Differential Gene Expression in Brain and Liver Tissue of Wistar Rats after Rapid Eye Movement Sleep Deprivation
Abstract
1. Introduction
2. Material and Methods
2.1. REM Sleep Deprivation Procedure
2.2. RNA Extraction and Quality Analysis
2.3. Microarray: Labeling, Hybridization, and Data Analysis
2.4. Gene Ontology Analysis
2.5. Pathway Analysis
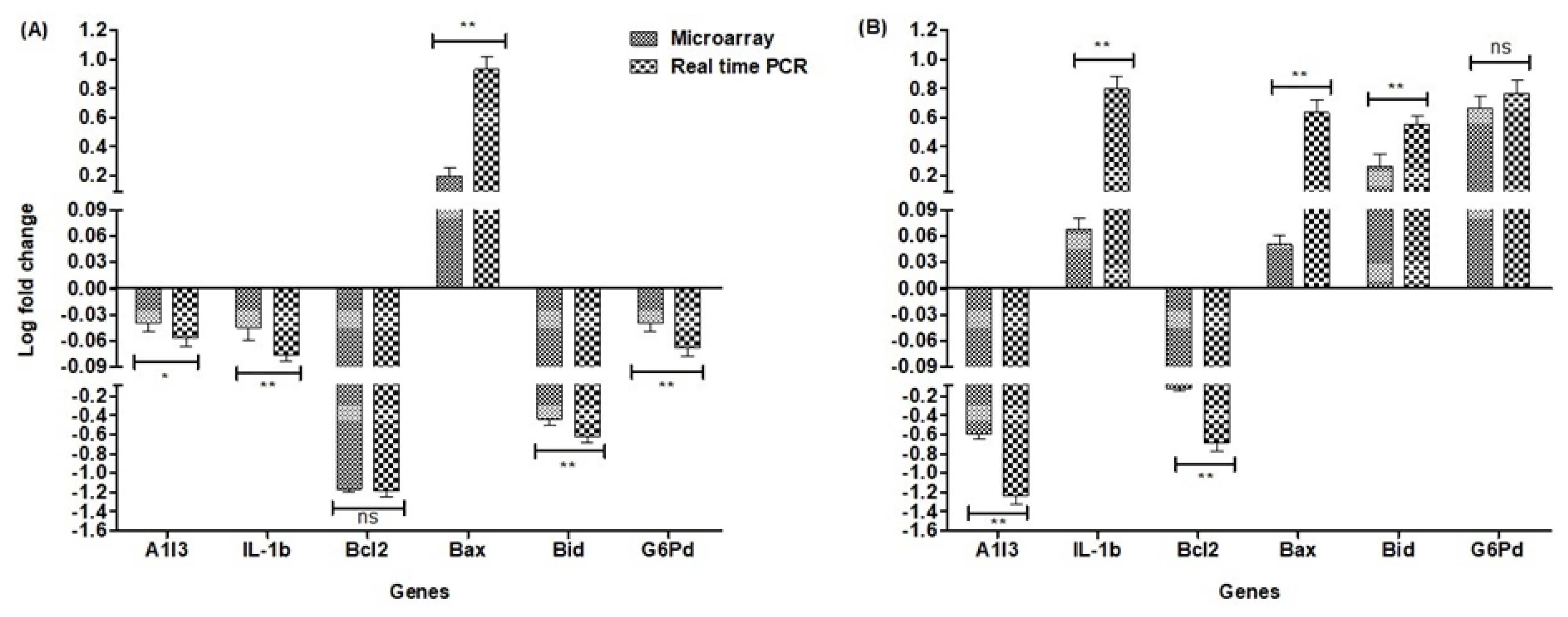
2.6. Validation of Array Expression with Real-Time Quantitative qPCR
2.7. Statistics
3. Results
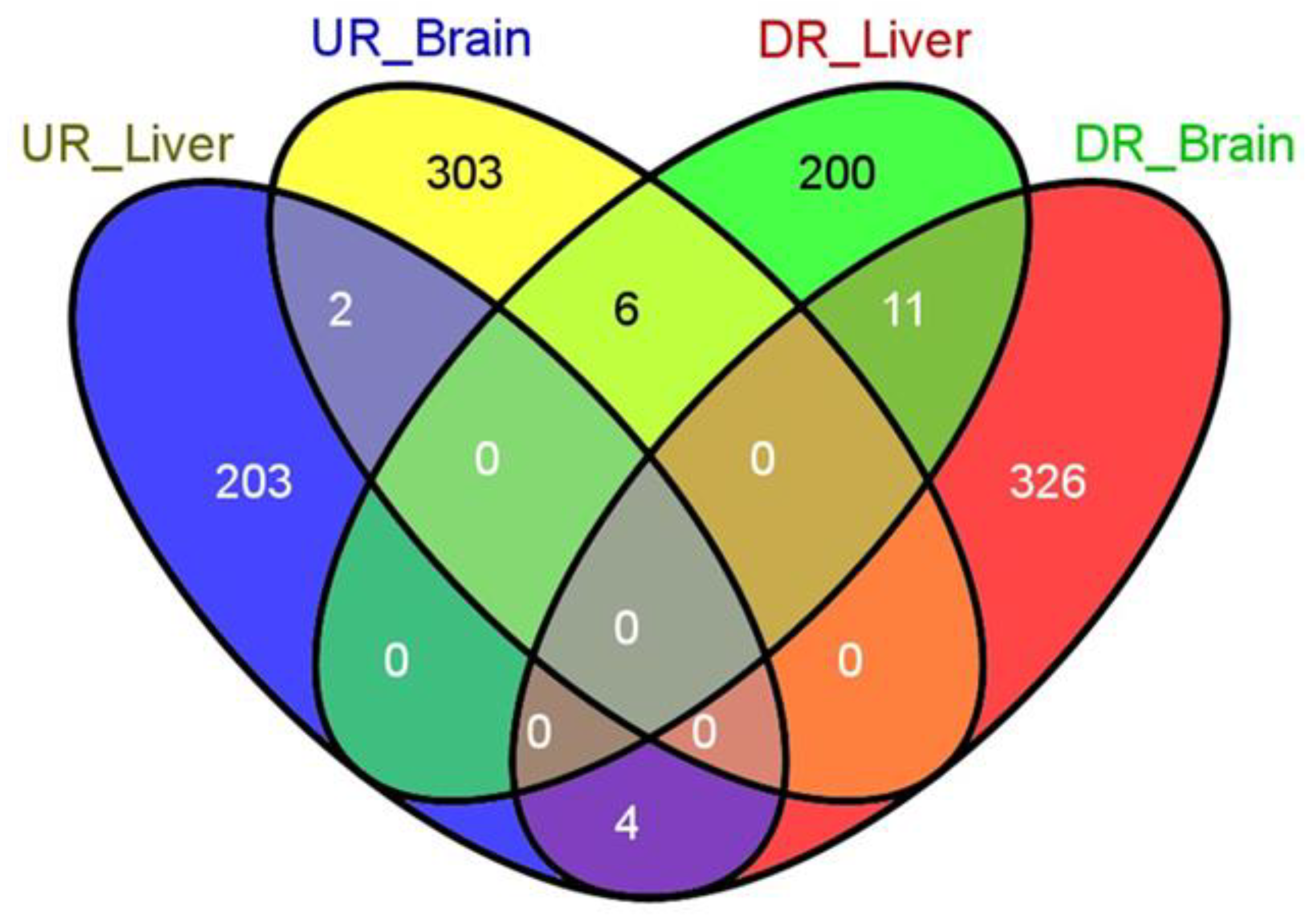
3.1. General Results
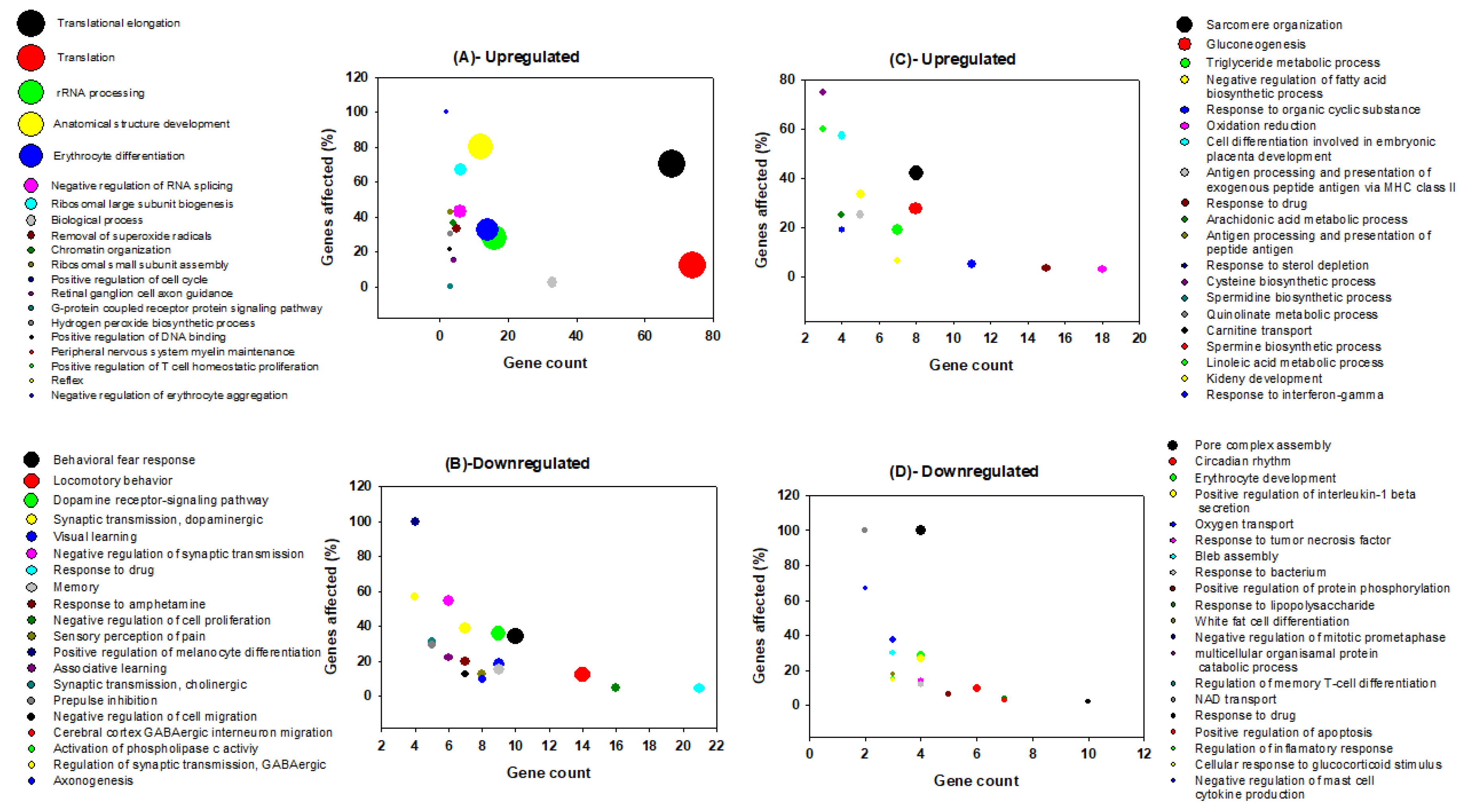
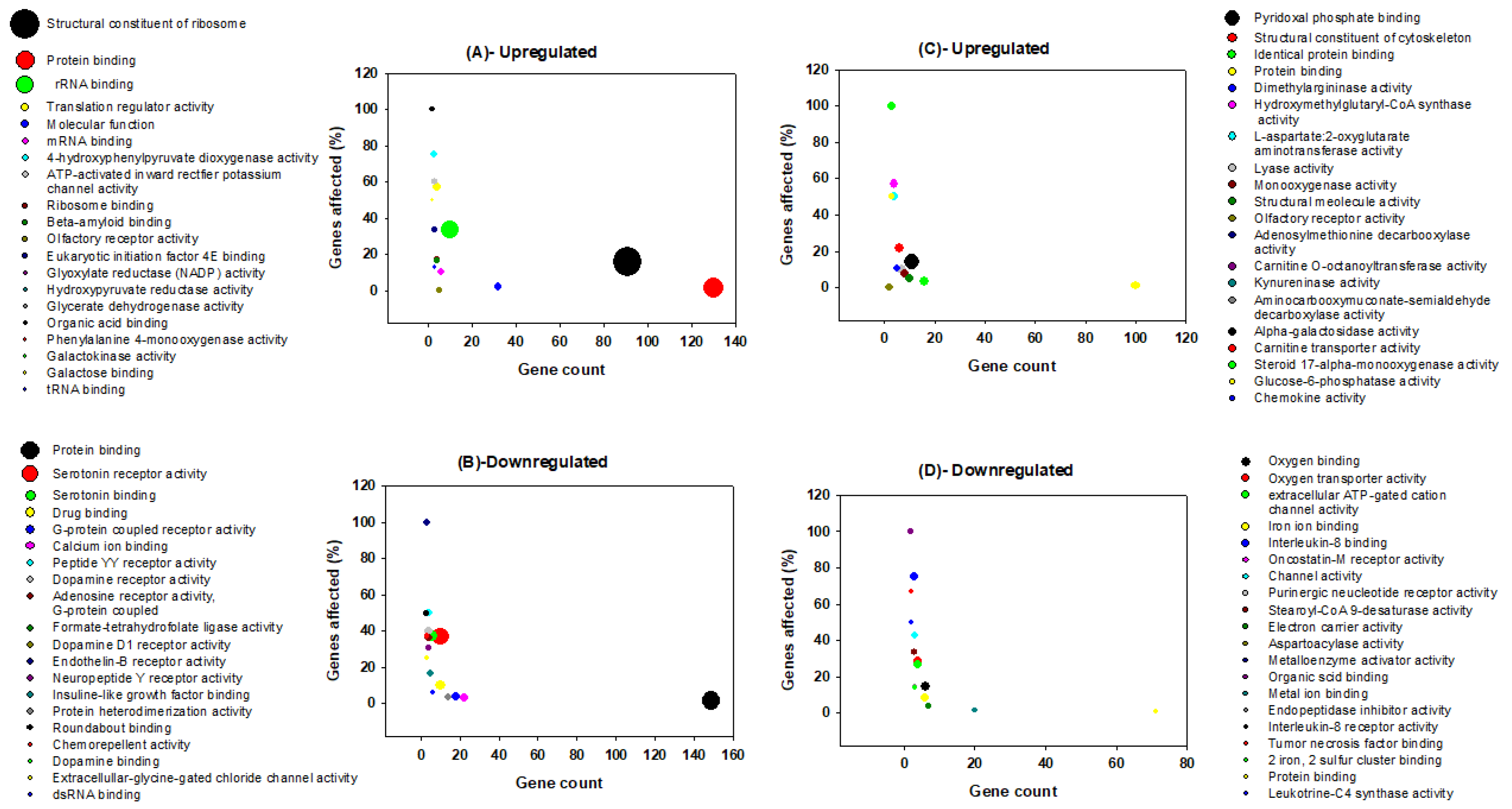
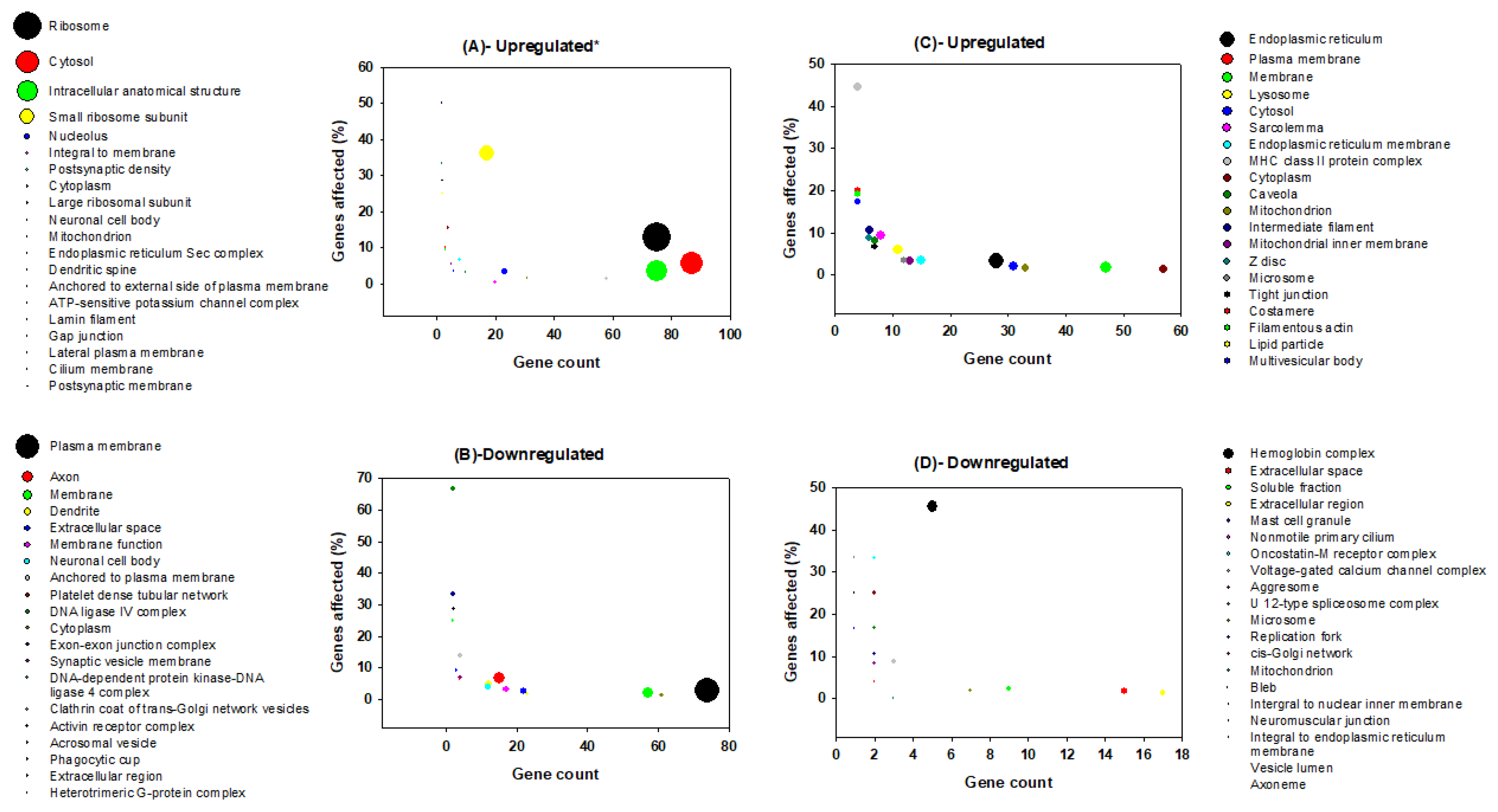
3.2. Gene Ontology Analysis
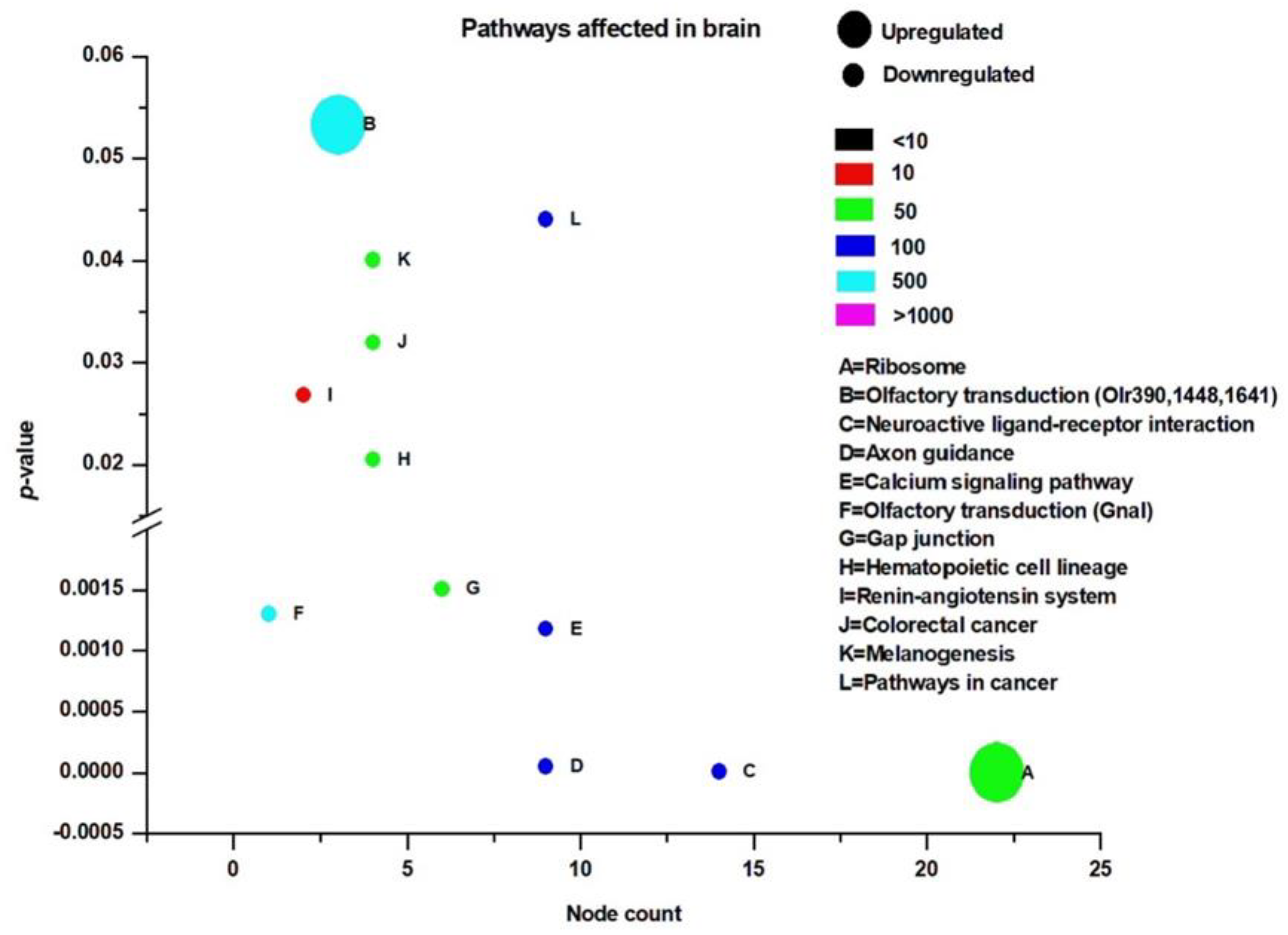
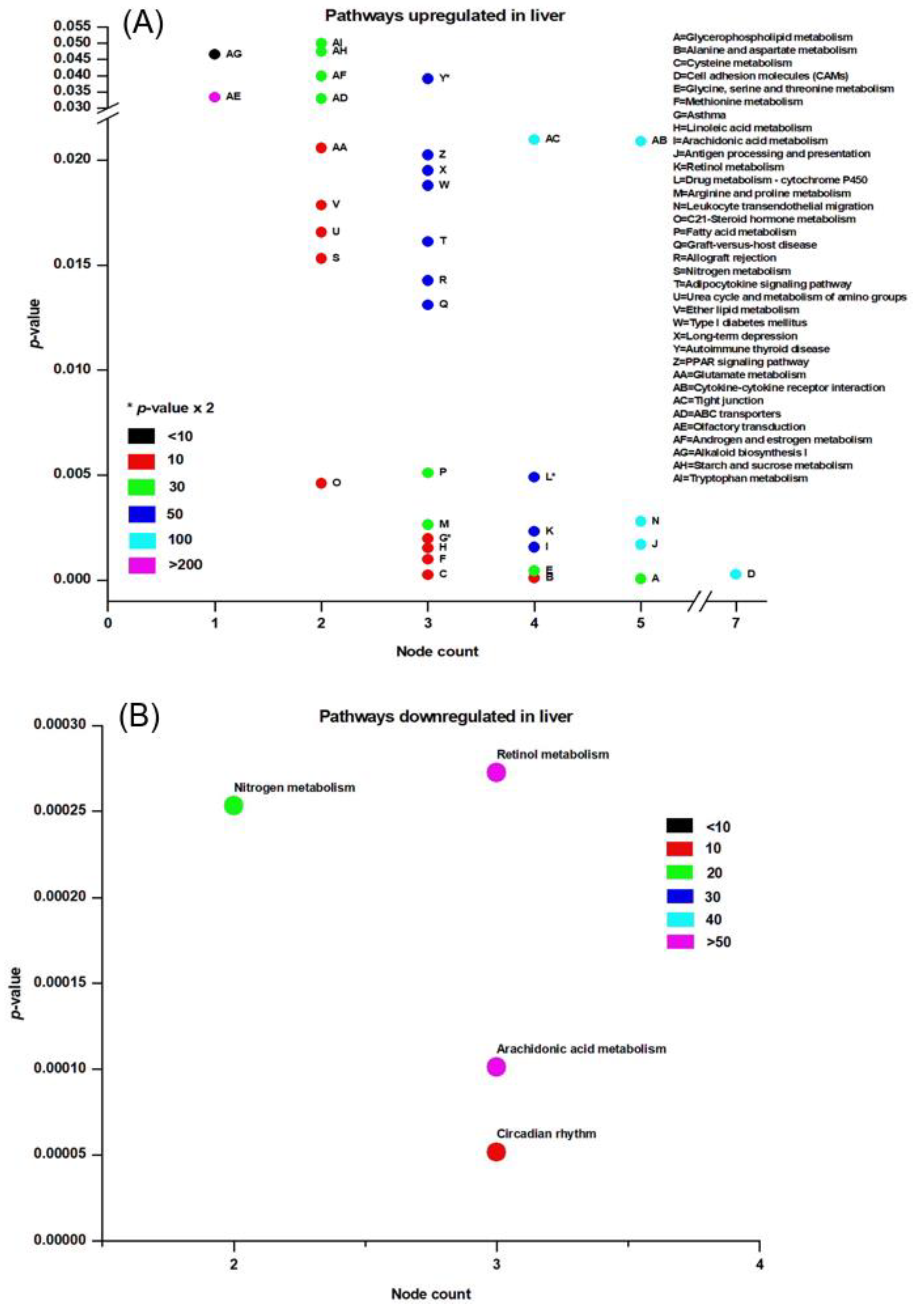
3.3. Pathway Analysis
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Kelly, M.L.; Collin, S.P.; Hemmi, J.M.; Lesku, J.A. Evidence for Sleep in Sharks and Rays: Behavioural, Physiological, and Evolutionary Considerations. Brain Behav. Evol. 2020. [Google Scholar] [CrossRef] [PubMed]
- Hendricks, J.C.; Finn, S.M.; Panckeri, K.A.; Chavkin, J.; Williams, J.A.; Sehgal, A.; Pack, A.I. Rest in Drosophila is a sleep-like state. Neuron 2000, 25, 129–138. [Google Scholar] [CrossRef]
- Shaw, P.J.; Cirelli, C.; Greensoan, R.J.; Tononi, G. Correlates of sleep and waking in Drosophila melanogaster. Science 2000, 287, 1834–1837. [Google Scholar] [CrossRef] [PubMed]
- Zimmerman, J.E.; Naidoo, N.; Raizen, D.M.; Pack, A.I. Conservation of sleep: Insights from non-mammalian model systems. Trends Neurosci. 2008, 31, 371–376. [Google Scholar] [CrossRef]
- Yokogawa, T.; Marin, W.; Faraco, J.; Pézeron, G.; Appelbaum, L.; Zhang, J.; Rosa, F.; Mourrain, F.; Mignot, E. Characterization of sleep in zebrafish and insomnia in hypocretin receptor mutants. PLoS Biol. 2007, 5, 2379–2397. [Google Scholar] [CrossRef]
- Prober, D.A.; Rihel, J.; Onah, A.A.; Sung, R.-J.; Schier, A.F. Hypocretin/Orexin Overexpression Induces An Insomnia-Like Phenotype in Zebrafish. J. Neurosci. 2006, 26, 13400–13410. [Google Scholar] [CrossRef]
- Zhdanova, I.V.; Wang, S.Y.; Leclair, O.U.; Danilova, N.P. Melatonin promotes sleep-like state in zebrafish. Brain Res. 2001, 903, 263–268. [Google Scholar] [CrossRef]
- Raizen, D.M.; Zimmerman, J.E.; Maycock, M.H.; Ta, U.D.; You, Y.J.; Sundaram, M.V.; Pack, A.I. Lethargus is a Caenorhabditis elegans sleep-like state. Nature 2008, 451, 569–572. [Google Scholar] [CrossRef]
- Eban-Rothschild, A.; Bloch, G. Circadian rhythms and sleep in honey bees. Honeybee Neurobiol. Behav. 2012, 31–45. [Google Scholar] [CrossRef]
- Eban-Rothschild, A.D.; Bloch, G. Differences in the sleep architecture of forager and young honeybees (Apis mellifera). J. Exp. Biol. 2008, 211, 2408–2416. [Google Scholar] [CrossRef]
- Klein, B.A.; Seeley, T.D. Work or sleep? Honeybee foragers opportunistically nap during the day when forage is not available. Anim. Behav. 2011, 82, 77–83. [Google Scholar] [CrossRef]
- Nagari, M.; Gera, A.; Jonsson, S.; Bloch, G. Bumble Bee Workers Give Up Sleep to Care for Offspring that Are Not Their Own. Curr. Biol. 2019, 29, 3488–3493. [Google Scholar] [CrossRef] [PubMed]
- Shaw, P.J.; Tortoni, G.; Greenspan, R.J.; Robinson, D.F. Stress response genes protect against lethal effects of sleep deprivation in Drosophila. Nature 2002. [Google Scholar] [CrossRef] [PubMed]
- Stephenson, R.; Chu, K.M.; Lee, J. Prolonged deprivation of sleep-like rest raises metabolic rate in the Pacific beetle cockroach, Diploptera punctata (Eschscholtz). J. Exp. Biol. 2007. [Google Scholar] [CrossRef] [PubMed]
- Montagna, P.; Lugaresi, E. Agrypnia Excitata: A generalized overactivity syndrome and a useful concept in the neurophysiopathology of sleep. Clin. Neurophysiol. 2002. [Google Scholar] [CrossRef]
