Immunofluorescence-Based Assay for High-Throughput Analysis of Multidrug Resistance Markers in Non-Small Cell Lung Carcinoma Patient-Derived Cells
Abstract
:1. Introduction
2. Materials and Methods
2.1. The Establishment of Primary Cultures from NSCLC Tissue Samples
2.2. Cell Lines
2.3. Drugs and Treatments
2.4. Immunofluorescence Assay
2.5. Statistical Analysis
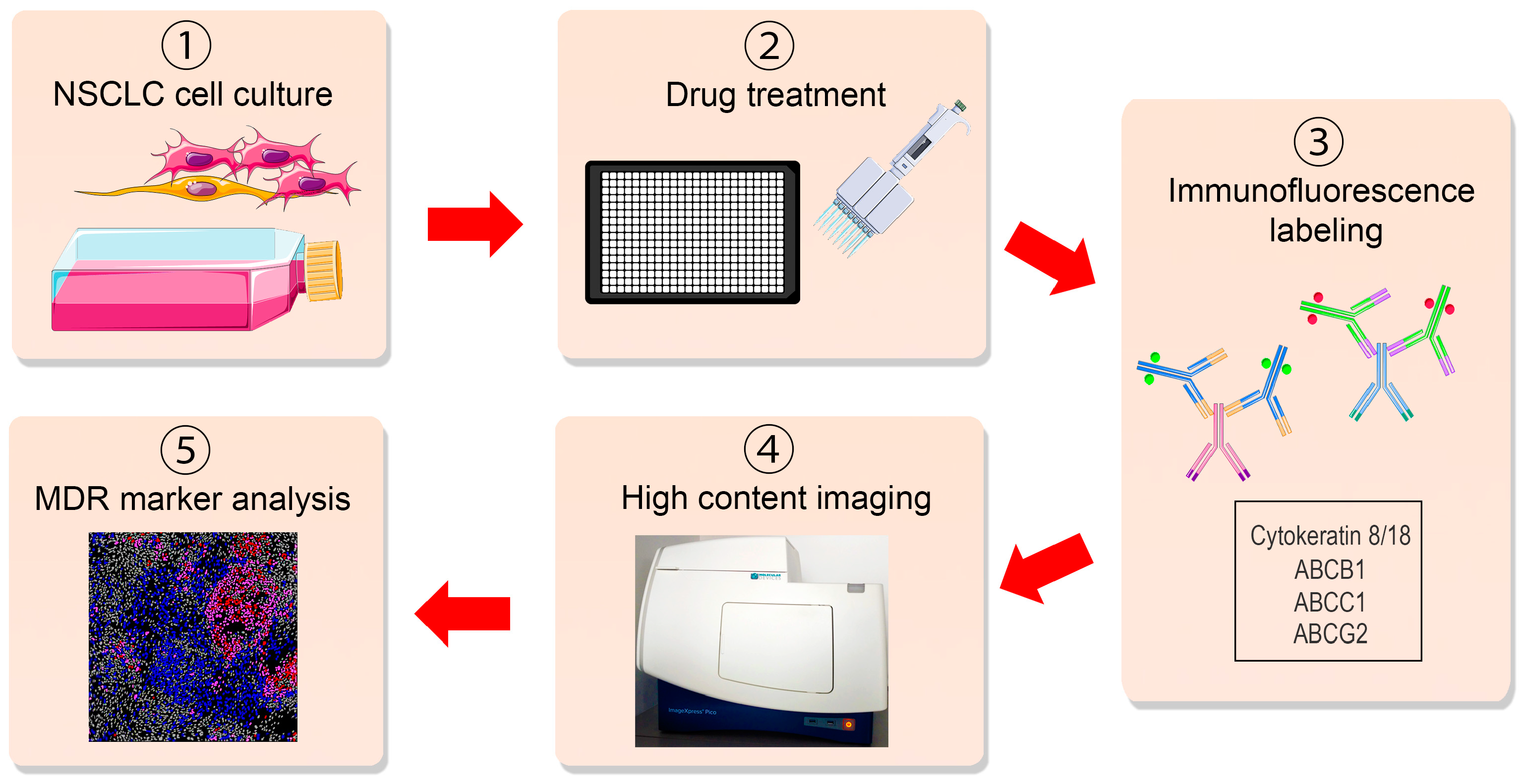
3. Results and Discussion
3.1. Immunoassay Principle
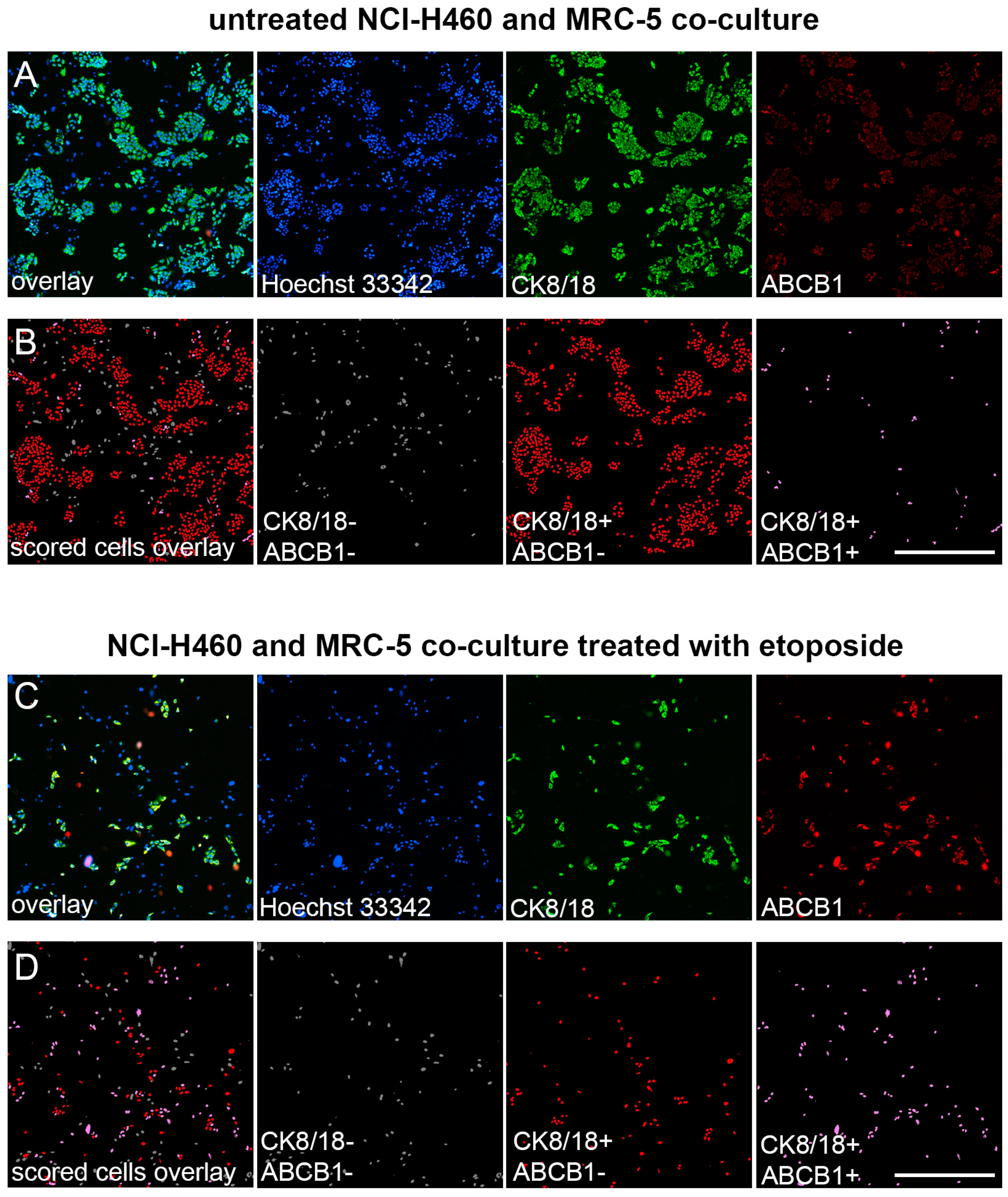
3.2. Validation of Immunofluorescence Assay for the Assessment of MDR Markers in Mixed Cell Populations
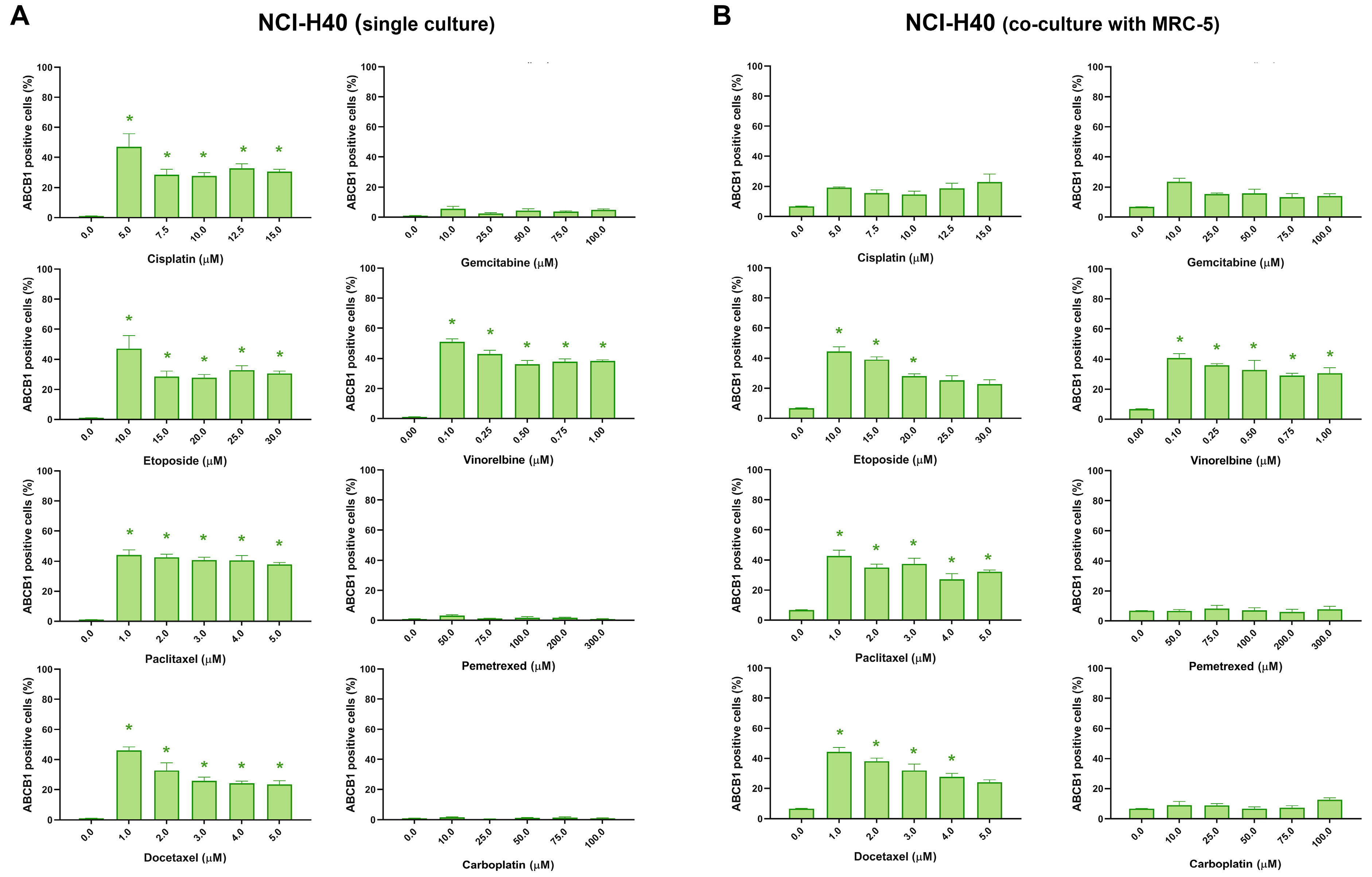
3.3. Assessment of MDR Marker Expression after Chemotherapeutics Treatment in NSCLC Cell Lines through Immunofluorescence Assay
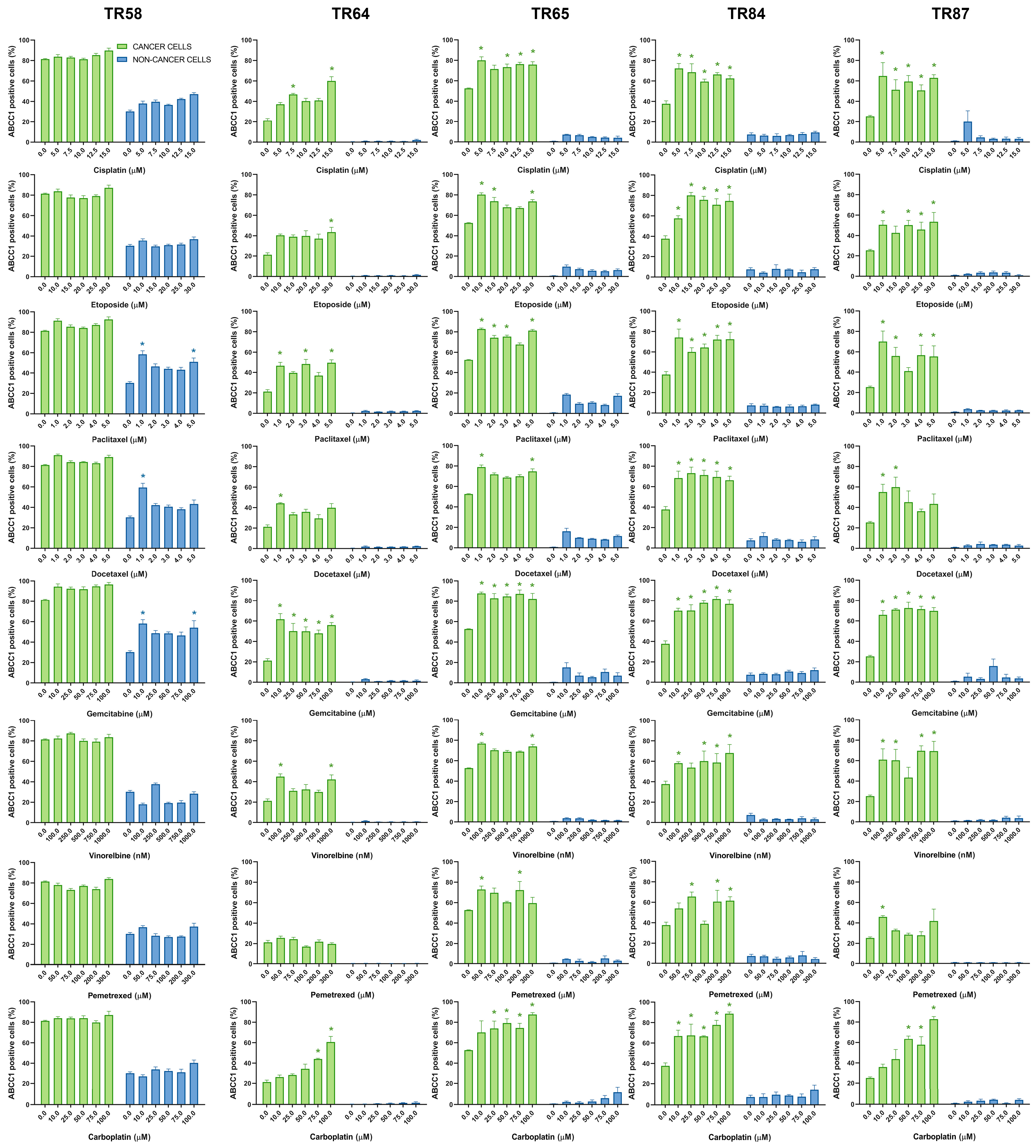
3.4. Assessment of MDR Marker Expression after Chemotherapeutics Treatment in Patient-Derived NSCLC Cultures through Immunofluorescence Assay
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Howlader, N.; Forjaz, G.; Mooradian, M.J.; Meza, R.; Kong, C.Y.; Cronin, K.A.; Mariotto, A.B.; Lowy, D.R.; Feuer, E.J. The Effect of Advances in Lung-Cancer Treatment on Population Mortality. N. Engl. J. Med. 2020, 383, 640–649. [Google Scholar] [CrossRef] [PubMed]
- Xiao, H.; Zheng, Y.; Ma, L.; Tian, L.; Sun, Q. Clinically-Relevant ABC Transporter for Anti-Cancer Drug Resistance. Front. Pharmacol. 2021, 12, 648407. [Google Scholar] [CrossRef] [PubMed]
- Emran, T.B.; Shahriar, A.; Mahmud, A.R.; Rahman, T.; Abir, M.H.; Siddiquee, M.F.; Ahmed, H.; Rahman, N.; Nainu, F.; Wahyudin, E.; et al. Multidrug Resistance in Cancer: Understanding Molecular Mechanisms, Immunoprevention and Therapeutic Approaches. Front. Oncol. 2022, 12, 891652. [Google Scholar] [CrossRef] [PubMed]
