. [Google Scholar] [CrossRef]
- Segu, A.; Kannan, N.N. The duration of caffeine treatment plays an essential role in its effect on sleep and circadian rhythm. Sleep. Adv. 2023, 4, zpad014. [Google Scholar] [CrossRef]
- Reichert, C.F.; Deboer, T.; Landolt, H.P. Adenosine, caffeine, and sleep-wake regulation: State of the science and perspectives. J. Sleep. Res. 2022, 31, e13597. [Google Scholar] [CrossRef]
- Porkka-Heiskanen, T.; Kalinchuk, A.V. Adenosine, energy metabolism and sleep homeostasis. Sleep Med. Rev. 2011, 15, 123–135. [Google Scholar] [CrossRef]
- Gandhi, A. The Regulation of Sleep and Circadian Rhythms: The Role of Melatonin and Adenosine in Zebrafish; California Institute of Technology: Pasadena, CA, USA, 2015. [Google Scholar]
- Rosemberg, D.B.; Rico, E.P.; Guidoti, M.R.; Dias, R.D.; Souza, D.O.; Bonan, C.D.; Bogo, M.R. Adenosine deaminase-related genes: Molecular identification, tissue expression pattern and truncated alternative splice isoform in adult zebrafish (Danio rerio). Life Sci. 2007, 81, 1526–1534. [Google Scholar] [CrossRef] [PubMed]
- Maximino, C.; Lima, M.G.; Olivera, K.R.; Picanco-Diniz, D.L.; Herculano, A.M. Adenosine A1, but not A2, receptor blockade increases anxiety and arousal in Zebrafish. Basic Clin. Pharmacol. Toxicol. 2011, 109, 203–207. [Google Scholar] [CrossRef] [PubMed]
- Casale, C.E.; Goel, N. Genetic Markers of Differential Vulnerability to Sleep Loss in Adults. Genes 2021, 12, 1317. [Google Scholar] [CrossRef] [PubMed]
- Lazarus, M.; Oishi, Y.; Bjorness, T.E.; Greene, R.W. Gating and the Need for Sleep: Dissociable Effects of Adenosine A1 and A2A Receptors. Front. Neurosci. 2019, 13, 740. [Google Scholar] [CrossRef]
- Jagannath, A.; Varga, N.; Dallmann, R.; Rando, G.; Gosselin, P.; Ebrahimjee, F.; Taylor, L.; Mosneagu, D.; Stefaniak, J.; Walsh, S.; et al. Adenosine integrates light and sleep signalling for the regulation of circadian timing in mice. Nat. Commun. 2021, 12, 2113. [Google Scholar] [CrossRef]
- Gonzales, E.D.; Tanenhaus, A.K.; Zhang, J.; Chaffee, R.P.; Yin, J.C. Early-onset sleep defects in Drosophila models of Huntington’s disease reflect alterations of PKA/CREB signaling. Hum. Mol. Genet. 2016, 25, 837–852. [Google Scholar] [CrossRef]
- Mayr, B.; Montminy, M. Transcriptional regulation by the phosphorylation-dependent factor CREB. Nat. Rev. Mol. Cell Biol. 2001, 2, 599–609. [Google Scholar] [CrossRef]
- Tran, T.-D.T.; Park, J.; Kim, D.Y.; Han, I.-O. Caffeine-induced protein kinase A activation restores cognitive deficits induced by sleep deprivation by regulating O-GlcNAc cycling in adult zebrafish. Am. J. Physiol.-Cell Physiol. 2024, 326, C978–C989. [Google Scholar] [CrossRef]
- An, X.; Lv, J.; Wang, F. Pterostilbene inhibits melanogenesis, melanocyte dendricity and melanosome transport through cAMP/PKA/CREB pathway. Eur. J. Pharmacol. 2022, 932, 175231. [Google Scholar] [CrossRef]