- Newman, S.M.; Paletz, E.M.; Rattenborg, N.C.; Obermeyer, W.H.; Benca, R.M. Sleep deprivation in the pigeon using the Disk-Over-Water method. Physiol. Behav. 2008. [Google Scholar] [CrossRef] [PubMed]
- Rechtschaffen, A.; Gilliland, M.A.; Bergmann, B.M.; Winter, J.B. Physiological correlates of prolonged sleep deprivation in rats. Science 1983, 221, 182–184. [Google Scholar] [CrossRef]
- Webb, W.B.; Agnew, H.W. Sleep deprivation, age, and exhaustion time in the rat. Science 1962, 136, 1122. [Google Scholar] [CrossRef] [PubMed]
- Cirelli, C.; Tononi, G. Is sleep essential? PLoS Biol. 2008, 6, 1605–1611. [Google Scholar] [CrossRef]
- Mirsky, A.F.; Cardon, P.V. A comparison of the behavioral and physiological changes accompanying sleep deprivation and chlorpromazine administration in man. Electroencephalogr. Clin. Neurophysiol. 1962. [Google Scholar] [CrossRef]
- Kayser, M.S.; Mainwaring, B.; Yue, Z.; Sehgal, A. Sleep deprivation suppresses aggression in Drosophila. Elife 2015. [Google Scholar] [CrossRef] [PubMed]
- Rial, R.V.; Nicolau, M.C.; Gamundí, A.; Akaârir, M.; Aparicio, S.; Garau, C.; Tejada, S.; Roca, C.; Gené, L.; Moranta, D.; et al. The trivial function of sleep. Sleep Med. Rev. 2007. [Google Scholar] [CrossRef] [PubMed]
- Rattenborg, N.C.; Lesku, J.A.; Martinez-Gonzalez, D.; Lima, S.L. The non-trivial functions of sleep. Sleep Med. Rev. 2007. [Google Scholar] [CrossRef]
- Tononi, G.; Cirelli, C. Sleep and synaptic down-selection. Eur. J. Neurosci. 2020. [Google Scholar] [CrossRef] [PubMed]
- Tononi, G.; Cirelli, C. Sleep and the Price of Plasticity: From Synaptic and Cellular Homeostasis to Memory Consolidation and Integration. Neuron 2014. [Google Scholar] [CrossRef]
- Mendelsohn, A.R.; Larrick, J.W. Sleep Facilitates Clearance of Metabolites from the Brain: Glymphatic Function in Aging and Neurodegenerative Diseases. Rejuvenation Res. 2013. [Google Scholar] [CrossRef]
- Opp, M.R.; Baracchi, F. Sleep and immune function. Curr. Adv. Sleep Biol. 2009. [Google Scholar] [CrossRef]
- Opp, M.R.; Krueger, J.M. Sleep and immunity: A growing field with clinical impact. Brain Behav. Immun. 2015. [Google Scholar] [CrossRef]
- Mullington, J.M.; Haack, M.; Toth, M.; Serrador, J.M.; Meier-Ewert, H.K. Cardiovascular, Inflammatory, and Metabolic Consequences of Sleep Deprivation. Prog. Cardiovasc. Dis. 2009. [Google Scholar] [CrossRef]
- Cirelli, C. Cellular consequences of sleep deprivation in the brain. Sleep Med. Rev. 2006. [Google Scholar] [CrossRef]
- Geissmann, Q.; Beckwith, E.J.; Gilestro, G.F. Most sleep does not serve a vital function. Evidence from Drosophila melanogaster. BioRxiv 2018. [Google Scholar] [CrossRef] [PubMed]
- Shein-Idelson, M.; Ondracek, J.M.; Liaw, H.P.; Reiter, S.; Laurent, G. Slow waves, sharp waves, ripples, and REM in sleeping dragons. Science 2016. [Google Scholar] [CrossRef] [PubMed]
- Boyce, R.; Glasgow, S.D.; Williams, S.; Adamantidis, A. Sleep research: Causal evidence for the role of REM sleep theta rhythm in contextual memory consolidation. Science 2016, 352, 812–816. [Google Scholar] [CrossRef] [PubMed]
- Graves, L.; Heller, E.; Pack, A.; Abel, T. Sleep deprivation selectively impairs memory consolidation for contextual fear conditioning. Learn. Mem. 2003, 10, 168–176. [Google Scholar] [CrossRef]
- Kumar, T.; Jha, S.K. Sleep Deprivation Impairs Consolidation of Cued Fear Memory in Rats. PLoS ONE 2012, 7. [Google Scholar] [CrossRef] [PubMed]
- Youngblood, B.D.; Zhou, J.; Smagin, G.N.; Ryan, D.H.; Harris, R.B.S. Sleep deprivation by the “flower pot” technique and spatial reference memory. Physiol. Behav. 1997, 61, 249–256. [Google Scholar] [CrossRef]
- Chokroverty, S. Physiological changes of sleep. Sleep Disord. Med. Basic Sci. Tech. Consid. Clin. 2017, 153–194. [Google Scholar] [CrossRef]