- Sissung, T.M.; Baum, C.E.; Kirkland, C.T.; Gao, R.; Gardner, E.R.; Figg, W.D. Pharmacogenetics of membrane transporters: An update on current approaches. Mol. Biotechnol. 2010, 44, 152–167. [Google Scholar] [CrossRef] [PubMed]
- He, J.; Fortunati, E.; Liu, D.X.; Li, Y. Pleiotropic Roles of ABC Transporters in Breast Cancer. Int. J. Mol. Sci. 2021, 22, 3199. [Google Scholar] [CrossRef] [PubMed]
- Vesel, M.; Rapp, J.; Feller, D.; Kiss, E.; Jaromi, L.; Meggyes, M.; Miskei, G.; Duga, B.; Smuk, G.; Laszlo, T.; et al. ABCB1 and ABCG2 drug transporters are differentially expressed in non-small cell lung cancers (NSCLC) and expression is modified by cisplatin treatment via altered Wnt signaling. Respir. Res. 2017, 18, 52. [Google Scholar] [CrossRef] [PubMed]
- Melguizo, C.; Prados, J.; Luque, R.; Ortiz, R.; Caba, O.; Alvarez, P.J.; Gonzalez, B.; Aranega, A. Modulation of MDR1 and MRP3 gene expression in lung cancer cells after paclitaxel and carboplatin exposure. Int. J. Mol. Sci. 2012, 13, 16624–16635. [Google Scholar] [CrossRef] [PubMed]
- Zhang, L.; Ou, W.; Liu, Q.; Li, N.; Liu, L.; Wang, S. Pemetrexed plus carboplatin as adjuvant chemotherapy in patients with curative resected non-squamous non-small cell lung cancer. Thorac. Cancer 2014, 5, 50–56. [Google Scholar] [CrossRef]
- Omori, M.; Noro, R.; Seike, M.; Matsuda, K.; Hirao, M.; Fukuizumi, A.; Takano, N.; Miyanaga, A.; Gemma, A. Inhibitors of ABCB1 and ABCG2 overcame resistance to topoisomerase inhibitors in small cell lung cancer. Thorac. Cancer 2022, 13, 2142–2151. [Google Scholar] [CrossRef]
- Berger, W.; Setinek, U.; Hollaus, P.; Zidek, T.; Steiner, E.; Elbling, L.; Cantonati, H.; Attems, J.; Gsur, A.; Micksche, M. Multidrug resistance markers P-glycoprotein, multidrug resistance protein 1, and lung resistance protein in non-small cell lung cancer: Prognostic implications. J. Cancer Res. Clin. Oncol. 2005, 131, 355–363. [Google Scholar] [CrossRef]
