Portability of Polygenic Risk Scores for Sleep Duration, Insomnia and Chronotype in 33,493 Individuals
Abstract
1. Introduction
2. Results
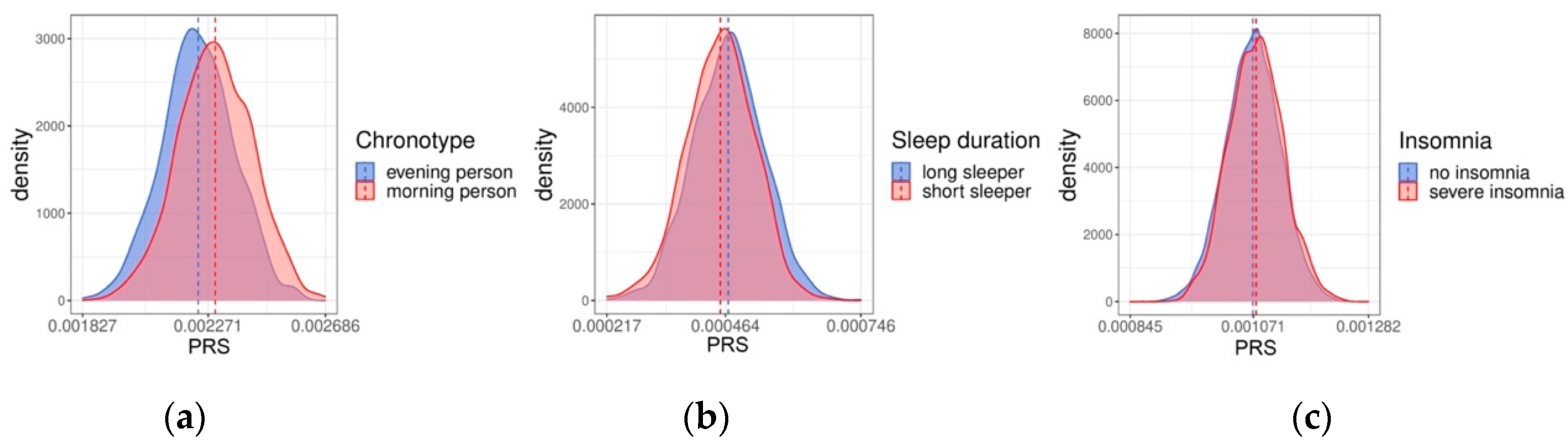
2.1. Polygenic Risk Scores for the FINRISK Cohorts
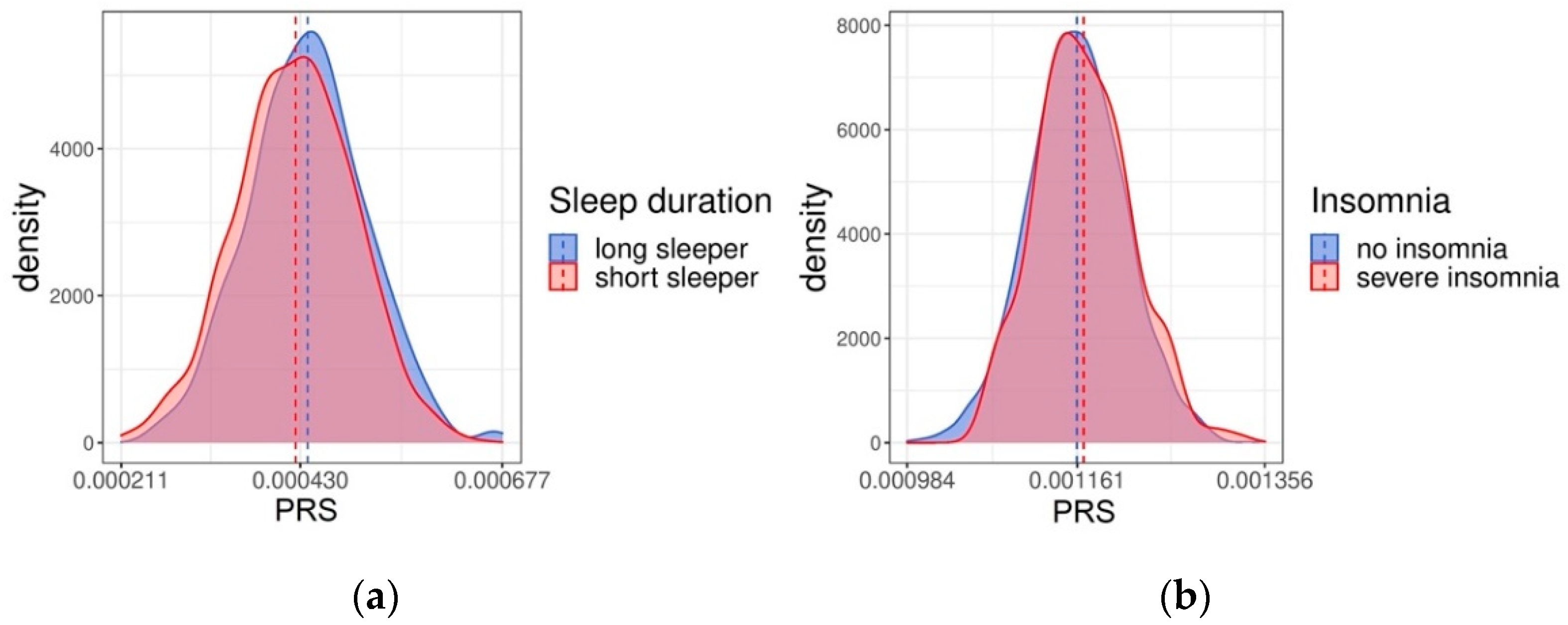
2.2. Validation of the PRSs in Health 2000/2011 Cohort
3. Discussion
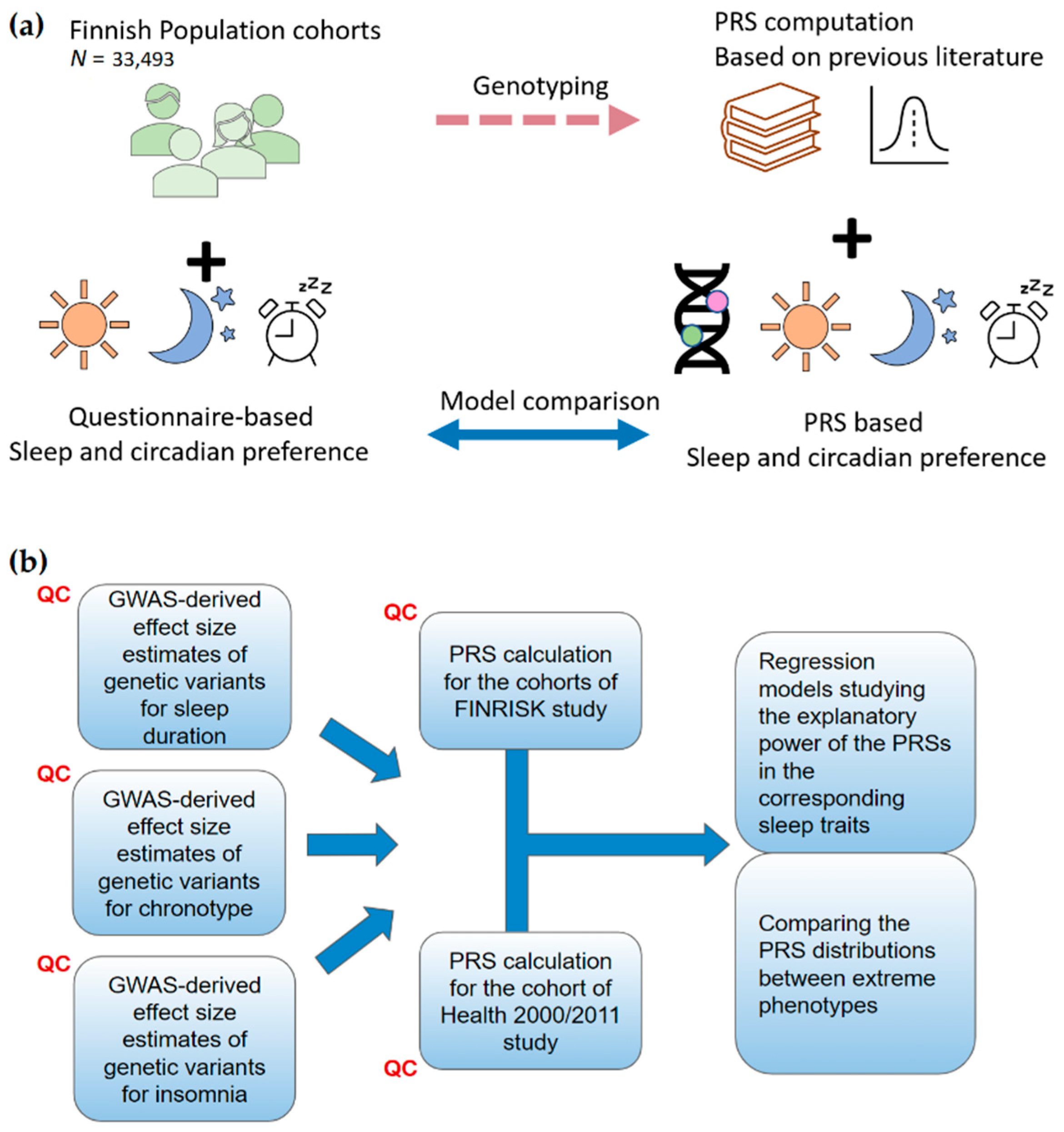
4. Materials and Methods
4.1. GWAS Summary Statistics
4.2. Target Data
4.3. Assessing Sleep Traits
4.4. Generating Risk Scores
4.5. Estimating the Explanatory Power of the Risk Scores
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
Appendix A
| Sleep Duration | Chronotype | Insomnia | |
|---|---|---|---|
| FINRISK ‘92 | unavailable | unavailable | Do you suffer from insomnia?
|
| FINRISK ‘97 | unavailable | unavailable | Do you suffer from insomnia?
|
| FINRISK ‘02 | unavailable | unavailable | Do you suffer from insomnia?
|
| FINRISK ‘07 | How many hours on average do you sleep in 24 h?
| There are so-called “morning people” (early to rise, early to bed) and “evening people” (late to rise, late to bed). Which are you?
| Do you suffer from insomnia?
|
| FINRISK ’12 | How many hours on average do you sleep in 24 h?
| There are so-called “morning people” (early to rise, early to bed) and “evening people” (late to rise, late to bed). Which are you?
| Do you suffer from insomnia?
|
| Health 2000 | How many hours on average do you sleep in 24 h?
| unavailable | Sleeping
|
| Health 2011 | How many hours on average do you sleep in 24 h?