- Mallick, B.N.; Singh, S.; Pal, D. Role of alpha and beta adrenoceptors in locus coeruleus stimulation-induced reduction in rapid eye movement sleep in freely moving rats. Behav. Brain Res. 2005, 158, 9–21. [Google Scholar] [CrossRef]
- Mônico-Neto, M.; Dáttilo, M.; Ribeiro, D.A.; Lee, K.S.; de Mello, M.T.; Tufik, S.; Antunes, H.K.M. REM sleep deprivation impairs muscle regeneration in rats. Growth Factors 2017. [Google Scholar] [CrossRef]
- Van Der Helm, E.; Yao, J.; Dutt, S.; Rao, V.; Saletin, J.M.; Walker, M.P. REM sleep depotentiates amygdala activity to previous emotional experiences. Curr. Biol. 2011, 21, 2029–2032. [Google Scholar] [CrossRef]
- Grosmark, A.D.; Mizuseki, K.; Pastalkova, E.; Diba, K.; Buzsáki, G. REM Sleep Reorganizes Hippocampal Excitability. Neuron 2012, 75, 1001–1007. [Google Scholar] [CrossRef]
- Li, W.; Ma, L.; Yang, G.; Gan, W.B. REM sleep selectively prunes and maintains new synapses in development and learning. Nat. Neurosci. 2017, 20, 427–437. [Google Scholar] [CrossRef]
- Somarajan, B.I.; Khanday, M.A.; Mallick, B.N. Rapid eye movement sleep deprivation induces neuronal apoptosis by noradrenaline acting on alpha1 adrenoceptor and by triggering mitochondrial intrinsic pathway. Front. Neurol. 2016, 7. [Google Scholar] [CrossRef]
- Pandey, A.; Kumar, D.; Ray, G.; Kar, S. Rapid eye movement sleep deprivation causes apoptotic cell-death of the hepatocytes in rat. BioRxiv 2018. [Google Scholar] [CrossRef]
- Biswas, S.; Mishra, P.; Mallick, B.N. Increased apoptosis in rat brain after rapid eye movement sleep loss. Neuroscience 2006, 142, 315–331. [Google Scholar] [CrossRef]
- Pandey, A.K.; Kar, S.K. REM sleep deprivation of rats induces acute phase response in liver. Biochem. Biophys. Res. Commun. 2011, 410, 242–246. [Google Scholar] [CrossRef]
- Yehuda, S.; Sredni, B.; Carasso, R.L.; Kenigsbuch-Sredni, D. REM sleep deprivation in rats results in inflammation and interleukin-17 elevation. J. Interf. Cytokine Res. 2009. [Google Scholar] [CrossRef]
- Pandey, A.; Kar, S.K. Rapid Eye Movement sleep deprivation of rat generates ROS in the hepatocytes and makes them more susceptible to oxidative stress. Sleep Sci. 2018. [Google Scholar] [CrossRef]
- Balestrieri, S.; D’Onofrio, G.; Giuditta, A. Deprivation of paradoxical sleep effect on weight and nucleic acid content of liver and brain. Neurochem. Res. 1980. [Google Scholar] [CrossRef] [PubMed]
- Li, T.; Cao, R.; Xia, R.; Xia, Z. Effects of 72 Hours Sleep Deprivation on Liver Circadian Clock Gene Expression and Oxidative Stress in Rats. Yangtze Med. 2017. [Google Scholar] [CrossRef]
- Koban, M.; Swinson, K.L. Chronic REM-sleep deprivation of rats elevates metabolic rate and increases UCP1 gene expression in brown adipose tissue. Am. J. Physiol. Endocrinol. Metab. 2005. [Google Scholar] [CrossRef] [PubMed]
- Gómez-González, B.; Hurtado-Alvarado, G.; Esqueda-León, E.; Santana-Miranda, R.; Rojas-Zamorano, J.Á.; Velázquez-Moctezuma, J. REM sleep loss and recovery regulates blood-brain barrier function. Curr. Neurovasc. Res. 2013, 10, 197–207. [Google Scholar] [CrossRef] [PubMed]
- Venancio, D.P.; Suchecki, D. Prolonged REM sleep restriction induces metabolic syndrome-related changes: Mediation by pro-inflammatory cytokines. Brain Behav. Immun. 2015. [Google Scholar] [CrossRef] [PubMed]
- Cirelli, C.; Gutierrez, C.M.; Tononi, G. Extensive and Divergent Effects of Sleep and Wakefulness on Brain Gene Expression. Neuron 2004, 41, 35–43. [Google Scholar] [CrossRef]
- Cirelli, C.; Tononi, G. Gene expression in the brain across the sleep-waking cycle. Brain Res. 2000. [Google Scholar] [CrossRef]
- Vyazovskiy, V.V.; Cirelli, C.; Pfister-Genskow, M.; Faraguna, U.; Tononi, G. Molecular and electrophysiological evidence for net synaptic potentiation in wake and depression in sleep. Nat. Neurosci. 2008. [Google Scholar] [CrossRef]