- Stankovic, T.; Stojsic, J.; Dragoj, M.; Milovanovic, Z.; Milosevic, Z.; Milinkovic, V.; Skodric-Trifunovic, V.; Denic-Markovic, L.; Pesic, M.; Stojkovic Buric, S.; et al. Chemosensitivity and survival of non-small cell lung carcinoma patients receiving neoadjuvant therapy depend on the expression of multidrug efflux transporters. JMCM 2019, 2, 129–135. [Google Scholar] [CrossRef]
- Kodack, D.P.; Farago, A.F.; Dastur, A.; Held, M.A.; Dardaei, L.; Friboulet, L.; von Flotow, F.; Damon, L.J.; Lee, D.; Parks, M.; et al. Primary Patient-Derived Cancer Cells and Their Potential for Personalized Cancer Patient Care. Cell Rep. 2017, 21, 3298–3309. [Google Scholar] [CrossRef] [PubMed]
- Morand du Puch, C.B.; Vanderstraete, M.; Giraud, S.; Lautrette, C.; Christou, N.; Mathonnet, M. Benefits of functional assays in personalized cancer medicine: More than just a proof-of-concept. Theranostics 2021, 11, 9538–9556. [Google Scholar] [CrossRef] [PubMed]
- Rodrigues Lopes, I.; Silva, R.J.; Caramelo, I.; Eulalio, A.; Mano, M. Shedding light on microRNA function via microscopy-based screening. Methods 2019, 152, 55–64. [Google Scholar] [CrossRef] [PubMed]
- Hynes, J.; Carey, C.; Will, Y. Fluorescence-Based Microplate Assays for In Vitro Assessment of Mitochondrial Toxicity, Metabolic Perturbation, and Cellular Oxygenation. Curr. Protoc. Toxicol. 2016, 70, 2–16. [Google Scholar] [CrossRef] [PubMed]
- Cribbes, S.; Kessel, S.; McMenemy, S.; Qiu, J.; Chan, L.L. A Novel Multiparametric Drug-Scoring Method for High-Throughput Screening of 3D Multicellular Tumor Spheroids Using the Celigo Image Cytometer. SLAS Discov. Adv. Life Sci. R D 2017, 22, 547–557. [Google Scholar] [CrossRef] [PubMed]
- Gorshkov, K.; Pradhan, M.; Xu, M.; Yang, S.; Lee, E.M.; Chen, C.Z.; Shen, M.; Zheng, W. Cell-Based No-Wash Fluorescence Assays for Compound Screens Using a Fluorescence Cytometry Plate Reader. J. Pharmacol. Exp. Ther. 2020, 374, 500–511. [Google Scholar] [CrossRef]
