Genomic Diversity in the Endosymbiotic Bacterium Rhizobium leguminosarum
Abstract
1. Introduction
2. Materials and Methods
2.1. Growth Conditions & DNA Preparation
2.2. Genome Sequencing, Assembly, & Quality Check
2.3. Genome Annotation
2.4. Comparative Genomics
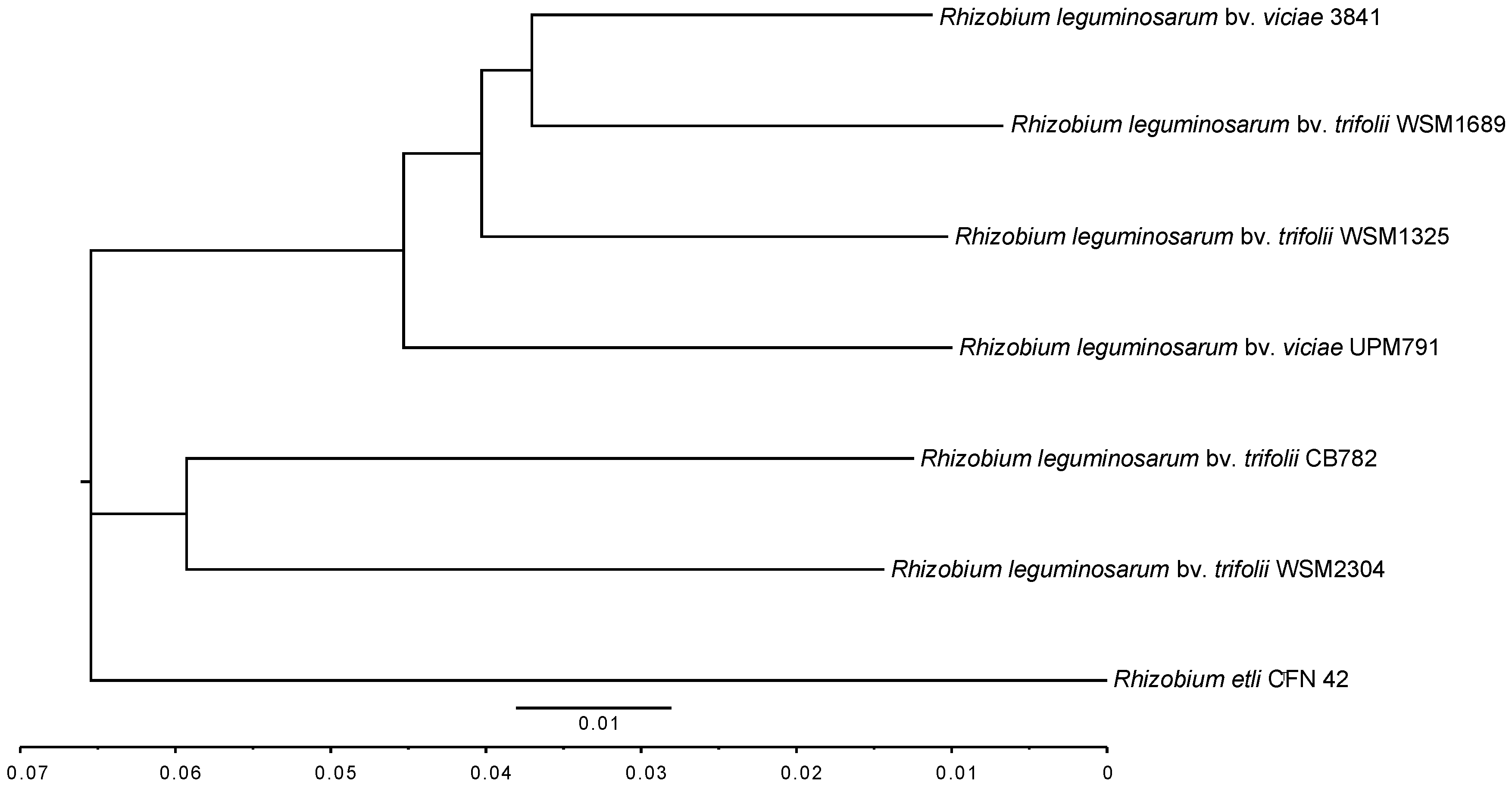
2.4.1. 16S Phylogeny and Average Nucleotide Identity
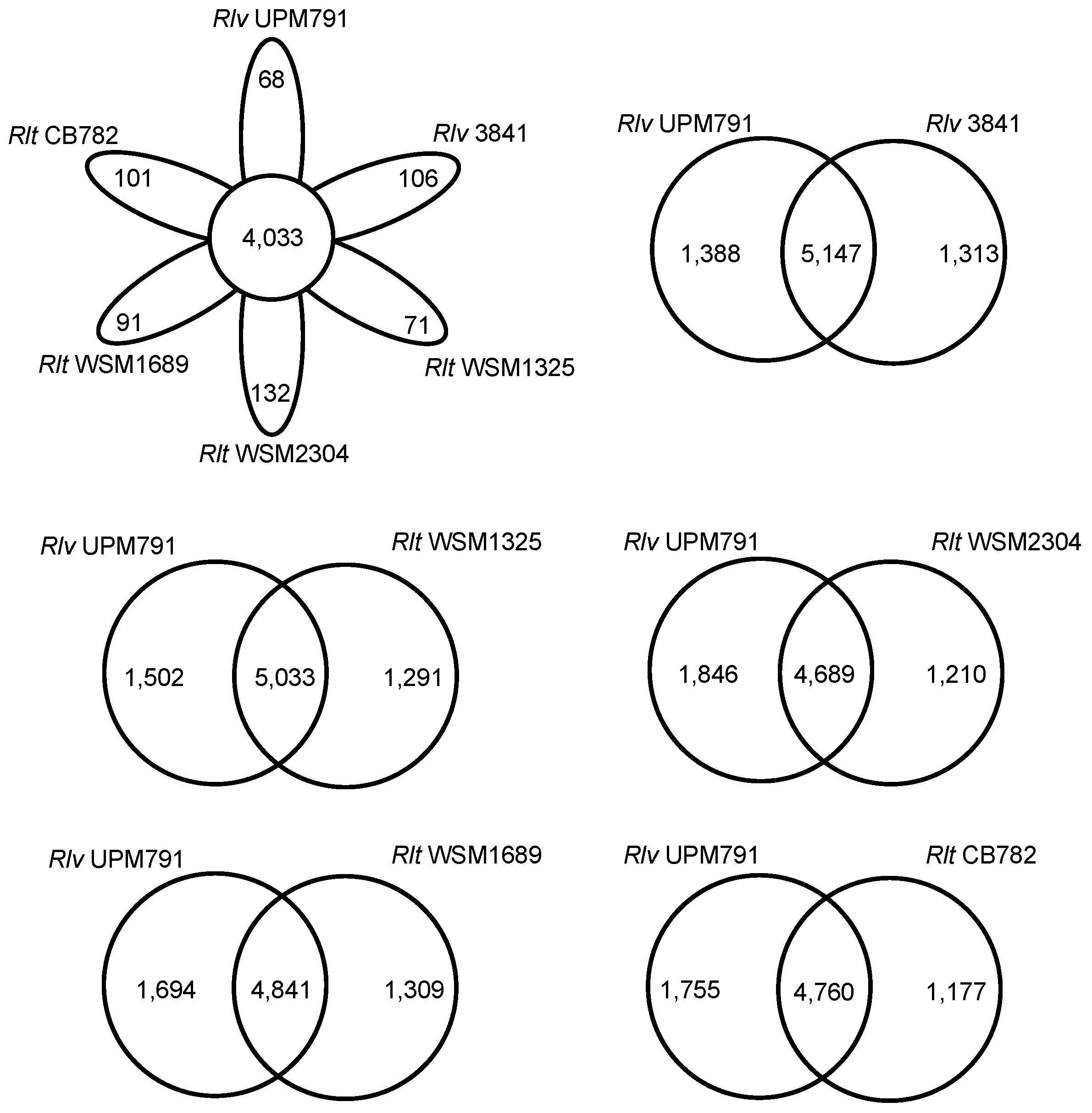
2.4.2. Protein Cluster Analysis
3. Results
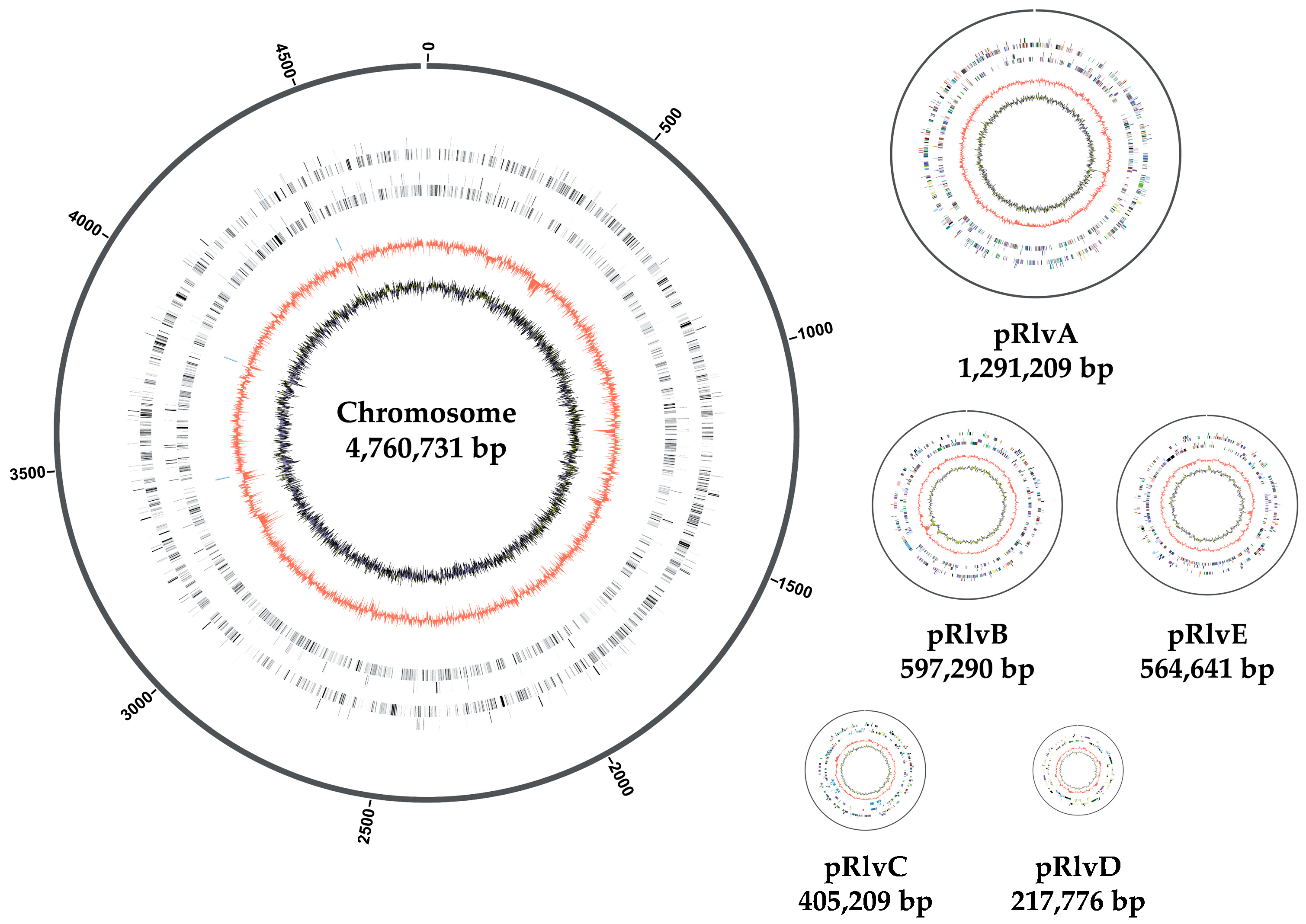
3.1. Rhizobium leguminosarum bv. viciae UPM791 Complete Genome Is a New Reference Sequence for Endosymbiotic Bacteria
3.2. Rhizobium leguminosarum bv. viciae UPM791 Global Genome Comparisons