- Tononi, G.; Cirelli, C. Sleep and synaptic homeostasis: A hypothesis. Brain Res. Bull. 2003, 62, 143–150. [Google Scholar] [CrossRef] [PubMed]
- Tononi, G.; Cirelli, C. Time to be SHY? Some comments on sleep and synaptic homeostasis. Neural. Plast. 2012. [Google Scholar] [CrossRef]
- Bliss, T.V.P.; Collingridge, G.L. Expression of NMDA receptor-dependent LTP in the hippocampus: Bridging the divide. Mol. Brain 2013. [Google Scholar] [CrossRef]
- Alberini, C.M. Transcription factors in long-term memory and synaptic plasticity. Physiol. Rev. 2009. [Google Scholar] [CrossRef] [PubMed]
- Bozon, B.; Kelly, Á.; Josselyn, S.A.; Silva, A.J.; Davis, S.; Laroche, S. MAPK, CREB and zif268 are all required for the consolidation of recognition memory. Philos. Trans. R. Soc. B Biol. Sci. 2003. [Google Scholar] [CrossRef]
- Chowdhury, S.; Shepherd, J.D.; Okuno, H.; Lyford, G.; Petralia, R.S.; Plath, N.; Kuhl, D.; Huganir, R.L.; Worley, P.F. Arc/Arg3.1 Interacts with the Endocytic Machinery to Regulate AMPA Receptor Trafficking. Neuron 2006. [Google Scholar] [CrossRef]
- Messaoudi, E.; Kanhema, T.; Soulé, J.; Tiron, A.; Dagyte, G.; Da Silva, B.; Bramham, C.R. Sustained Arc/Arg3.1 synthesis controls long-term potentiation consolidation through regulation of local actin polymerization in the dentate gyrus in vivo. J. Neurosci. 2007. [Google Scholar] [CrossRef] [PubMed]
- Racaniello, M.; Cardinale, A.; Mollinari, C.; D’Antuono, M.; De Chiara, G.; Tancredi, V.; Merlo, D. Phosphorylation Changes of CaMKII, ERK1/2, PKB/Akt Kinases and CREB Activation during Early Long-Term Potentiation at Schaffer Collateral-CA1 Mouse Hippocampal Synapses. Neurochem. Res. 2010. [Google Scholar] [CrossRef]
- Shepherd, J.D.; Rumbaugh, G.; Wu, J.; Chowdhury, S.; Plath, N.; Kuhl, D.; Huganir, R.L.; Worley, P.F. Arc/Arg3.1 Mediates Homeostatic Synaptic Scaling of AMPA Receptors. Neuron 2006. [Google Scholar] [CrossRef] [PubMed]
- Tzingounis, A.V.; Nicoll, R.A. Arc/Arg3.1: Linking Gene Expression to Synaptic Plasticity and Memory. Neuron 2006. [Google Scholar] [CrossRef]
- Pompeiano, M.; Cirelli, C.; Tononi, G. Immediate-early genes in spontaneous wakefulness and sleep: Expression of c-fos and NGFI-A mRNA and protein. J. Sleep Res. 1994. [Google Scholar] [CrossRef] [PubMed]
- Calais, J.B.; Ojopi, E.B.; Morya, E.; Sameshima, K.; Ribeiro, S. Experience-dependent upregulation of multiple plasticity factors in the hippocampus during early REM sleep. Neurobiol. Learn. Mem. 2015. [Google Scholar] [CrossRef]
- Smith-Hicks, C.; Xiao, B.; Deng, R.; Ji, Y.; Zhao, X.; Shepherd, J.D.; Posern, G.; Kuhl, D.; Huganir, R.L.; Ginty, D.D.; et al. SRF binding to SRE 6.9 in the Arc promoter is essential for LTD in cultured Purkinje cells. Nat. Neurosci. 2010. [Google Scholar] [CrossRef]
- Waung, M.W.; Pfeiffer, B.E.; Nosyreva, E.D.; Ronesi, J.A.; Huber, K.M. Rapid Translation of Arc/Arg3.1 Selectively Mediates mGluR-Dependent LTD through Persistent Increases in AMPAR Endocytosis Rate. Neuron 2008. [Google Scholar] [CrossRef]
- Korb, E.; Wilkinson, C.L.; Delgado, R.N.; Lovero, K.L.; Finkbeiner, S. Arc in the nucleus regulates PML-dependent GluA1 transcription and homeostatic plasticity. Nat. Neurosci. 2013. [Google Scholar] [CrossRef] [PubMed]
- Okuno, H.; Akashi, K.; Ishii, Y.; Yagishita-Kyo, N.; Suzuki, K.; Nonaka, M.; Kawashima, T.; Fujii, H.; Takemoto-Kimura, S.; Abe, M.; et al. Inverse synaptic tagging of inactive synapses via dynamic interaction of Arc/Arg3.1 with CaMKIIβ. Cell 2012. [Google Scholar] [CrossRef] [PubMed]
- Lambrecht, C.; Haesen, D.; Sents, W.; Ivanova, E.; Janssens, V. Structure, regulation, and pharmacological modulation of PP2A phosphatases. Methods Mol. Biol. 2013. [Google Scholar] [CrossRef]