- Hayford, C.E.; Tyson, D.R.; Robbins, C.J., 3rd; Frick, P.L.; Quaranta, V.; Harris, L.A. An in vitro model of tumor heterogeneity resolves genetic, epigenetic, and stochastic sources of cell state variability. PLoS Biol. 2021, 19, e3000797. [Google Scholar] [CrossRef]
- Bergin, C.J.; Benoit, Y.D. Protocol for serial organoid formation assay using primary colorectal cancer tissues to evaluate cancer stem cell activity. STAR Protoc. 2022, 3, 101218. [Google Scholar] [CrossRef]
- Vunnam, N.; Young, M.C.; Liao, E.E.; Lo, C.H.; Huber, E.; Been, M.; Thomas, D.D.; Sachs, J.N. Nimesulide, a COX-2 inhibitor, sensitizes pancreatic cancer cells to TRAIL-induced apoptosis by promoting DR5 clustering dagger. Cancer Biol. Ther. 2023, 24, 2176692. [Google Scholar] [CrossRef]
- Pesic, M.; Markovic, J.Z.; Jankovic, D.; Kanazir, S.; Markovic, I.D.; Rakic, L.; Ruzdijic, S. Induced resistance in the human non small cell lung carcinoma (NCI-H460) cell line in vitro by anticancer drugs. J. Chemother. 2006, 18, 66–73. [Google Scholar] [CrossRef] [PubMed]
- Chen, Z.; Fillmore, C.M.; Hammerman, P.S.; Kim, C.F.; Wong, K.K. Non-small-cell lung cancers: A heterogeneous set of diseases. Nat. Rev. Cancer 2014, 14, 535–546. [Google Scholar] [CrossRef] [PubMed]
- Dinic, J.; Podolski-Renic, A.; Jovanovic, M.; Musso, L.; Tsakovska, I.; Pajeva, I.; Dallavalle, S.; Pesic, M. Novel Heat Shock Protein 90 Inhibitors Suppress P-Glycoprotein Activity and Overcome Multidrug Resistance in Cancer Cells. Int. J. Mol. Sci. 2019, 20, 4575. [Google Scholar] [CrossRef] [PubMed]