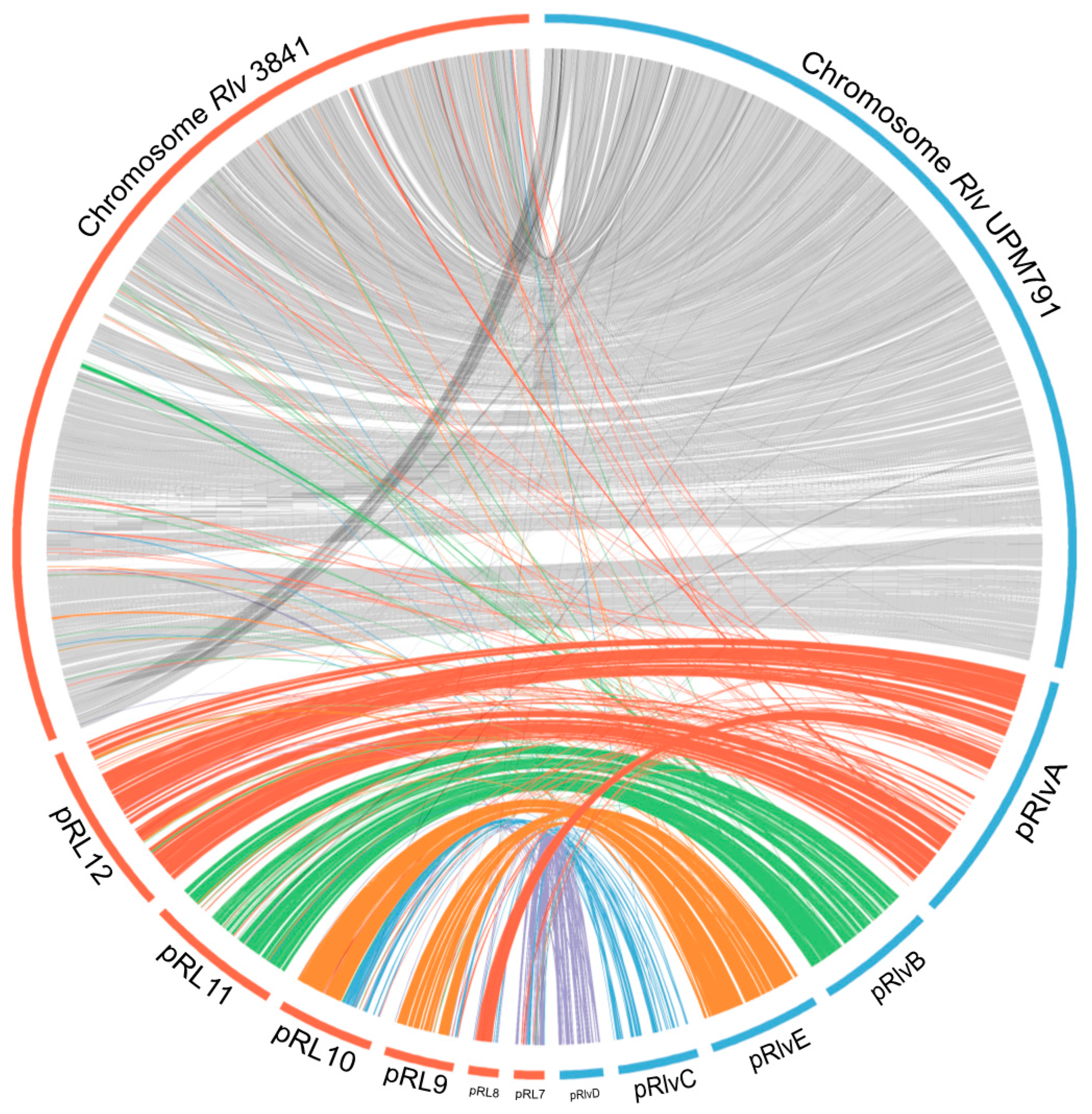
3.3. High Diversity of the Plasmid-Associated Genome in Rhizobium leguminosarum
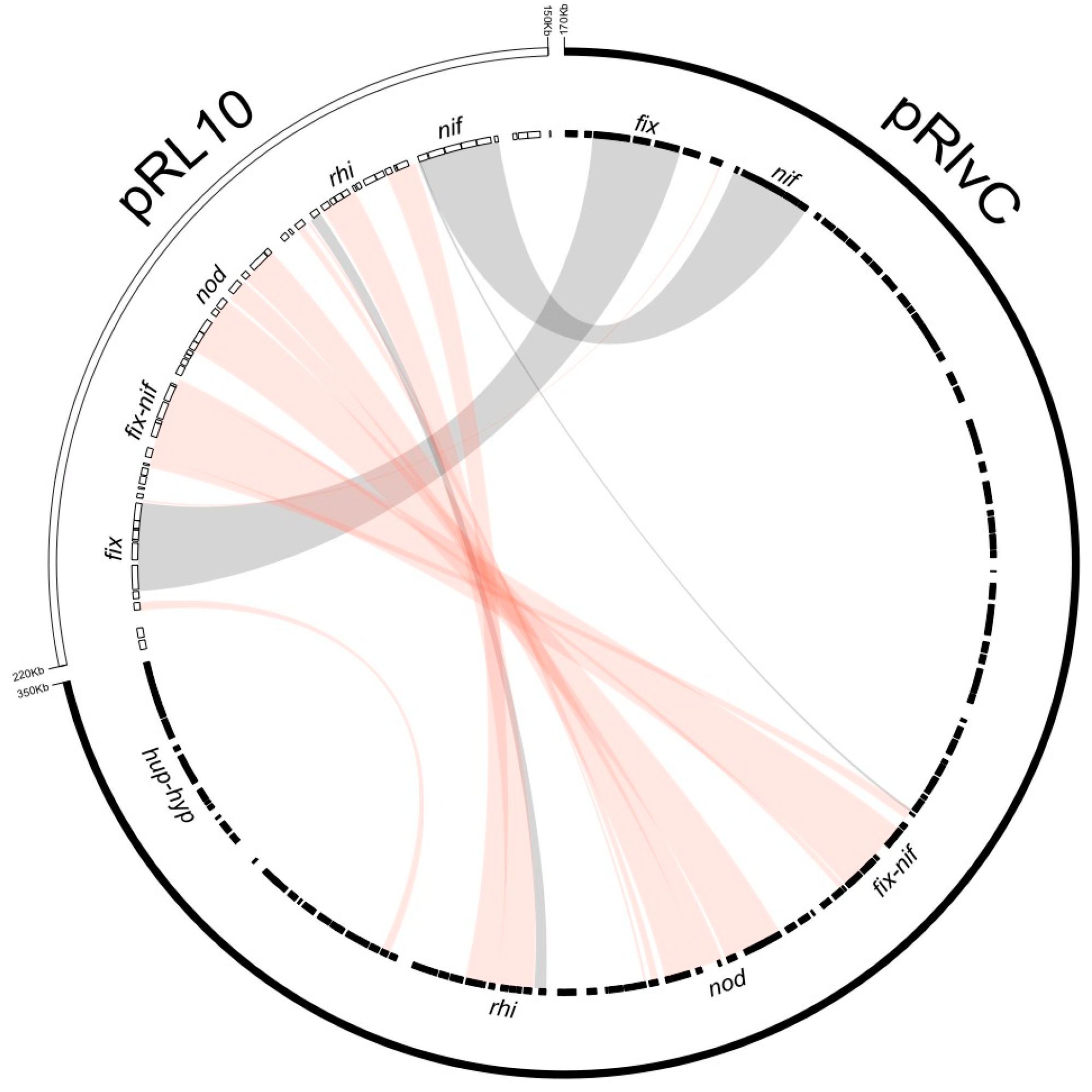
3.4. Rhizobium leguminosarum bv. viciae UPM791 Harbours a Unique Symbiotic Plasmid
3.5. Specific Traits Define the Physiology of Each Rhizobial Strain
3.5.1. Metabolic Pathways
3.5.2. Transport Systems
3.5.3. Motility and Chemotaxis Proteins
3.5.4. Cell-Surface Polysaccharide Production
3.5.5. Secretion Systems
3.5.6. Control of Microoxic Metabolism
3.5.7. Regulatory Proteins
3.5.8. Stress Proteins
4. Discussion
5. Conclusions
Supplementary Materials
Acknowledgments
Author Contributions
Conflicts of Interest
Data Deposition
References
- Reeve, W.; O’Hara, G.; Chain, P.; Ardley, J.; Brau, L.; Nandesena, K.; Tiwari, R.; Malfatti, S.; Kiss, H.; Lapidus, A.; et al. Complete genome sequence of Rhizobium leguminosarum bv. trifolii strain WSM2304, an effective microsymbiont of the South American clover Trifolium polymorphum. Stand. Genom. Sci. 2010, 2, 66–76. [Google Scholar] [CrossRef] [PubMed]
- Downie, J.A. The roles of extracellular proteins, polysaccharides and signals in the interactions of rhizobia with legume roots. FEMS Microbiol. Rev. 2010, 34, 150–170. [Google Scholar] [CrossRef] [PubMed]
- Genomes OnLine Database. Available online: http://gold.jgi.doe.gov (accessed on 21 November 2017).