- Ravassard, P.; Pachoud, B.; Comte, J.C.; Mejia-Perez, C.; Scoté-Blachon, C.; Gay, N.; Claustrat, B.; Touret, M.; Luppi, P.H.; Salin, P.A. Paradoxical (REM) sleep deprivation causes a large and rapidly reversible decrease in long-term potentiation, synaptic transmission, glutamate receptor protein levels, and ERK/MAPK activation in the dorsal hippocampus. Sleep 2009. [Google Scholar] [CrossRef]
- Mignot, E. Why we sleep: The temporal organization of recovery. PLoS Biol. 2008, 6, 661–669. [Google Scholar] [CrossRef]
- Cirelli, C.; Bushey, D. Sleep and wakefulness in Drosophila melanogaster. Ann. N. Y. Acad. Sci. 2008, 1129, 323–329. [Google Scholar] [CrossRef]
- Cirelli, C.; LaVaute, T.M.; Tononi, G. Sleep and wakefulness modulate gene expression in Drosophila. J. Neurochem. 2005, 94, 1411–1419. [Google Scholar] [CrossRef]
- Cirelli, C. Locus Ceruleus Control of State-Dependent Gene Expression. J. Neurosci. 2004, 24, 5410–5419. [Google Scholar] [CrossRef]
- Jones, S.; Pfister-Genskow, M.; Benca, R.M.; Cirelli, C. Molecular correlates of sleep and wakefulness in the brain of the white-crowned sparrow. J. Neurochem. 2008, 105, 46–62. [Google Scholar] [CrossRef]
- Mackiewicz, M.; Shockley, K.R.; Romer, M.A.; Galante, R.J.; Zimmerman, J.E.; Naidoo, N.; Baldwin, D.A.; Jensen, S.T.; Churchill, G.A.; Pack, A.I. Macromolecule biosynthesis: A key function of sleep. Physiol. Genom. 2007, 31, 441–457. [Google Scholar] [CrossRef]
- Maret, S.; Dorsaz, S.; Gurcel, L.; Pradervand, S.; Petit, B.; Pfister, C.; Hagenbuchle, O.; O’Hara, B.F.; Franken, P.; Tafti, M. Homer1a is a core brain molecular correlate of sleep loss. Proc. Natl. Acad. Sci. USA 2007, 104, 20090–20095. [Google Scholar] [CrossRef] [PubMed]
- Zimmerman, J.E.; Rizzo, W.; Shockley, K.R.; Raizen, D.M.; Naidoo, N.; Mackiewicz, M.; Churchill, G.A.; Pack, A.I. Multiple mechanisms limit the duration of wakefulness in Drosophila brain. Physiol. Genom. 2006, 27, 337–350. [Google Scholar] [CrossRef]
- Volkow, N.D.; Tomasi, D.; Wang, G.J.; Telang, F.; Fowler, J.S.; Logan, J.; Benveniste, H.; Kim, R.; Thanos, P.K.; Ferré, S. Evidence that sleep deprivation down regulates dopamine D2R in ventral striatum in the human brain. J. Neurosci. 2012. [Google Scholar] [CrossRef] [PubMed]
- França, A.S.C.; Lobão-Soares, B.; Muratori, L.; Nascimento, G.; Winne, J.; Pereira, C.M.; Jeronimo, S.M.B.; Ribeiro, S. D2 dopamine receptor regulation of learning, sleep and plasticity. Eur. Neuropsychopharmacol. 2015. [Google Scholar] [CrossRef] [PubMed]
- Nicholson, A.N.; Pascoe, P.A. Dopaminergic transmission and the sleep-wakefulness continuum in man. Neuropharmacology 1990. [Google Scholar] [CrossRef]
- Marlene Oscar, K.B. Dopaminergic Neurogenetics of Sleep Disorders in Reward Deficiency Syndrome (RDS). J. Sleep Disord. Ther. 2014. [Google Scholar] [CrossRef]
- Parameswari, P.; Chethan, N.; Saravana Babu, C. Neurochemicals and Behavioural Alterations in Sleep Deprivation: A Revisit. J. Dement. 2017, 1, 104. [Google Scholar]
- Peever, J.; Fuller, P.M. The Biology of REM Sleep. Curr. Biol. 2017. [Google Scholar] [CrossRef]
- Yamada, R.G.; Ueda, H.R. Molecular Mechanisms of REM Sleep. Front. Neurosci. 2020. [Google Scholar] [CrossRef]
- Cirelli, C.; Faraguna, U.; Tononi, G. Changes in brain gene expression after long-term sleep deprivation. J. Neurochem. 2006. [Google Scholar] [CrossRef]
- Hicks, R.A.; Okuda, A.; Thomsen, D. Depriving rats of REM sleep: The identification of a methodological problem. Am. J. Psychol. 1977, 90, 95–102. [Google Scholar] [CrossRef]
- van Hulzen, Z.J.M.; Coenen, A.M.L. Paradoxical sleep deprivation and locomotor activity in rats. Physiol. Behav. 1981, 27, 741–744. [Google Scholar] [CrossRef]
- R Development Core Team. Computing R: A language and environment for statistical. R Found. Stat. Comput. 2013. Available online: https://www.R-project.org (accessed on 23 October 2020).