- Podolski-Renic, A.; Dinic, J.; Stankovic, T.; Jovanovic, M.; Ramovic, A.; Pustenko, A.; Zalubovskis, R.; Pesic, M. Sulfocoumarins, specific carbonic anhydrase IX and XII inhibitors, interact with cancer multidrug resistant phenotype through pH regulation and reverse P-glycoprotein mediated resistance. Eur. J. Pharm. Sci. Off. J. Eur. Fed. Pharm. Sci. 2019, 138, 105012. [Google Scholar] [CrossRef] [PubMed]
- Dragoj, M.; Milosevic, Z.; Bankovic, J.; Tanic, N.; Pesic, M.; Stankovic, T. Targeting CXCR4 and FAK reverses doxorubicin resistance and suppresses invasion in non-small cell lung carcinoma. Cell. Oncol. 2017, 40, 47–62. [Google Scholar] [CrossRef]
- de Wit, M.; Gao, Y.; Mercieca, D.; de Heer, I.; Valkenburg, B.; van Royen, M.E.; Aerts, J.; Sillevis Smitt, P.; French, P. Mutation and drug-specific intracellular accumulation of EGFR predict clinical responses to tyrosine kinase inhibitors. EBioMedicine 2020, 56, 102796. [Google Scholar] [CrossRef]
- Fukuhara, T.; Maemondo, M.; Inoue, A.; Kobayashi, K.; Sugawara, S.; Oizumi, S.; Isobe, H.; Gemma, A.; Harada, M.; Yoshizawa, H.; et al. Factors associated with a poor response to gefitinib in the NEJ002 study: Smoking and the L858R mutation. Lung Cancer 2015, 88, 181–186. [Google Scholar] [CrossRef]



| Cell Lines | IC50 (µM) | |||||||
|---|---|---|---|---|---|---|---|---|
| Cisplatin | Etoposide | Paclitaxel | Docetaxel | Gemcitabine | Vinorelbine | Pemetrexed | Carboplatin | |
| NCI-H460 | 5.705 | 4.809 | 0.3808 | 0.5541 | 41.565 | 0.03656 | >300 | >100 |
| NCI-H460 (co-culture with MRC-5) | 9.791 | 6.074 | 0.3391 | 0.6416 | 34.588 | 0.03332 | >300 | >100 |
| NCI-H460/R | 5.334 | 7.922 | >5 | 0.7041 | 33.588 | 0.9902 | >300 | >100 |
| NCI-H460/R (co-culture with MRC-5) | 5.599 | 7.500 | >5 | 0.8008 | 29.388 | 0.8385 | >300 | >100 |
| MRC-5 | 14.682 s | >30 s | >5 s | >5 s | >100 s | >1 s | >300 | >100 |
| MRC-5 (co-culture with NCI-H460) | >15 s | >30 s | >5 s | >5 s | >100 s | 0.4773 s | >300 | >100 |