| unavailable | Sleeping
|
References
- Partinen, M.; Kaprio, J.; Koskenvuo, M.; Putkonen, P.; Langinvainio, H. Genetic and Environmental Determination of Human Sleep. Sleep 1983, 6, 179–185. [Google Scholar] [CrossRef]
- Vink, J.M.; Groot, A.S.; Kerkhof, G.A.; Boomsma, D.I. Genetic analysis of morningness and eveningness. Chronobiol. Int. 2001, 18, 809–822. [Google Scholar] [CrossRef]
- Koskenvuo, M.; Hublin, C.; Partinen, M.; Heikkilä, K.; Kaprio, J. Heritability of diurnal type: A nationwide study of 8753 adult twin pairs. J. Sleep Res. 2007, 16, 156–162. [Google Scholar] [CrossRef]
- Madrid-Valero, J.J.; Rubio-Aparicio, M.; Gregory, A.M.; Sánchez-Meca, J.; Ordoñana, J.R. The heritability of insomnia: Systematic review and meta-analysis of twin studies. Sleep Med. Rev. 2021, 58, 101437. [Google Scholar] [CrossRef]
- Dashti, H.S.; Jones, S.E.; Wood, A.R.; Lane, J.M.; van Hees, V.T.; Wang, H.; Rhodes, J.A.; Song, Y.; Patel, K.; Anderson, S.G.; et al. Genome-wide association study identifies genetic loci for self-reported habitual sleep duration supported by accelerometer-derived estimates. Nat. Commun. 2019, 10, 1100. [Google Scholar] [CrossRef]
- Jones, S.E.; Lane, J.M.; Wood, A.R.; van Hees, V.T.; Tyrrell, J.; Beaumont, R.N.; Jeffries, A.R.; Dashti, H.S.; Hillsdon, M.; Ruth, K.S.; et al. Genome-wide association analyses of chronotype in 697,828 individuals provides insights into circadian rhythms. Nat. Commun. 2019, 10, 343. [Google Scholar] [CrossRef]
- Lane, J.M.; Sleep, H.A.I.; Jones, S.E.; Dashti, H.S.; Wood, A.R.; Aragam, K.G.; van Hees, V.T.; Strand, L.B.; Winsvold, B.S.; Wang, H.; et al. Biological and clinical insights from genetics of insomnia symptoms. Nat. Genet. 2019, 51, 387–393. [Google Scholar] [CrossRef]
- Merikanto, I.; Kantojärvi, K.; Partonen, T.; Pesonen, A.-K.; Paunio, T. Genetic variants for morningness in relation to habitual sleep-wake behavior and diurnal preference in a population-based sample of 17,243 adults. Sleep Med. 2021, 80, 322–332. [Google Scholar] [CrossRef]
- Lane, J.M.; Liang, J.; Vlasac, I.; Anderson, S.G.; A Bechtold, D.; Bowden, J.; Emsley, R.; Gill, S.; A Little, M.; I Luik, A.; et al. Genome-wide association analyses of sleep disturbance traits identify new loci and highlight shared genetics with neuropsychiatric and metabolic traits. Nat. Genet. 2016, 49, 274–281. [Google Scholar] [CrossRef]
- Knutson, K.L.; von Schantz, M. Associations between chronotype, morbidity and mortality in the UK Biobank cohort. Chronobiol. Int. 2018, 35, 1045–1053. [Google Scholar] [CrossRef]
- Patterson, F.; Malone, S.K.; Grandner, M.A.; Lozano, A.; Perkett, M.; Hanlon, A. Interactive effects of sleep duration and morning/evening preference on cardiovascular risk factors. Eur. J. Public Health 2018, 28, 155–161. [Google Scholar] [CrossRef]
- Merikanto, I.; Lahti, T.; Puolijoki, H.; Vanhala, M.; Peltonen, M.; Laatikainen, T.; Vartiainen, E.; Salomaa, V.; Kronholm, E.; Partonen, T. Associations of Chronotype and Sleep with Cardiovascular Diseases and Type 2 Diabetes. Chronobiol. Int. 2013, 30, 470–477. [Google Scholar] [CrossRef]
- Lewis, C.M.; Vassos, E. Polygenic risk scores: From research tools to clinical instruments. Genome Med. 2020, 12, 44. [Google Scholar] [CrossRef]
- Choi, S.W.; Mak, T.S.-H.; O’Reilly, P.F. Tutorial: A guide to performing polygenic risk score analyses. Nat. Protoc. 2020, 15, 2759–2772. [Google Scholar] [CrossRef]