- R Foundation for Statistical Computing. R: A Language and Environment for Statistical Computing. R Found. Stat. Comput. 2016. Available online: https://www.R-project.org (accessed on 23 October 2020).
- Ashburner, M.; Ball, C.A.; Blake, J.A.; Butler, H.; Cherry, J.M.; Eppig, J.T.; Harris, M.; Hill, D.P.; Mungall, C.; Reiser, L.; et al. Creating the Gene Ontology resource: Design and implementation. Genome Res. 2001. [Google Scholar] [CrossRef]
- Kanehisa, M.; Goto, S. KEGG: Kyoto Encyclopedia of Genes and Genomes. Nucleic Acids Res. 2000. [Google Scholar] [CrossRef]
- Kanehisa, M.; Sato, Y.; Furumichi, M.; Morishima, K.; Tanabe, M. New approach for understanding genome variations in KEGG. Nucleic Acids Res. 2019. [Google Scholar] [CrossRef]
- Kanehisa, M. Toward understanding the origin and evolution of cellular organisms. Protein Sci. 2019. [Google Scholar] [CrossRef]
- Core Team R. R: A Language and Environment for Statistical Computing. R Found. Stat. Comput. 2020. Available online: http://softlibre.unizar.es/manuales/aplicaciones/r/fullrefman.pdf (accessed on 23 October 2020).
- Yu, G.; Wang, L.G.; Han, Y.; He, Q.Y. ClusterProfiler: An R package for comparing biological themes among gene clusters. Omi. J. Integr. Biol. 2012. [Google Scholar] [CrossRef]
- Wang, H.; Liu, Y.; Briesemann, M.; Yan, J. Computational analysis of gene regulation in animal sleep deprivation. Physiol. Genom. 2010. [Google Scholar] [CrossRef]
- Martyn-Nemeth, P.; Phillips, S.A.; Mihailescu, D.; Farabi, S.S.; Park, C.; Lipton, R.; Idemudia, E.; Quinn, L. Poor sleep quality is associated with nocturnal glycaemic variability and fear of hypoglycaemia in adults with type 1 diabetes. J. Adv. Nurs. 2018. [Google Scholar] [CrossRef]
- Kim, J.H.; Kim, J.H.; Cho, Y.E.; Baek, M.C.; Jung, J.Y.; Lee, M.G.; Jang, I.S.; Lee, H.O.; Suk, K. Chronic sleep deprivation-induced proteome changes in astrocytes of the rat hypothalamus. J. Proteome Res. 2014. [Google Scholar] [CrossRef]
- Obholzer, N.; Wolfson, S.; Trapani, J.G.; Mo, W.; Nechiporuk, A.; Busch-Nentwich, E.; Seiler, C.; Sidi, S.; Söllner, C.; Duncan, R.N.; et al. Vesicular glutamate transporter 3 is required for synaptic transmission in zebrafish hair cells. J. Neurosci. 2008. [Google Scholar] [CrossRef]
- McGaugh, S.E.; Passow, C.N.; Jaggard, J.B.; Stahl, B.A.; Keene, A.C. Unique transcriptional signatures of sleep loss across independently evolved cavefish populations. J. Exp. Zool. Part B Mol. Dev. Evol. 2020. [Google Scholar] [CrossRef]
- Wei, Y. Comparative transcriptome analysis of the hippocampus from sleep-deprived and alzheimer’s disease mice. Genet. Mol. Biol. 2020. [Google Scholar] [CrossRef]
- Dyken, M.E.; Yamada, T.; Glenn, C.L.; Berger, H.A. Obstructive sleep apnea associated with cerebral hypoxemia and death. Neurology 2004. [Google Scholar] [CrossRef]
- Muller, N.L.; Francis, P.W.; Gurwitz, D.; Levison, H.; Bryan, A.C. Mechanism of hemoglobin desaturation during rapid-eye-movement sleep in normal subjects and in patients with cystic fibrosis. Am. Rev. Respir. Dis. 1980. [Google Scholar] [CrossRef]
- Gaudet, P.; Livstone, M.S.; Lewis, S.E.; Thomas, P.D. Phylogenetic-based propagation of functional annotations within the Gene Ontology consortium. Brief. Bioinform. 2011. [Google Scholar] [CrossRef]
- Schwenk, B.M.; Lang, C.M.; Hogl, S.; Tahirovic, S.; Orozco, D.; Rentzsch, K.; Lichtenthaler, S.F.; Hoogenraad, C.C.; Capell, A.; Haass, C.; et al. The FTLD risk factor TMEM106B and MAP6 control dendritic trafficking of lysosomes. EMBO J. 2014. [Google Scholar] [CrossRef]
- Chen-Plotkin, A.S.; Unger, T.L.; Gallagher, M.D.; Bill, E.; Kwong, L.K.; Volpicelli-Daley, L.; Busch, J.I.; Akle, S.; Grossman, M.; Van Deerlin, V.; et al. TMEM106B, the Risk Gene for Frontotemporal Dementia, Is Regulated by the microRNA-132/212 Cluster and Affects Progranulin Pathways. J. Neurosci. 2012. [Google Scholar] [CrossRef]
- Niethard, N.; Burgalossi, A.; Born, J. Plasticity during sleep is linked to specific regulation of cortical circuit activity. Front. Neural. Circuits 2017. [Google Scholar] [CrossRef]
- González-Rueda, A.; Pedrosa, V.; Feord, R.C.; Clopath, C.; Paulsen, O. Activity-Dependent Downscaling of Subthreshold Synaptic Inputs during Slow-Wave-Sleep-like Activity In Vivo. Neuron 2018. [Google Scholar] [CrossRef]
- Seibt, J.; Frank, M.G. Primed to sleep: The dynamics of synaptic plasticity across brain states. Front. Syst. Neurosci. 2019. [Google Scholar] [CrossRef]
- Wang, G.; Grone, B.; Colas, D.; Appelbaum, L.; Mourrain, P. Synaptic plasticity in sleep: Learning, homeostasis and disease. Trends Neurosci. 2011. [Google Scholar] [CrossRef]
- Havekes, R.; Vecsey, C.G.; Abel, T. The impact of sleep deprivation on neuronal and glial signaling pathways important for memory and synaptic plasticity. Cell. Signal. 2012. [Google Scholar] [CrossRef]
- Cirelli, C. Sleep and synaptic changes. Curr. Opin. Neurobiol. 2013. [Google Scholar] [CrossRef]
- Shaffery, J.P.; Lopez, J.; Bissette, G.; Roffwarg, H.P. Rapid eye movement sleep deprivation revives a form of developmentally regulated synaptic plasticity in the visual cortex of post-critical period rats. Neurosci. Lett. 2006. [Google Scholar] [CrossRef]
- Shaffery, J.P.; Sinton, C.M.; Bissette, G.; Roffwarg, H.P.; Marks, G.A. Rapid eye movement sleep deprivation modifies expression of long-term potentiation in visual cortex of immature rats. Neuroscience 2002. [Google Scholar] [CrossRef]
- Katayose, Y.; Tasaki, M.; Ogata, H.; Nakata, Y.; Tokuyama, K.; Satoh, M. Metabolic rate and fuel utilization during sleep assessed by whole-body indirect calorimetry. Metabolism 2009. [Google Scholar] [CrossRef]
- Spiegel, K.; Leproult, R.; Van Cauter, E. Impact of sleep debt on metabolic and endocrine function. Lancet 1999, 354, 1435–1439. [Google Scholar] [CrossRef]