| MRC-5 (co-culture with NCI-H460/R) | >15 s | >30 s | >5 | >5 s | >100 s | >1 s | >300 | >100 |
| Cell Lines | ABCB1 Expression * | |||||||
|---|---|---|---|---|---|---|---|---|
| Cisplatin | Etoposide | Paclitaxel | Docetaxel | Gemcitabine | Vinorelbine | Pemetrexed | Carboplatin | |
| NCI-H460 | increase | increase | increase | increase | no change | increase | no change | no change |
| NCI-H460 (co-culture with MRC-5) | no change | increase | increase | increase | no change | increase | no change | no change |
| NCI-H460/R | no change | no change | no change | no change | no change | no change | no change | no change |
| NCI-H460/R (co-culture with MRC-5) | no change | no change | no change | no change | no change | no change | no change | no change |
| MRC-5 | no change | no change | no change | no change | no change | no change | no change | no change |
| MRC-5 (co-culture with NCI-H460) | no change | no change | no change | no change | no change | no change | no change | no change |
| MRC-5 (co-culture with NCI-H460/R) | no change | no change | no change | no change | no change | no change | no change | no change |
| NSCLC Cultures | IC50 (µM) | |||||||
|---|---|---|---|---|---|---|---|---|
| Cisplatin | Etoposide | Paclitaxel | Docetaxel | Gemcitabine | Vinorelbine | Pemetrexed | Carboplatin | |
| Cancer cells (CK8/18+) | ||||||||
| TR58 | 5.780 | 7.131 | 1.180 | 1.312 | 2.550 | 0.1291 | 180.739 | 60.842 |
| TR64 | 6.067 | 11.919 | 1.354 | 1.560 | 4.924 | 0.1716 | 251.855 | 38.982 |
| TR65 | 14.636 | 25.393 | 1.180 | >5 | 4.912 | >1 | 231.473 | 22.492 |
| TR84 | 2.112 | 2.983 | 2.568 | 1.850 | 2.683 | 0.1763 | >300 | 8.147 |
| TR87 | 4.821 | 12.980 | 4.787 | >5 | 3.407 | >1 | >300 | 43.253 |
| Non-cancer cells (CK8/18-) | ||||||||
| TR58 | 5.104 | 6.490 | 1.399 | 1.594 | 3.074 s | 0.1601 | >300 s | 27.334 |