- Jukarainen, S.; Kiiskinen, T.; Kuitunen, S.; Havulinna, A.S.; Karjalainen, J.; Cordioli, M.; Rämö, J.T.; Mars, N.; Samocha, K.E.; Ollila, H.M.; et al. Genetic risk factors have a substantial impact on healthy life years. Nat. Med. 2022, 28, 1893–1901. [Google Scholar] [CrossRef]
- Grove, J.; Ripke, S.; Als, T.D.; Mattheisen, M.; Walters, R.K.; Won, H.; Pallesen, J.; Agerbo, E.; Andreassen, O.A.; Anney, R.; et al. Identification of common genetic risk variants for autism spectrum disorder. Nat. Genet. 2019, 51, 431–444. [Google Scholar] [CrossRef]
- Neumann, A.; Jolicoeur-Martineau, A.; Szekely, E.; Sallis, H.M.; O’Donnel, K.; Greenwood, C.M.; Levitan, R.; Meaney, M.J.; Wazana, A.; Evans, J.; et al. Combined polygenic risk scores of different psychiatric traits predict general and specific psychopathology in childhood. J. Child Psychol. Psychiatry 2021, 63, 636–664. [Google Scholar] [CrossRef]
- Mars, N.; Kerminen, S.; Feng, Y.-C.A.; Kanai, M.; Läll, K.; Thomas, L.F.; Skogholt, A.H.; Parolo, P.D.B.; Neale, B.M.; Smoller, J.W.; et al. Genome-wide risk prediction of common diseases across ancestries in one million people. Cell Genom. 2022, 2, 100118. [Google Scholar] [CrossRef]
- Duncan, L.; Shen, H.; Gelaye, B.; Meijsen, J.; Ressler, K.; Feldman, M.; Peterson, R.; Domingue, B. Analysis of polygenic risk score usage and performance in diverse human populations. Nat. Commun. 2019, 10, 3328. [Google Scholar] [CrossRef]
- Evans, D.G.; van Veen, E.M.; Byers, H.; Roberts, E.; Howell, A.; Howell, S.J.; Harkness, E.F.; Brentnall, A.; Cuzick, J.; Newman, W.G. The importance of ethnicity: Are breast cancer polygenic risk scores ready for women who are not of White European origin? Int. J. Cancer 2021, 150, 73–79. [Google Scholar] [CrossRef]
- Dikilitas, O.; Schaid, D.J.; Kosel, M.L.; Carroll, R.J.; Chute, C.G.; Denny, J.C.; Fedotov, A.; Feng, Q.; Hakonarson, H.; Jarvik, G.P.; et al. Predictive Utility of Polygenic Risk Scores for Coronary Heart Disease in Three Major Racial and Ethnic Groups. Am. J. Hum. Genet. 2020, 106, 707–716. [Google Scholar] [CrossRef]
- Kassam, I.; Foo, L.-L.; Lanca, C.; Xu, L.; Hoang, Q.V.; Cheng, C.-Y.; Hysi, P.; Saw, S.-M. The Potential of Current Polygenic Risk Scores to Predict High Myopia and Myopic Macular Degeneration in Multiethnic Singapore Adults. Ophthalmology 2022, 129, 890–902. [Google Scholar] [CrossRef]
- Sudlow, C.; Gallacher, J.; Allen, N.; Beral, V.; Burton, P.; Danesh, J.; Downey, P.; Elliott, P.; Green, J.; Landray, M.; et al. UK Biobank: An Open Access Resource for Identifying the Causes of a Wide Range of Complex Diseases of Middle and Old Age. PLoS Med. 2015, 12, e1001779. [Google Scholar] [CrossRef]
- Hu, Y.; Shmygelska, A.; Tran, D.; Eriksson, N.; Tung, J.Y.; Hinds, D.A. GWAS of 89,283 individuals identifies genetic variants associated with self-reporting of being a morning person. Nat. Commun. 2016, 7, 10448. [Google Scholar] [CrossRef]
- Borodulin, K.; Tolonen, H.; Jousilahti, P.; Jula, A.; Juolevi, A.; Koskinen, S.; Kuulasmaa, K.; Laatikainen, T.; Männistö, S.; Peltonen, M.; et al. Cohort Profile: The National FINRISK Study. Leuk. Res. 2017, 47, 696. [Google Scholar] [CrossRef]
- Horne, J.A.; Ostberg, O. A self-Assessment Questionnaire to Determine Morningness-Eveningness in Human Circadian Rhythms. 1976. Available online: https://pubmed.ncbi.nlm.nih.gov/1027738/ (accessed on 23 January 2022).