- Donga, E.; Van Dijk, M.; Van Dijk, J.G.; Biermasz, N.R.; Lammers, G.J.; Van Kralingen, K.W.; Corssmit, E.P.M.; Romijn, J. A single night of partial sleep deprivation induces insulin resistance in multiple metabolic pathways in healthy subjects. J. Clin. Endocrinol. Metab. 2010. [Google Scholar] [CrossRef]
- Pamidi, S.; Aronsohn, R.S.; Tasali, E. Obstructive sleep apnea: Role in the risk and severity of diabetes. Best Pract. Res. Clin. Endocrinol. Metab. 2010. [Google Scholar] [CrossRef]
- Iiyori, N.; Alonso, L.C.; Li, J.; Sanders, M.H.; Garcia-Ocana, A.; O’Doherty, R.M.; Polotsky, V.Y.; O’Donnell, C.P. Intermittent hypoxia causes insulin resistance in lean mice independent of autonomic activity. Am. J. Respir. Crit. Care Med. 2007. [Google Scholar] [CrossRef]
- Asif, N.; Iqbal, R.; Nazir, C.F. Human immune system during sleep. Am. J. Clin. Exp. Immunol. 2017, 6, 92. [Google Scholar]
- Besedovsky, L.; Lange, T.; Haack, M. The sleep-immune crosstalk in health and disease. Physiol. Rev. 2019. [Google Scholar] [CrossRef]
- Ruiz, F.S.; Andersen, M.L.; Martins, R.C.S.; Zager, A.; Lopes, J.D.; Tufik, S. Immune alterations after selective rapid eye movement or total sleep deprivation in healthy male volunteers. Innate Immun. 2012. [Google Scholar] [CrossRef]
- Grønli, J.; Soulé, J.; Bramham, C.R. Sleep and protein synthesis-dependent synaptic plasticity: Impacts of sleep loss and stress. Front. Behav. Neurosci. 2014. [Google Scholar] [CrossRef]
- Hoffmann, M.S.; Singh, P.; Wolk, R.; Romero-Corral, A.; Raghavakaimal, S.; Somers, V.K. Microarray studies of genomic oxidative stress and cell cycle responses in obstructive sleep apnea. Antioxid. Redox Signal. 2007. [Google Scholar] [CrossRef]
- Fiorentino, L.; Ancoli-Israel, S. Sleep dysfunction in patients with cancer. Curr. Treat. Options Neurol. 2007. [Google Scholar] [CrossRef]
- Fang, H.F.; Miao, N.F.; Chen, C.D.; Sithole, T.; Chung, M.H. Risk of cancer in patients with insomnia, parasomnia, and obstructive sleep apnea: A nationwide nested case-control study. J. Cancer 2015. [Google Scholar] [CrossRef]
- Sateia, M.J.; Lang, B.J. Sleep and cancer: Recent developments. Curr. Oncol. Rep. 2008. [Google Scholar] [CrossRef] [PubMed]
- Shi, T.; Min, M.; Sun, C.; Zhang, Y.; Liang, M.; Sun, Y. Does insomnia predict a high risk of cancer? A systematic review and meta-analysis of cohort studies. J. Sleep Res. 2020. [Google Scholar] [CrossRef] [PubMed]
- Martínez-Garćia, M.Á.; Martorell-Calatayud, A.; Nagore, E.; Valero, I.; Selma, M.J.; Chiner, E.; Landete, P.; Montserrat, J.M.; Carrera, C.; Peŕez-Gil, A.; et al. Association between sleep disordered breathing and aggressiveness markers of malignant cutaneous melanoma. Eur. Respir. J. 2014. [Google Scholar] [CrossRef] [PubMed]
- Lin, C.L.; Liu, T.C.; Wang, Y.N.; Chung, C.H.; Chien, W.C. The association between sleep disorders and the risk of colorectal cancer in patients: A Population-based Nested Case–Control Study. In Vivo 2019. [Google Scholar] [CrossRef]
- Sillah, A.; Watson, N.F.; Schwartz, S.M.; Gozal, D.; Phipps, A.I. Sleep apnea and subsequent cancer incidence. Cancer Causes Control 2018. [Google Scholar] [CrossRef]
- Chiu, H.Y.; Huang, C.J.; Fan, Y.C.; Tsai, P.S. Insomnia but Not Hypnotics Use Associates with the Risk of Breast Cancer: A Population-Based Matched Cohort Study. J. Womens Health 2018. [Google Scholar] [CrossRef]
- Sturgeon, S.R.; Luisi, N.; Balasubramanian, R.; Reeves, K.W. Sleep duration and endometrial cancer risk. Cancer Causes Control 2012. [Google Scholar] [CrossRef]
- Liu, R.; Xu, K.P.; Tan, G.S. Cyclooxygenase-2 inhibitors in lung cancer treatment: Bench to bed. Eur. J. Pharmacol. 2015. [Google Scholar] [CrossRef]
- Lin, X.M.; Luo, W.; Wang, H.; Li, R.Z.; Huang, Y.S.; Chen, L.K.; Wu, X.P. The role of prostaglandin-endoperoxide synthase-2 in chemoresistance of non-small cell lung cancer. Front. Pharmacol. 2019. [Google Scholar] [CrossRef]
- Mueller, S.; Engleitner, T.; Maresch, R.; Zukowska, M.; Lange, S.; Kaltenbacher, T.; Konukiewitz, B.; Öllinger, R.; Zwiebel, M.; Strong, A.; et al. Evolutionary routes and KRAS dosage define pancreatic cancer phenotypes. Nature 2018. [Google Scholar] [CrossRef]
- LoRusso, P.M.; Herbst, R.S.; Rischin, D.; Ranson, M.; Calvert, H.; Raymond, E.; Kieback, D.; Kaye, S.; Gianni, L.; Harris, A.; et al. Improvements in quality of life and disease-related symptoms in phase I trials of the selective oral epidermal growth factor receptor tyrosine kinase inhibitor ZD1839 in non-small cell lung cancer and other solid tumors. Clin. Cancer. Res. 2003, 9, 2040–2048. [Google Scholar] [PubMed]
- Thomadaki, H.; Scorilas, A. BCL2 family of apoptosis-related genes: Functions and clinical implications in cancer. Crit. Rev. Clin. Lab. Sci. 2006. [Google Scholar] [CrossRef]
- Cruse, G.; Metcalfe, D.D.; Olivera, A. Functional deregulation of KIT: Link to mast cell proliferative diseases and other neoplasms. Immunol. Allergy Clin. N. Am. 2014. [Google Scholar] [CrossRef] [PubMed]
- Redline, S.; Tishler, P.V. The genetics of sleep apnea. Sleep Med. Rev. 2000. [Google Scholar] [CrossRef] [PubMed]
- Cirelli, C.; Tononi, G. On the Functional Significance of c-fos Induction During the Sleep-waking Cycle. Sleep 2000. [Google Scholar] [CrossRef]
- Cirelli, C.; Pompeiano, M.; Tononi, G. Sleep deprivation and c-fos expression in the rat brain. J. Sleep Res. 1995. [Google Scholar] [CrossRef]
- Gregory, A.; Kurian, M.A.; Maher, E.R.; Hogarth, P.; Hayflick, S.J. PLA2G6-Associated Neurodegeneration. In GeneReviews; Adam, M.P., Ardinger, H.H., Pagon, R.A., Wallace, S.E., Bean, L.J.H., Stephens, K., Amemiya, A., Eds.; GeneReviews® [Internet]; University of Washington: Seattle, WA, USA, 2008. Available online: https://www.ncbi.nlm.nih.gov/books/ (accessed on 23 October 2020).