- Dworkin, S.; Heath, J.K.; deJong-Curtain, T.A.; Hogan, B.M.; Lieschke, G.J.; Malaterre, J.; Ramsay, R.G.; Mantamadiotis, T. CREB activity modulates neural cell proliferation, midbrain–hindbrain organization and patterning in zebrafish. Dev. Biol. 2007, 307, 127–141. [Google Scholar] [CrossRef]
- Goodwin, N.; Karp, N.A.; Blackledge, S.; Clark, B.; Keeble, R.; Kovacs, C.; Murray, K.N.; Price, M.; Thompson, P.; Bussell, J. Standardized welfare terms for the zebrafish community. Zebrafish 2016, 13, S-164–S-168. [Google Scholar] [CrossRef] [PubMed]
- Rihel, J.; Prober, D.A.; Arvanites, A.; Lam, K.; Zimmerman, S.; Jang, S.; Haggarty, S.J.; Kokel, D.; Rubin, L.L.; Peterson, R.T.; et al. Zebrafish behavioral profiling links drugs to biological targets and rest/wake regulation. Science 2010, 327, 348–351. [Google Scholar] [CrossRef] [PubMed]


| Locus | Sequence (3′ → 5′) | Size Range (bp) |
|---|---|---|
| cry1a | F: CCGTGGAGACCTGTGGAT R: GTGGAAGAACTGCTGGAAGAAG | 136 |
| cry2 | F: CACCACCGCTGTCTGAATC R: TCTTCTGTTTGCTGGGGGT | 114 |
| Bmal1a | F: GAAGACATTACGAGGGGCCA R: AGAGGAAACCATCAGCAGCC | 113 |
| per1b | F: AGGAAGGCTGACAGATGATGAATG R: CCAGAGTGGGCTAAAGCGAAGTA | 149 |
| per2 | F: ACGAGGACAAGCCAGAGGAACG R: GCACTGGCTGGTGATGGAGA | 194 |
| per3 | F: GTTCTGGCGGAGTAATGGAG R: TGACGACGTTTTACTGGTGC | 116 |
| adora1a | F: GGCTGGAACAATCTGGACAAG R: AGGATGAGTGGTGGCAACA | 146 |
| adora1b | F: GAACAAGAAGGTGTCCAGTCAT R: CGTGAGCAGAATGGCAATGTA | 196 |
| adora2aa | F: ACGCCTACAGAATACGAGAGT R: CGCACCAGACCATTGACTT | 160 |
| adora2ab | F: CTCTTCATCGCTTGCTTTGTG R: CTGTGACGAGGCTGTTGT | 121 |
| adora2b | F: GCTCAAGAACGCCACCAAT R: ACCACAGACCGATGCTTATAGT | 107 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Wei, Y.; Miao, Z.; Ye, H.; Wu, M.; Wei, X.; Zhang, Y.; Cai, L. The Effect of Caffeine Exposure on Sleep Patterns in Zebrafish Larvae and Its Underlying Mechanism. Clocks & Sleep 2024, 6, 749-763. https://doi.org/10.3390/clockssleep6040048
Wei Y, Miao Z, Ye H, Wu M, Wei X, Zhang Y, Cai L. The Effect of Caffeine Exposure on Sleep Patterns in Zebrafish Larvae and Its Underlying Mechanism. Clocks & Sleep. 2024; 6(4):749-763. https://doi.org/10.3390/clockssleep6040048
Chicago/Turabian StyleWei, Yuanzheng, Zongyu Miao, Huixin Ye, Meihui Wu, Xinru Wei, Yu Zhang, and Lei Cai. 2024. "The Effect of Caffeine Exposure on Sleep Patterns in Zebrafish Larvae and Its Underlying Mechanism" Clocks & Sleep 6, no. 4: 749-763. https://doi.org/10.3390/clockssleep6040048
APA StyleWei, Y., Miao, Z., Ye, H., Wu, M., Wei, X., Zhang, Y., & Cai, L. (2024). The Effect of Caffeine Exposure on Sleep Patterns in Zebrafish Larvae and Its Underlying Mechanism. Clocks & Sleep, 6(4), 749-763. https://doi.org/10.3390/clockssleep6040048