- Segovia, L.; Young, J.P.; Martinez-Romero, E. Reclassification of American Rhizobium leguminosarum biovar phaseoli Type I strains as Rhizobium etli sp. nov. Int. J. Syst. Bacteriol. 1993, 43, 374–377. [Google Scholar] [CrossRef] [PubMed]
- Johnston, A.W.; Beringer, J.E. Identification of the Rhizobium strains in pea root nodules using genetic markers. J. Gen. Microbiol. 1975, 87, 343–350. [Google Scholar] [CrossRef] [PubMed]
- Young, J.P.; Crossman, L.C.; Johnston, A.W.; Thomson, N.R.; Ghazoui, Z.F.; Hull, K.H.; Wexler, M.; Curson, A.R.; Todd, J.D.; Poole, P.S.; et al. The genome of Rhizobium leguminosarum has recognizable core and accessory components. Genome Biol. 2006, 7, R34. [Google Scholar] [CrossRef] [PubMed]
- Reeve, W.; O’Hara, G.; Chain, P.; Ardley, J.; Brau, L.; Nandesena, K.; Tiwari, R.; Copeland, A.; Nolan, M.; Han, C.; et al. Complete genome sequence of Rhizobium leguminosarum bv. trifolii strain WSM1325, an effective microsymbiont of annual mediterranean clovers. Stand. Genom. Sci. 2010, 2, 347–356. [Google Scholar] [CrossRef] [PubMed]
- Terpolilli, J.; Rui, T.; Yates, R.; Howieson, J.; Poole, P.; Munk, C.; Tapia, R.; Han, C.; Markowitz, V.; Tatiparthi, R.; et al. Genome sequence of Rhizobium leguminosarum bv trifolii strain WSM1689, the microsymbiont of the one flowered clover Trifolium uniflorum. Stand. Genom. Sci. 2014, 9, 527–539. [Google Scholar] [CrossRef] [PubMed]
- Downie, J.A.; Young, J.P. Genome sequencing. The ABC of symbiosis. Nature 2001, 412, 597–598. [Google Scholar] [CrossRef] [PubMed]
- diCenzo, G.C.; Finan, T.M. The divided bacterial genome: Structure, function, and evolution. Microbiol. Mol. Biol. Rev. 2017, 81, e00019-17. [Google Scholar] [CrossRef] [PubMed]
- Cevallos, M.A.; Cervantes-Rivera, R.; Gutierrez-Rios, R.M. The repABC plasmid family. Plasmid 2008, 60, 19–37. [Google Scholar] [CrossRef] [PubMed]
- MacLean, A.M.; Finan, T.M.; Sadowsky, M.J. Genomes of the symbiotic nitrogen-fixing bacteria of legumes. Plant Physiol. 2007, 144, 615–622. [Google Scholar] [CrossRef] [PubMed]
- Pini, F.; Galardini, M.; Bazzicalupo, M.; Mengoni, A. Plant-bacteria association and symbiosis: Are there common genomic traits in alphaproteobacteria? Genes 2011, 2, 1017–1032. [Google Scholar] [CrossRef] [PubMed]
- Black, M.; Moolhuijzen, P.; Chapman, B.; Barrero, R.; Howieson, J.; Hungria, M.; Bellgard, M. The genetics of symbiotic nitrogen fixation: Comparative genomics of 14 rhizobia strains by resolution of protein clusters. Genes 2012, 3, 138–166. [Google Scholar] [CrossRef] [PubMed]
- Tian, C.F.; Zhou, Y.J.; Zhang, Y.M.; Li, Q.Q.; Zhang, Y.Z.; Li, D.F.; Wang, S.; Wang, J.; Gilbert, L.B.; Li, Y.R.; et al. Comparative genomics of rhizobia nodulating soybean suggests extensive recruitment of lineage-specific genes in adaptations. Proc. Natl. Acad. Sci. USA 2012, 109, 8629–8634. [Google Scholar] [CrossRef] [PubMed]
- Sugawara, M.; Epstein, B.; Badgley, B.D.; Unno, T.; Xu, L.; Reese, J.; Gyaneshwar, P.; Denny, R.; Mudge, J.; Bharti, A.K.; et al. Comparative genomics of the core and accessory genomes of 48 Sinorhizobium strains comprising five genospecies. Genome Biol. 2013, 14, R17. [Google Scholar] [CrossRef] [PubMed]
- Leyva, A.; Palacios, J.M.; Mozo, T.; Ruiz-Argüeso, T. Cloning and characterization of hydrogen uptake genes from Rhizobium leguminosarum. J. Bacteriol. 1987, 169, 4929–4934. [Google Scholar] [CrossRef] [PubMed]
- Ruiz-Argüeso, T.; Hanus, F.J.; Evans, H.J. Hydrogen production and uptake by pea nodules as affected by strains of Rhizobium leguminosarum. Arch. Microbiol. 1978, 116, 113–118. [Google Scholar] [CrossRef]
- Beringer, J.E. R factor transfer in Rhizobium leguminosarum. Microbiology 1974, 84, 188–198. [Google Scholar] [CrossRef] [PubMed]
- Wilson, K. Preparation of genomic DNA from bacteria. Curr. Protoc. Mol. Biol. 2001. [Google Scholar] [CrossRef]
- Chin, C.; Alexander, D.; Marks, P.; Klammer, A.A.; Drake, J.; Heiner, C.; Clum, A.; Copeland, A.; Huddleston, J.; Eichler, E.; et al. Nonhybrid, finished microbial genome assemblies from long-read SMRT sequencing data. Nat. Methods 2013, 10, 563–569. [Google Scholar] [CrossRef] [PubMed]
- Bankevich, A.; Nurk, S.; Antipov, D.; Gurevich, A.A.; Dvorkin, M.; Kulikov, A.S.; Lesin, V.M.; Nikolenko, S.I.; Pham, S.; Prjibelski, A.D.; et al. SPAdes: A new genome assembly algorithm and its applications to single-cell sequencing. J. Comput. Biol. 2012, 19, 455–477. [Google Scholar] [CrossRef] [PubMed]
- Myers, E.W.; Sutton, G.G.; Delcher, A.L.; Dew, I.M.; Fasulo, D.P.; Flanigan, M.J.; Kravitz, S.A.; Mobarry, C.M.; Reinert, K.H.J.; Remington, K.A.; et al. A whole-genome assembly of Drosophila. Science 2000, 287, 2196–2204. [Google Scholar] [CrossRef] [PubMed]
- Chevreux, B.; Wetter, T.; Suhai, S. Genome sequence assembly using trace signals and additional sequence information. In Computer Science and Biology: Proceedings of the German Conference on Bioinformatics, GCB’99, Hannover, Germany, 4–6 October 1999; GBF-Braunschweig, Department of Bioinformatics: Braunschweig, Germany, 1999; pp. 45–56. [Google Scholar]
- Assefa, S.; Keane, T.M.; Otto, T.D.; Newbold, C.; Berriman, M. ABACAS: Algorithm-based automatic contiguation of assembled sequences. Bioinformatics 2009, 25, 1968–1969. [Google Scholar] [CrossRef] [PubMed]
- Bonfield, J.K.; Smith, K.; Staden, R. A new DNA sequence assembly program. Nucl. Acids Res. 1995, 23, 4992–4999. [Google Scholar] [CrossRef] [PubMed]
- Staden, R. The staden sequence analysis package. Mol. Biotechnol. 1996, 5, 233–241. [Google Scholar] [CrossRef] [PubMed]
- Hunt, M.; Silva, N.D.; Otto, T.; Parkhill, J.; Keane, J.; Harris, S. Circlator: Automated circularization of genome assemblies using long sequencing reads. Genome Biol. 2015. [Google Scholar] [CrossRef] [PubMed]
- Treangen, T.; Sommer, D.; Angly, F.; Koren, S.; Pop, M. Next generation sequence assembly with AMOS. Curr. Protoc. Bioinform. 2011, 33, 11.8.2–11.8.18. [Google Scholar]