- Gregory, A.; Westaway, S.K.; Holm, I.E.; Kotzbauer, P.T.; Hogarth, P.; Sonek, S.; Coryell, J.C.; Nguyen, T.M.; Nardocci, N.; Zorzi, G.; et al. Neurodegeneration associated with genetic defects in phospholipase A2. Neurology 2008. [Google Scholar] [CrossRef]
- Kinghorn, K.J.; Castillo-Quan, J.I.; Bartolome, F.; Angelova, P.R.; Li, L.; Pope, S.; Cochemé, H.M.; Khan, S.; Asghari, S.; Bhatia, K.P.; et al. Loss of PLA2G6 leads to elevated mitochondrial lipid peroxidation and mitochondrial dysfunction. Brain 2015. [Google Scholar] [CrossRef]
- Ollila, H. Genetics of Sleep, Sleep and Comorbidities: Connection at the Genetic Level; National Institute for Health and Welfare: Helsinki, Finland, 2013. [Google Scholar]
- Gao, Y.; Yi, X.; Ding, Y. Combined transcriptomic analysis revealed AKR1B10 played an important role in psoriasis through the dysregulated lipid pathway and overproliferation of keratinocyte. Biomed. Res. Int. 2017. [Google Scholar] [CrossRef]
- Barge-Schaapveld, D.Q.C.M.; Ofman, R.; Knegt, A.C.; Alders, M.; Höhne, W.; Kemp, S.; Hennekam, R.C.M. Intellectual Disability and Hemizygous GPD2 Mutation. Am. J. Med. Genet. Part A 2013. [Google Scholar] [CrossRef]
- Barclay, J.L.; Husse, J.; Bode, B.; Naujokat, N.; Meyer-Kovac, J.; Schmid, S.M.; Lehnert, H.; Oster, H. Circadian desynchrony promotes metabolic disruption in a mouse model of shiftwork. PLoS ONE 2012. [Google Scholar] [CrossRef]
- Arriaga, F.; Dugovic, C.; Wauquier, A. Effects of lithium on dopamine behavioural supersensitivity induced by rapid eye movement sleep deprivation. Neuropsychobiology 1988. [Google Scholar] [CrossRef]
- Albert, I.; Cicala, G.A.; Siegel, J. The behavioral effects of REM sleep deprivation in rats. Psychophysiology 1970. [Google Scholar] [CrossRef]
- Asakura, W.; Matsumoto, K.; Ohta, H.; Watanabe, H. REM sleep deprivation decreases apomorphine-induced stimulation of locomotor activity but not stereotyped behavior in mice. Gen. Pharmacol. 1992. [Google Scholar] [CrossRef]
- Maquet, P. Functional neuroimaging of normal human sleep by positron emission tomography. J. Sleep Res. 2000. [Google Scholar] [CrossRef]
- Van Luutelaar, E.L.J.M.; Coenen, A.M.L. Differential behavioural effects of two instrumental paradoxical sleep deprivation techniques in rats. Biol. Psychol. 1982. [Google Scholar] [CrossRef]
- Drewes, A.M.; Svendsen, L.; Taagholt, S.J.; Bjerregård, K.; Nielsen, K.D.; Hansen, B. Sleep in rheumatoid arthritis: A comparison with healthy subjects and studies of sleep/wake interactions. Br. J. Rheumatol. 1998. [Google Scholar] [CrossRef]
- Aǧargün, M.Y.; Tekeoǧlu, I.; Güneş, A.; Adak, B.; Kara, H.; Ercan, M. Sleep quality and pain threshold in patients with fibromyalgia. Compr. Psychiatry 1999. [Google Scholar] [CrossRef]
- Raymond, I.; Nielsen, T.A.; Lavigne, G.; Manzini, C.; Choinière, M. Quality of sleep and its daily relationship to pain intensity in hospitalized adult burn patients. Pain 2001. [Google Scholar] [CrossRef]
- Schrimpf, M.; Liegl, G.; Boeckle, M.; Leitner, A.; Geisler, P.; Pieh, C. The effect of sleep deprivation on pain perception in healthy subjects: A meta-analysis. Sleep Med. 2015. [Google Scholar] [CrossRef]
- Stroemel-Scheder, C.; Kundermann, B.; Lautenbacher, S. The effects of recovery sleep on pain perception: A systematic review. Neurosci. Biobehav. Rev. 2020. [Google Scholar] [CrossRef]
- Roehrs, T.; Hyde, M.; Blaisdell, B.; Greenwald, M.; Roth, T. Sleep loss and REM sleep loss are hyperalgesic. Sleep 2006. [Google Scholar] [CrossRef] [PubMed]
- Staffe, A.T.; Bech, M.W.; Clemmensen, S.L.K.; Nielsen, H.T.; Larsen, D.B.; Petersen, K.K. Total sleep deprivation increases pain sensitivity, impairs conditioned pain modulation and facilitates temporal summation of pain in healthy participants. PLoS ONE 2019. [Google Scholar] [CrossRef] [PubMed]
- Kim, S.H.; Park, J.Y.; Shin, H.E.; Lee, S.B.; Ryu, D.W.; Kim, T.W.; Park, J.W. The influence of rapid eye movement sleep deprivation on nociceptive transmission and the duration of facial allodynia in rats: A behavioral and Fos immunohistochemical study. J. Headache Pain 2019. [Google Scholar] [CrossRef] [PubMed]
- Azevedo, E.; Manzano, G.M.; Silva, A.; Martins, R.; Andersen, M.L.; Tufik, S. The effects of total and REM sleep deprivation on laser-evoked potential threshold and pain perception. Pain 2011. [Google Scholar] [CrossRef]
- Onen, S.H.; Alloui, A.; Gross, A.; Eschallier, A.; Dubray, C. The effects of total sleep deprivation, selective sleep interruption and sleep recovery on pain tolerance thresholds in healthy subjects. J. Sleep Res. 2001. [Google Scholar] [CrossRef]
- Chouchou, F.; Chauny, J.M.; Rainville, P.; Lavigne, G.J. Selective REM sleep deprivation improves expectation-related placebo analgesia. PLoS ONE 2015. [Google Scholar] [CrossRef]
- Smith, M.T.; Edwards, R.R.; Stonerock, G.L.; McCann, U.D. Individual variation in rapid eye movement sleep is associated with pain perception in healthy women: Preliminary data. Sleep 2005. [Google Scholar] [CrossRef]
- Onen, S.H.; Alloui, A.; Jourdan, D.; Eschalier, A.; Dubray, C. Effects of rapid eye movement (REM) sleep deprivation on pain sensitivity in the rat. Brain Res. 2001. [Google Scholar] [CrossRef]
- Hoekstra, M.M.B.; Emmenegger, Y.; Hubbard, J.; Franken, P. Cold-inducible RNA-binding protein (CIRBP) adjusts clock-gene expression and REM-sleep recovery following sleep deprivation. Elife 2019. [Google Scholar] [CrossRef]
- Gaudet, P.; Dessimoz, C. Gene ontology: Pitfalls, biases, and remedies. Methods Mol. Biol. 2017. [Google Scholar] [CrossRef]
- Tomczak, A.; Mortensen, J.M.; Winnenburg, R.; Liu, C.; Alessi, D.T.; Swamy, V.; Vallania, F.; Lofgren, S.; Haynes, W.; Shah, N.H.; et al. Interpretation of biological experiments changes with evolution of the Gene Ontology and its annotations. Sci. Rep. 2018. [Google Scholar] [CrossRef] [PubMed]
- Carbon, S.; Douglass, E.; Dunn, N.; Good, B.; Harris, N.L.; Lewis, S.E.; Mungall, C.J.; Basu, S.; Chisholm, R.L.; Dodson, R.J.; et al. The Gene Ontology Resource: 20 years and still GOing strong. Nucleic Acids Res. 2019. [Google Scholar] [CrossRef]
- Du, J.; Li, M.; Yuan, Z.; Guo, M.; Song, J.; Xie, X.; Chen, Y. A decision analysis model for KEGG pathway analysis. BMC Bioinform. 2016. [Google Scholar] [CrossRef]
- Du, J.; Yuan, Z.; Ma, Z.; Song, J.; Xie, X.; Chen, Y. KEGG-PATH: Kyoto encyclopedia of genes and genomes-based pathway analysis using a path analysis model. Mol. Biosyst. 2014. [Google Scholar] [CrossRef]
- Bauer-Mehren, A.; Furlong, L.I.; Sanz, F. Pathway databases and tools for their exploitation: Benefits, current limitations and challenges. Mol. Syst. Biol. 2009. [Google Scholar] [CrossRef]
- Khatri, P.; Sirota, M.; Butte, A.J. Ten years of pathway analysis: Current approaches and outstanding challenges. PLoS Comput. Biol. 2012. [Google Scholar] [CrossRef] [PubMed]
- Kittleson, M.M.; Hare, J.M. Molecular signature analysis: Using the myocardial transcriptome as a biomarker in cardiovascular disease. Trends Cardiovasc. Med. 2005. [Google Scholar] [CrossRef] [PubMed]
- Mohr, S.; Liew, C.C. The peripheral-blood transcriptome: New insights into disease and risk assessment. Trends Mol. Med. 2007. [Google Scholar] [CrossRef] [PubMed]
- Seugnet, L.; Boero, J.; Gottschalk, L.; Duntley, S.P.; Shaw, P.J. Identification of a biomarker for sleep drive in flies and humans. Proc. Natl. Acad. Sci. USA 2006. [Google Scholar] [CrossRef]
- Thimgan, M.; Gottschalk, L.; Duntley, S.; Shaw, P.J. New biomarkers of sleepiness identified in humans and Drosophila. In Proceedings of the Sleep 2009, 23rd Annual Meeting of the Associated Professional Sleep Societies, Seattle, WA, USA, 6–11 June 2009. [Google Scholar]
- Irwin, M.R.; Wang, M.; Campomayor, C.O.; Collado-Hidalgo, A.; Cole, S. Sleep deprivation and activation of morning levels of cellular and genomic markers of inflammation. Arch. Intern. Med. 2006. [Google Scholar] [CrossRef]
- Khalyfa, A.; Capdevila, O.S.; Buazza, M.O.; Serpero, L.D.; Kheirandish-Gozal, L.; Gozal, D. Genome-wide gene expression profiling in children with non-obese obstructive sleep apnea. Sleep Med. 2009. [Google Scholar] [CrossRef] [PubMed]

Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Pandey, A.; Oliver, R.; Kar, S.K. Differential Gene Expression in Brain and Liver Tissue of Wistar Rats after Rapid Eye Movement Sleep Deprivation. Clocks & Sleep 2020, 2, 442-465. https://doi.org/10.3390/clockssleep2040033
Pandey A, Oliver R, Kar SK. Differential Gene Expression in Brain and Liver Tissue of Wistar Rats after Rapid Eye Movement Sleep Deprivation. Clocks & Sleep. 2020; 2(4):442-465. https://doi.org/10.3390/clockssleep2040033
Chicago/Turabian StylePandey, Atul, Ryan Oliver, and Santosh K Kar. 2020. "Differential Gene Expression in Brain and Liver Tissue of Wistar Rats after Rapid Eye Movement Sleep Deprivation" Clocks & Sleep 2, no. 4: 442-465. https://doi.org/10.3390/clockssleep2040033
APA StylePandey, A., Oliver, R., & Kar, S. K. (2020). Differential Gene Expression in Brain and Liver Tissue of Wistar Rats after Rapid Eye Movement Sleep Deprivation. Clocks & Sleep, 2(4), 442-465. https://doi.org/10.3390/clockssleep2040033