- Chang, C.C.; Chow, C.C.; Tellier, L.C.; Vattikuti, S.; Purcell, S.M.; Lee, J.J. Second-generation PLINK: Rising to the challenge of larger and richer datasets. Gigascience 2015, 4, 7. [Google Scholar] [CrossRef]
- Cardon, L.R.; Palmer, L.J. Population stratification and spurious allelic association. Lancet 2003, 361, 598–604. [Google Scholar] [CrossRef]
- Mineta, K.; Goto, K.; Gojobori, T.; Alkuraya, F.S. Population structure of indigenous inhabitants of Arabia. PLoS Genet. 2021, 17, e1009210. [Google Scholar] [CrossRef]
- Takeuchi, F.; Katsuya, T.; Kimura, R.; Nabika, T.; Isomura, M.; Ohkubo, T.; Tabara, Y.; Yamamoto, K.; Yokota, M.; Liu, X.; et al. The fine-scale genetic structure and evolution of the Japanese population. PLoS ONE 2017, 12, e0185487. [Google Scholar] [CrossRef]
- Yepiskoposyan, L.; Hovhannisyan, A.; Khachatryan, Z. Genetic Structure of the Armenian Population. Arch. Immunol. Ther. Exp. 2016, 64, 113–116. [Google Scholar] [CrossRef]
| PRS in Linear Model | PRS in Logistic Model | |||||
|---|---|---|---|---|---|---|
| N | R2 | p | N | pR2 | p | |
| Chronotype | 8530 | 0.021 | <0.001 | 2993 | 0.033 | <0.001 |
| Sleep duration | 7647 | 0.004 | <0.001 | 2016 | 0.010 | <0.001 |
| Insomnia | - | - | - | 15,843 | 0.003 | <0.001 |
| PRS in Linear Model | PRS in Logistic Model | |||||
|---|---|---|---|---|---|---|
| N | R2 | p | N | pR2 | p | |
| Sleep duration | 6389 | 0.004 | <0.001 | 1812 | 0.008 | <0.001 |
| Insomnia | - | - | - | 3335 | 0.003 | 0.03 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Perkiö, A.; Merikanto, I.; Kantojärvi, K.; Paunio, T.; Sinnott-Armstrong, N.; Jones, S.E.; Ollila, H.M. Portability of Polygenic Risk Scores for Sleep Duration, Insomnia and Chronotype in 33,493 Individuals. Clocks & Sleep 2023, 5, 10-20. https://doi.org/10.3390/clockssleep5010002
Perkiö A, Merikanto I, Kantojärvi K, Paunio T, Sinnott-Armstrong N, Jones SE, Ollila HM. Portability of Polygenic Risk Scores for Sleep Duration, Insomnia and Chronotype in 33,493 Individuals. Clocks & Sleep. 2023; 5(1):10-20. https://doi.org/10.3390/clockssleep5010002
Chicago/Turabian StylePerkiö, Anna, Ilona Merikanto, Katri Kantojärvi, Tiina Paunio, Nasa Sinnott-Armstrong, Samuel E. Jones, and Hanna M. Ollila. 2023. "Portability of Polygenic Risk Scores for Sleep Duration, Insomnia and Chronotype in 33,493 Individuals" Clocks & Sleep 5, no. 1: 10-20. https://doi.org/10.3390/clockssleep5010002
APA StylePerkiö, A., Merikanto, I., Kantojärvi, K., Paunio, T., Sinnott-Armstrong, N., Jones, S. E., & Ollila, H. M. (2023). Portability of Polygenic Risk Scores for Sleep Duration, Insomnia and Chronotype in 33,493 Individuals. Clocks & Sleep, 5(1), 10-20. https://doi.org/10.3390/clockssleep5010002