- Jorrin, B.; Imperial, J. Population genomics analysis of legume host preference for specific rhizobial genotypes in the Rhizobium leguminosarum bv. viciae symbioses. Mol. Plant-Microbe Interact. 2015, 28, 310–318. [Google Scholar] [CrossRef] [PubMed]
- Langmead, B.; Salzberg, S.L. Fast gapped-read alignment with Bowtie 2. Nat. Methods 2012, 9, 357–359. [Google Scholar] [CrossRef] [PubMed]
- Li, H.; Handsaker, B.; Wysoker, A.; Fennell, T.; Ruan, J.; Homer, N.; Marth, G.; Abecasis, G.; Durbin, R.; Genome Project Data, P. The sequence Alignment/Map format and SAMtools. Bioinformatics 2009, 25, 2078–2079. [Google Scholar] [CrossRef] [PubMed]
- Koboldt, D.C.; Zhang, Q.Y.; Larson, D.E.; Shen, D.; McLellan, M.D.; Lin, L.; Miller, C.A.; Mardis, E.R.; Ding, L.; Wilson, R.K. VarScan 2: Somatic mutation and copy number alteration discovery in cancer by exome sequencing. Genome Res. 2012, 22, 568–576. [Google Scholar] [CrossRef] [PubMed]
- Robinson, J.T.; Thorvaldsdottir, H.; Winckler, W.; Guttman, M.; Lander, E.S.; Getz, G.; Mesirov, J.P. Integrative genomics viewer. Nat. Biotech. 2011, 29, 24–26. [Google Scholar] [CrossRef] [PubMed]
- Galens, K.; Orvis, J.; Daugherty, S.; Creasy, H.H.; Angiuoli, S.; White, O.; Wortman, J.; Mahurkar, A.; Giglio, M.G. The IGS standard operating procedure for automated prokaryotic annotation. Stand. Genom. Sci. 2011, 4, 244–251. [Google Scholar] [CrossRef] [PubMed]
- Delcher, A.L.; Harmon, D.; Kasif, S.; White, O.; Salzberg, S.L. Improved microbial gene identification with GLIMMER. Nucl. Acids Res. 1999, 27, 4636–4641. [Google Scholar] [CrossRef] [PubMed]
- Lowe, T.M.; Chan, P.P. tRNAscan-SE On-line: Integrating search and context for analysis of transfer RNA genes. Nucleic Acids Res. 2016, 44, W54–W57. [Google Scholar] [CrossRef] [PubMed]
- Lagesen, K.; Hallin, P.; Rodland, E.A.; Staerfeldt, H.H.; Rognes, T.; Ussery, D.W. RNAmmer: Consistent and rapid annotation of ribosomal RNA genes. Nucleic Acids Res, 2007, 35, 3100–3108. [Google Scholar] [CrossRef] [PubMed]
- Tamura, K.; Stecher, G.; Peterson, D.; Filipski, A.; Kumar, S. MEGA6: Molecular evolutionary genetics analysis version 6.0. Mol. Biol. Evol. 2013, 30, 2725–2729. [Google Scholar] [CrossRef] [PubMed]
- Delcher, A.L.; Salzberg, S.L.; Phillippy, A.M. Using MUMmer to identify similar regions in large sequence sets. Curr Protoc Bioinformatics 2003, 10. Unit 10.3. [Google Scholar] [PubMed]
- Richter, M.; Rossello-Mora, R. Shifting the genomic gold standard for the prokaryotic species definition. Proc. Natl. Acad. Sci. USA 2009, 106, 19126–19131. [Google Scholar] [CrossRef] [PubMed]
- Sneath, P.; Sokal, R. Numerical Taxonomy: The Principles and Practice of Numerical Classification; Freeman: San Francisco, CA, USA, 1973. [Google Scholar]
- Richter, M.; Rossello-Mora, R.; Oliver Glockner, F.; Peplies, J. JSpeciesWS: A web server for prokaryotic species circumscription based on pairwise genome comparison. Bioinformatics 2016, 32, 929–931. [Google Scholar] [CrossRef] [PubMed]
- Vesth, T.; Lagesen, K.; Acar, N.; Ussery, D. CMG-Biotools, a free workbench for basic comparative microbial genomics. PLoS ONE 2013, 8, e60120. [Google Scholar] [CrossRef] [PubMed]
- Reeve, W.; Ardley, J.; Tian, R.; Eshragi, L.; Yoon, J.W.; Ngamwisetkun, P.; Seshadri, R.; Ivanova, N.N.; Kyrpides, N.C. A genomic encyclopedia of the root nodule bacteria: Assessing genetic diversity through a systematic biogeographic survey. Stand. Genom. Sci. 2015, 10, 14. [Google Scholar] [CrossRef] [PubMed]
- Rhoads, A.; Au, K.F. PacBio sequencing and its applications. Genom. Proteom. Bioinform. 2015, 13, 278–289. [Google Scholar] [CrossRef] [PubMed]
- Ricker, N.; Qian, H.; Fulthorpe, R.R. The limitations of draft assemblies for understanding prokaryotic adaptation and evolution. Genomics 2012, 100, 167–175. [Google Scholar] [CrossRef] [PubMed]
- Loman, N.J.; Quick, J.; Simpson, J.T. A complete bacterial genome assembled de novo using only nanopore sequencing data. Nat. Methods 2015, 12, 733–735. [Google Scholar] [CrossRef] [PubMed]
- Bautista-Zapanta, J.N.; Yoshida, K.; Suzuki, K. Precise characterization of rDNA genes by intraspecies and inter-loci comparison of rdna sequences and biochemical analysis of ribosomal RNA molecules in Agrobacterium tumefaciens. Genes Genet. Syst. 2005, 80, 9–17. [Google Scholar] [CrossRef] [PubMed]
- Carter, A.P.; Clemons, W.M.; Brodersen, D.E.; Morgan-Warren, R.J.; Wimberly, B.T.; Ramakrishnan, V. Functional insights from the structure of the 30S ribosomal subunit and its interactions with antibiotics. Nature 2000, 407, 340–348. [Google Scholar] [PubMed]
- Durfee, T.; Nelson, R.; Baldwin, S.; Plunkett, G., 3rd; Burland, V.; Mau, B.; Petrosino, J.F.; Qin, X.; Muzny, D.M.; Ayele, M.; et al. The complete genome sequence of Escherichia coli DH10B: Insights into the biology of a laboratory workhorse. J. Bacteriol. 2008, 190, 2597–2606. [Google Scholar] [CrossRef] [PubMed]
- Sreevatsan, S.; Pan, X.; Stockbauer, K.E.; Williams, D.L.; Kreiswirth, B.N.; Musser, J.M. Characterization of rpsL and rrs mutations in streptomycin-resistant Mycobacterium tuberculosis isolates from diverse geographic localities. Antimicrob. Agents Chemother. 1996, 40, 1024–1026. [Google Scholar] [PubMed]
- Goris, J.; Konstantinidis, K.T.; Klappenbach, J.A.; Coenye, T.; Vandamme, P.; Tiedje, J.M. DNA-DNA hybridization values and their relationship to whole-genome sequence similarities. Int. J. Syst. Evol. Microbiol. 2007, 57, 81–91. [Google Scholar] [CrossRef] [PubMed]
- Kumar, N.; Lad, G.; Giuntini, E.; Kaye, M.E.; Udomwong, P.; Shamsani, N.J.; Young, J.P.W.; Bailly, X. Bacterial genospecies that are not ecologically coherent: Population genomics of Rhizobium leguminosarum. Open Biol. 2015, 5, 140133. [Google Scholar] [CrossRef] [PubMed]
- Leyva, A.; Palacios, J.M.; Ruiz-Argueso, T. Conserved plasmid hydrogen-uptake (hup)-specific sequences within Hup+ Rhizobium leguminosarum strains. Appl. Environ. Microbiol. 1987, 53, 2539–2543. [Google Scholar] [PubMed]