| TR64 | 8.255 s | 18.133 s | 1.953 | 2.725 s | 7.063 s | 0.1840 | >300 s | 12.755 |
| TR65 | 1.205 | 2.076 | 1.399 | 0.4610 | 0.3511 | 0.03281 | 25.177 | 2.161 |
| TR84 | 0.7891 | 1.013 | 0.312 | 0.1819 | 0.4909 | 0.03387 | >300 | 1.280 |
| TR87 | 3.381 | 10.205 | 1.866 | 3.051 | 1.128 | 0.3606 | >300 | 22.921 |
| NSCLC Cultures Cancer Cells (CK8/18+) | Cisplatin | Etoposide | Paclitaxel | Docetaxel | Gemcitabine | Vinorelbine | Pemetrexed | Carboplatin |
|---|---|---|---|---|---|---|---|---|
| ABCB1 expression * | ||||||||
| TR58 | no change | no change | increase | no change | no change | no change | no change | no change |
| TR64 | increase | increase | increase | increase | increase | increase | no change | increase |
| TR65 | increase | increase | increase | no change | increase | increase | no change | increase |
| TR84 | increase | increase | increase | increase | increase | increase | increase | increase |
| TR87 | increase | increase | increase | increase | increase | increase | increase | increase |
| ABCC1 expression * | ||||||||
| TR58 | no change | no change | no change | no change | no change | no change | no change | no change |
| TR64 | increase | increase | increase | increase | increase | increase | no change | increase |
| TR65 | increase | increase | increase | increase | increase | increase | increase | increase |
| TR84 | increase | increase | increase | increase | increase | increase | increase | increase |
| TR87 | increase | increase | increase | increase | increase | increase | increase | increase |
| ABCG2 expression * | ||||||||
| TR58 | increase | no change | increase | increase | increase | increase | no change | increase |
| TR64 | increase | increase | increase | increase | increase | increase | no change | increase |
| TR65 | increase | increase | increase | no change | increase | increase | no change | increase |