- Harrison, P.W.; Lower, R.P.; Kim, N.K.; Young, J.P. Introducing the bacterial ‘chromid’: Not a chromosome, not a plasmid. Trends Microbiol. 2010, 18, 141–148. [Google Scholar] [CrossRef] [PubMed]
- Ramachandran, V.K.; East, A.K.; Karunakaran, R.; Downie, J.A.; Poole, P.S. Adaptation of Rhizobium leguminosarum to pea, alfalfa and sugar beet rhizospheres investigated by comparative transcriptomics. Genome Biol. 2011, 12, R106. [Google Scholar] [CrossRef] [PubMed]
- De Boer, P.A.; Crossley, R.E.; Rothfield, L.I. Central role for the Escherichia coli minC gene product in two different cell division-inhibition systems. Proc. Natl. Acad. Sci. USA 1990, 87, 1129–1133. [Google Scholar] [CrossRef] [PubMed]
- Palacios, J.M.; Manyani, H.; Martinez, M.; Ureta, A.C.; Brito, B.; Bascones, E.; Rey, L.; Imperial, J.; Ruiz-Argueso, T. Genetics and biotechnology of the H2-uptake (NiFe) hydrogenase from Rhizobium leguminosarum bv. viciae, a legume endosymbiotic bacterium. Biochem. Soc. Trans. 2005, 33, 94–96. [Google Scholar] [CrossRef] [PubMed]
- Ruiz-Argüeso, T.; Palacios, J.M.; Imperial, J. Regulation of the hydrogenase system in Rhizobium leguminosarum. Plant Soil 2001, 230, 49–57. [Google Scholar] [CrossRef]
- Fernandez, D.; Toffanin, A.; Palacios, J.M.; Ruiz-Argueso, T.; Imperial, J. Hydrogenase genes are uncommon and highly conserved in Rhizobium leguminosarum bv. viciae. FEMS Microbiol. Lett. 2005, 253, 83–88. [Google Scholar] [CrossRef] [PubMed]
- Ruiz-Argueso, T.; Emerich, D.W.; Evans, H.J. Hydrogenase system in legume nodules: A mechanism of providing nitrogenase with energy and protection from oxygen damage. Biochem. Biophys. Res. Commun. 1979, 86, 259–264. [Google Scholar] [CrossRef]
- Albareda, M.; Rodrigue, A.; Brito, B.; Ruiz-Argueso, T.; Imperial, J.; Mandrand-Berthelot, M.A.; Palacios, J. Rhizobium leguminosarum HupE is a highly-specific diffusion facilitator for nickel uptake. Metallomics 2015, 7, 691–701. [Google Scholar] [CrossRef] [PubMed]
- Baginsky, C.; Brito, B.; Imperial, J.; Palacios, J.M.; Ruiz-Argueso, T. Diversity and evolution of hydrogenase systems in rhizobia. Appl. Environ. Microbiol. 2002, 68, 4915–4924. [Google Scholar] [CrossRef] [PubMed]
- Galibert, F.; Finan, T.M.; Long, S.R.; Puhler, A.; Abola, P.; Ampe, F.; Barloy-Hubler, F.; Barnett, M.J.; Becker, A.; Boistard, P.; et al. The composite genome of the legume symbiont Sinorhizobium meliloti. Science 2001, 293, 668–672. [Google Scholar] [CrossRef] [PubMed]
- Mulley, G.; Lopez-Gomez, M.; Zhang, Y.; Terpolilli, J.; Prell, J.; Finan, T.; Poole, P. Pyruvate is synthesized by two pathways in pea bacteroids with different efficiencies for nitrogen fixation. J. Bacteriol. 2010, 192, 4944–4953. [Google Scholar] [CrossRef] [PubMed]
- Terpolilli, J.J.; Masakapalli, S.K.; Karunakaran, R.; Webb, I.U.; Green, R.; Watmough, N.J.; Kruger, N.J.; Ratcliffe, R.G.; Poole, P.S. Lipogenesis and redox balance in nitrogen-fixing pea bacteroids. J. Bacteriol. 2016, 198, 2864–2875. [Google Scholar] [CrossRef] [PubMed]
- Mauchline, T.H.; Fowler, J.E.; East, A.K.; Sartor, A.L.; Zaheer, R.; Hosie, A.H.; Poole, P.S.; Finan, T.M. Mapping the Sinorhizobium meliloti 1021 solute-binding protein-dependent transportome. Proc. Natl. Acad. Sci. USA 2006, 103, 17933–17938. [Google Scholar] [CrossRef] [PubMed]
- Linton, K.J.; Higgins, C.F. The Escherichia coli ATP-binding cassette (ABC) proteins. Mol. Microbiol. 1998, 28, 5–13. [Google Scholar] [CrossRef] [PubMed]
- Prell, J.; Poole, P. Metabolic changes of rhizobia in legume nodules. Trends Microbiol. 2006, 14, 161–168. [Google Scholar] [CrossRef] [PubMed]
- Prell, J.; White, J.P.; Bourdes, A.; Bunnewell, S.; Bongaerts, R.J.; Poole, P.S. Legumes regulate Rhizobium bacteroid development and persistence by the supply of branched-chain amino acids. Proc. Natl. Acad. Sci. USA 2009, 106, 12477–12482. [Google Scholar] [CrossRef] [PubMed]
- Bruins, M.R.; Kapil, S.; Oehme, F.W. Microbial resistance to metals in the environment. Ecotoxicol. Environ. Saf. 2000, 45, 198–207. [Google Scholar] [CrossRef] [PubMed]
- Rubio-Sanz, L.; Prieto, R.I.; Imperial, J.; Palacios, J.M.; Brito, B. Functional and expression analysis of the metal-inducible DmeRF system from Rhizobium leguminosarum bv. viciae. Appl. Environ. Microbiol. 2013, 79, 6414–6422. [Google Scholar] [CrossRef] [PubMed]
- Brito, B.; Palacios, J.M.; Hidalgo, E.; Imperial, J.; Ruiz-Argueso, T. Nickel availability to pea (Pisum sativum L.) plants limits hydrogenase activity of Rhizobium leguminosarum bv. viciae bacteroids by affecting the processing of the hydrogenase structural subunits. J. Bacteriol. 1994, 176, 5297–5303. [Google Scholar] [CrossRef] [PubMed]
- Yost, C.K.; Rochepeau, P.; Hynes, M.F. Rhizobium leguminosarum contains a group of genes that appear to code for Methyl-Accepting Chemotaxis proteins. Microbiology 1998, 144 Pt 7, 1945–1956. [Google Scholar] [CrossRef] [PubMed]
- Ortega, A.; Zhulin, I.B.; Krell, T. Sensory repertoire of bacterial chemoreceptors. Microbiol. Mol. Biol. Rev. 2017, 81, e00033-17. [Google Scholar] [CrossRef] [PubMed]
- Lacal, J.; Garcia-Fontana, C.; Munoz-Martinez, F.; Ramos, J.L.; Krell, T. Sensing of environmental signals: Classification of chemoreceptors according to the size of their ligand binding regions. Environ. Microbiol. 2010, 12, 2873–2884. [Google Scholar] [CrossRef] [PubMed]
- Skorupska, A.; Janczarek, M.; Marczak, M.; Mazur, A.; Król, J. Rhizobial exopolysaccharides: Genetic control and symbiotic functions. Microb. Cell Fact. 2006, 5, 7. [Google Scholar] [CrossRef] [PubMed]
- Janczarek, M.; Urbanik-Sypniewska, T. Expression of the Rhizobium leguminosarum bv. trifolii pssA gene, involved in exopolysaccharide synthesis, is regulated by RosR, phosphate, and the carbon source. J. Bacteriol. 2013, 195, 3412–3423. [Google Scholar] [CrossRef] [PubMed]
- Janczarek, M. Environmental signals and regulatory pathways that influence exopolysaccharide production in rhizobia. Int. J. Mol. Sci. 2011, 12, 7898–7933. [Google Scholar] [CrossRef] [PubMed]
- Meloni, S.; Rey, L.; Sidler, S.; Imperial, J.; Ruiz-Argueso, T.; Palacios, J.M. The Twin-Arginine Translocation (TAT) system is essential for Rhizobium-legume symbiosis. Mol. Microbiol. 2003, 48, 1195–1207. [Google Scholar] [CrossRef] [PubMed]
- Nelson, M.S.; Sadowsky, M.J. Secretion systems and signal exchange between nitrogen-fixing rhizobia and legumes. Front. Plant Sci. 2015, 6, 491. [Google Scholar] [CrossRef] [PubMed]
- Kaneko, T.; Nakamura, Y.; Sato, S.; Asamizu, E.; Kato, T.; Sasamoto, S.; Watanabe, A.; Idesawa, K.; Ishikawa, A.; Kawashima, K.; et al. Complete genome structure of the nitrogen-fixing symbiotic bacterium Mesorhizobium loti. DNA Res. 2000, 7, 331–338. [Google Scholar] [CrossRef] [PubMed]
- Kaneko, T.; Nakamura, Y.; Sato, S.; Minamisawa, K.; Uchiumi, T.; Sasamoto, S.; Watanabe, A.; Idesawa, K.; Iriguchi, M.; Kawashima, K.; et al. Complete genomic sequence of nitrogen-fixing symbiotic bacterium Bradyrhizobium japonicum USDA110. DNA Res. 2002, 9, 189–197. [Google Scholar] [CrossRef] [PubMed]
- Schmeisser, C.; Liesegang, H.; Krysciak, D.; Bakkou, N.; Le Quere, A.; Wollherr, A.; Heinemeyer, I.; Morgenstern, B.; Pommerening-Roser, A.; Flores, M.; et al. Rhizobium sp. strain NGR234 possesses a remarkable number of secretion systems. Appl. Environ. Microbiol. 2009, 75, 4035–4045. [Google Scholar] [CrossRef] [PubMed]
- Ormeño-Orrillo, E.; Menna, P.; Almeida, L.G.; Ollero, F.J.; Nicolas, M.F.; Pains Rodrigues, E.; Shigueyoshi Nakatani, A.; Silva Batista, J.S.; Oliveira Chueire, L.M.; Souza, R.C.; et al. Genomic basis of broad host range and environmental adaptability of Rhizobium tropici CIAT 899 and Rhizobium sp. PRF 81 which are used in inoculants for common bean (Phaseolus vulgaris L.). BMC Genom. 2012, 13, 735. [Google Scholar] [CrossRef] [PubMed]
- Ramsay, J.P.; Sullivan, J.T.; Stuart, G.S.; Lamont, I.L.; Ronson, C.W. Excision and transfer of the Mesorhizobium loti R7A symbiosis island requires an integrase IntS, a novel recombination directionality factor RdfS, and a putative relaxase RlxS. Mol. Microbiol. 2006, 62, 723–734. [Google Scholar] [CrossRef] [PubMed]
- Juhas, M.; Crook, D.W.; Hood, D.W. Type IV secretion systems: Tools of bacterial horizontal gene transfer and virulence. Cell. Microbiol. 2008, 10, 2377–2386. [Google Scholar] [CrossRef] [PubMed]
- Tomich, M.; Planet, P.J.; Figurski, D.H. The tad locus: Postcards from the widespread colonization island. Nat. Rev. Microbiol. 2007, 5, 363–375. [Google Scholar] [CrossRef] [PubMed]
- Dixon, R.; Kahn, D. Genetic regulation of biological nitrogen fixation. Nat. Rev. Microbiol. 2004, 2, 621–631. [Google Scholar] [CrossRef] [PubMed]
- Schluter, A.; Patschkowski, T.; Quandt, J.; Selinger, L.B.; Weidner, S.; Kramer, M.; Zhou, L.; Hynes, M.F.; Priefer, U.B. Functional and regulatory analysis of the two copies of the fixNOQP operon of Rhizobium leguminosarum strain VF39. Mol. Plant-Microbe Interact. 1997, 10, 605–616. [Google Scholar] [CrossRef] [PubMed]
- Hernando, Y.; Palacios, J.M.; Imperial, J.; Ruiz-Argueso, T. The hypBFCDE operon from Rhizobium leguminosarum biovar viciae is expressed from an Fnr-type promoter that escapes mutagenesis of the FnrN gene. J. Bacteriol. 1995, 177, 5661–5669. [Google Scholar] [CrossRef] [PubMed]
- Gutierrez, D.; Hernando, Y.; Palacios, J.M.; Imperial, J.; Ruiz-Argueso, T. FnrN controls symbiotic nitrogen fixation and hydrogenase activities in Rhizobium leguminosarum biovar viciae UPM791. J. Bacteriol. 1997, 179, 5264–5270. [Google Scholar] [CrossRef] [PubMed]
- Colombo, M.V.; Gutierrez, D.; Palacios, J.M.; Imperial, J.; Ruiz-Argueso, T. A novel autoregulation mechanism of fnrn expression in Rhizobium leguminosarum bv. viciae. Mol. Microbiol. 2000, 36, 477–486. [Google Scholar] [CrossRef] [PubMed]
- Lopez, O.; Morera, C.; Miranda-Rios, J.; Girard, L.; Romero, D.; Soberon, M. Regulation of gene expression in response to oxygen in Rhizobium etli: Role of fnrN in fixNOQP expression and in symbiotic nitrogen fixation. J. Bacteriol. 2001, 183, 6999–7006. [Google Scholar] [CrossRef] [PubMed]
- Brito, B.; Martinez, M.; Fernandez, D.; Rey, L.; Cabrera, E.; Palacios, J.M.; Imperial, J.; Ruiz-Argueso, T. Hydrogenase genes from Rhizobium leguminosarum bv. viciae are controlled by the nitrogen fixation regulatory protein NifA. Proc. Natl. Acad. Sci. USA 1997, 94, 6019–6024. [Google Scholar] [CrossRef] [PubMed]
- Fuqua, W.C.; Winans, S.C.; Greenberg, E.P. Quorum sensing in bacteria: The LuxR-LuxI family of cell density-responsive transcriptional regulators. J. Bacteriol. 1994, 176, 269–275. [Google Scholar] [CrossRef] [PubMed]
- Cantero, L. Análisis Genético y Proteómico de la Regulación de Los Sistemas de Autoinducción en Rhizobium leguminosarum bv. viciae UPM791 y 3841. Ph.D. Thesis, Universidad Politécnica de Madrid, Madrid, Spain, 2005. [Google Scholar]
- Frederix, M.; Downie, A.J. Quorum sensing: Regulating the regulators. Adv. Microb. Physiol. 2011, 58, 23–80. [Google Scholar] [PubMed]
- Patankar, A.V.; Gonzalez, J.E. Orphan LuxR regulators of quorum sensing. FEMS Microbiol. Rev. 2009, 33, 739–756. [Google Scholar] [CrossRef] [PubMed]
- Rossbach, S.; Kulpa, D.A.; Rossbach, U.; de Bruijn, F.J. Molecular and genetic characterization of the rhizopine catabolism (mocABRC) genes of Rhizobium meliloti L5-30. Mol. Gen. Genet. 1994, 245, 11–24. [Google Scholar] [CrossRef] [PubMed]
- Pedrosa, F.O.; Zancan, G.T. L-arabinose metabolism in Rhizobium japonicum. J. Bacteriol. 1974, 119, 336–338. [Google Scholar] [PubMed]
- Hungria, M.; Joseph, C.M.; Phillips, D.A. Rhizobium nod gene inducers exuded naturally from roots of common bean (Phaseolus vulgaris L.). Plant Physiol. 1991, 97, 759–764. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Kiss, E.; Mergaert, P.; Olàh, B.; Kereszt, A.; Staehelin, C.; Davies, A.E.; Downie, J.A.; Kondorosi, A.; Kondorosi, E. Conservation of nolR in the Sinorhizobium and Rhizobium genera of the rhizobiaceae family. Mol. Plant-Microbe Interact. 1998, 11, 1186–1195. [Google Scholar] [CrossRef]