| TR84 | increase | increase | increase | increase | increase | increase | increase | increase |
| TR87 | increase | increase | increase | increase | increase | increase | no change | increase |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Dinić, J.; Podolski-Renić, A.; Dragoj, M.; Jovanović Stojanov, S.; Stepanović, A.; Lupšić, E.; Pajović, M.; Jovanović, M.; Petrović Rodić, D.; Marić, D.; et al. Immunofluorescence-Based Assay for High-Throughput Analysis of Multidrug Resistance Markers in Non-Small Cell Lung Carcinoma Patient-Derived Cells. Diagnostics 2023, 13, 3617. https://doi.org/10.3390/diagnostics13243617
Dinić J, Podolski-Renić A, Dragoj M, Jovanović Stojanov S, Stepanović A, Lupšić E, Pajović M, Jovanović M, Petrović Rodić D, Marić D, et al. Immunofluorescence-Based Assay for High-Throughput Analysis of Multidrug Resistance Markers in Non-Small Cell Lung Carcinoma Patient-Derived Cells. Diagnostics. 2023; 13(24):3617. https://doi.org/10.3390/diagnostics13243617
Chicago/Turabian StyleDinić, Jelena, Ana Podolski-Renić, Miodrag Dragoj, Sofija Jovanović Stojanov, Ana Stepanović, Ema Lupšić, Milica Pajović, Mirna Jovanović, Dušica Petrović Rodić, Dragana Marić, and et al. 2023. "Immunofluorescence-Based Assay for High-Throughput Analysis of Multidrug Resistance Markers in Non-Small Cell Lung Carcinoma Patient-Derived Cells" Diagnostics 13, no. 24: 3617. https://doi.org/10.3390/diagnostics13243617
APA StyleDinić, J., Podolski-Renić, A., Dragoj, M., Jovanović Stojanov, S., Stepanović, A., Lupšić, E., Pajović, M., Jovanović, M., Petrović Rodić, D., Marić, D., Ercegovac, M., & Pešić, M. (2023). Immunofluorescence-Based Assay for High-Throughput Analysis of Multidrug Resistance Markers in Non-Small Cell Lung Carcinoma Patient-Derived Cells. Diagnostics, 13(24), 3617. https://doi.org/10.3390/diagnostics13243617