- Janczarek, M.; Kutkowska, J.; Piersiak, T.; Skorupska, A. Rhizobium leguminosarum bv. trifolii RosR is required for interaction with clover, biofilm formation and adaptation to the environment. BMC Microbiol. 2010, 10, 284. [Google Scholar] [CrossRef] [PubMed]
- Schwartzman, J.A.; Ruby, E.G. Stress as a normal cue in the symbiotic environment. Trends Microbiol. 2016, 24, 414–424. [Google Scholar] [CrossRef] [PubMed]
- Kondorosi, E.; Mergaert, P.; Kereszt, A. A paradigm for endosymbiotic life: Cell differentiation of Rhizobium bacteria provoked by host plant factors. Annu. Rev. Microbiol. 2013, 67, 611–628. [Google Scholar] [CrossRef] [PubMed]
- Haslbeck, M.; Vierling, E. A first line of stress defense: Small Heat Shock proteins and their function in protein homeostasis. J. Mol. Biol. 2015, 427, 1537–1548. [Google Scholar] [CrossRef] [PubMed]
- Han, M.-J.; Yun, H.; Lee, S.Y. Microbial Small Heat Shock proteins and their use in biotechnology. Biotechnol. Adv. 2008, 26, 591–609. [Google Scholar] [CrossRef] [PubMed]
- Montiel, J.; Downie, J.A.; Farkas, A.; Bihari, P.; Herczeg, R.; Balint, B.; Mergaert, P.; Kereszt, A.; Kondorosi, E. Morphotype of bacteroids in different legumes correlates with the number and type of symbiotic NCR peptides. Proc. Natl. Acad. Sci. USA 2017, 114, 5041–5046. [Google Scholar] [CrossRef] [PubMed]
- Haag, A.F.; Baloban, M.; Sani, M.; Kerscher, B.; Pierre, O.; Farkas, A.; Longhi, R.; Boncompagni, E.; Herouart, D.; Dall’Angelo, S.; et al. Protection of Sinorhizobium against host cysteine-rich antimicrobial peptides is critical for symbiosis. PLoS Biol. 2011, 9, e1001169. [Google Scholar] [CrossRef] [PubMed]
- Karunakaran, R.; Haag, A.F.; East, A.K.; Ramachandran, V.K.; Prell, J.; James, E.K.; Scocchi, M.; Ferguson, G.P.; Poole, P.S. BacA is essential for bacteroid development in nodules of galegoid, but not phaseoloid, legumes. J. Bacteriol. 2010, 192, 2920–2928. [Google Scholar] [CrossRef] [PubMed]
- Gonzalez, V.; Santamaria, R.I.; Bustos, P.; Hernandez-Gonzalez, I.; Medrano-Soto, A.; Moreno-Hagelsieb, G.; Janga, S.C.; Ramirez, M.A.; Jimenez-Jacinto, V.; Collado-Vides, J.; et al. The partitioned Rhizobium etli genome: Genetic and metabolic redundancy in seven interacting replicons. Proc. Natl. Acad. Sci. USA 2006, 103, 3834–3839. [Google Scholar] [CrossRef] [PubMed]
- Bentley, S.D.; Parkhill, J. Comparative genomic structure of prokaryotes. Annu. Rev. Genet. 2004, 38, 771–792. [Google Scholar] [CrossRef] [PubMed]
- Mavingui, P.; Flores, M.; Guo, X.; Davila, G.; Perret, X.; Broughton, W.J.; Palacios, R. Dynamics of genome architecture in Rhizobium sp. strain NGR234. J. Bacteriol. 2002, 184, 171–176. [Google Scholar] [CrossRef] [PubMed]
- Guo, X.; Flores, M.; Mavingui, P.; Fuentes, S.I.; Hernandez, G.; Davila, G.; Palacios, R. Natural genomic design in Sinorhizobium meliloti: Novel genomic architectures. Genome Res. 2003, 13, 1810–1817. [Google Scholar] [PubMed]
- Flores, M.; Mavingui, P.; Perret, X.; Broughton, W.J.; Romero, D.; Hernández, G.; Dávila, G.; Palacios, R. Prediction, identification, and artificial selection of DNA rearrangements in Rhizobium: Toward a natural genomic design. Proc. Natl. Acad. Sci. USA 2000, 97, 9138–9143. [Google Scholar] [CrossRef] [PubMed]
- Krol, J.E.; Mazur, A.; Marczak, M.; Skorupska, A. Application of physical and genetic map of Rhizobium leguminosarum bv. trifolii TA1 to comparison of three closely related rhizobial genomes. Mol. Genet. Genom. 2008, 279, 107–121. [Google Scholar]
- Crossman, L.C.; Castillo-Ramirez, S.; McAnnula, C.; Lozano, L.; Vernikos, G.S.; Acosta, J.L.; Ghazoui, Z.F.; Hernandez-Gonzalez, I.; Meakin, G.; Walker, A.W.; et al. A common genomic framework for a diverse assembly of plasmids in the symbiotic nitrogen fixing bacteria. PLoS ONE 2008, 3, e2567. [Google Scholar] [CrossRef]
- Cubo, M.T.; Economou, A.; Murphy, G.; Johnston, A.W.; Downie, J.A. Molecular characterization and regulation of the rhizosphere-expressed genes rhiABCR that can influence nodulation by Rhizobium leguminosarum biovar viciae. J. Bacteriol. 1992, 174, 4026–4035. [Google Scholar] [CrossRef] [PubMed]
- Rigottier-Gois, L.; Turner, S.L.; Young, J.P.W.; Amarger, N. Distribution of repC plasmid-replication sequences among plasmids and isolates of Rhizobium leguminosarum bv. viciae from field populations. Microbiology 1998, 144, 771–780. [Google Scholar]
- Remigi, P.; Zhu, J.; Young, J.P.W.; Masson-Boivin, C. Symbiosis within symbiosis: Evolving nitrogen-fixing legume symbionts. Trends Microbiol. 2016, 24, 63–75. [Google Scholar] [CrossRef] [PubMed]
- Slater, S.C.; Goldman, B.S.; Goodner, B.; Setubal, J.C.; Farrand, S.K.; Nester, E.W.; Burr, T.J.; Banta, L.; Dickerman, A.W.; Paulsen, I.; et al. Genome sequences of three Agrobacterium biovars help elucidate the evolution of multichromosome genomes in bacteria. J. Bacteriol. 2009, 191, 2501–2511. [Google Scholar] [CrossRef] [PubMed]
- Landeta, C.; Davalos, A.; Cevallos, M.A.; Geiger, O.; Brom, S.; Romero, D. Plasmids with a chromosome-like role in rhizobia. J. Bacteriol. 2011, 193, 1317–1326. [Google Scholar] [CrossRef] [PubMed]
- Hwang, H.H.; Liu, Y.T.; Huang, S.C.; Tung, C.Y.; Huang, F.C.; Tsai, Y.L.; Cheng, T.F.; Lai, E.M. Overexpression of the hspL promotes Agrobacterium tumefaciens virulence in Arabidopsis under heat shock conditions. Phytopathology 2015, 105, 160–168. [Google Scholar] [CrossRef] [PubMed]
- Miller, L.D.; Yost, C.K.; Hynes, M.F.; Alexandre, G. The major chemotaxis gene cluster of Rhizobium leguminosarum bv. viciae is essential for competitive nodulation. Mol. Microbiol. 2007, 63, 348–362. [Google Scholar] [PubMed]
- Scharf, B.E.; Hynes, M.F.; Alexandre, G.M. Chemotaxis signaling systems in model beneficial plant-bacteria associations. Plant. Mol. Biol. 2016, 90, 549–559. [Google Scholar] [CrossRef] [PubMed]
- Briegel, A.; Wong, M.L.; Hodges, H.L.; Oikonomou, C.M.; Piasta, K.N.; Harris, M.J.; Fowler, D.J.; Thompson, L.K.; Falke, J.J.; Kiessling, L.L.; et al. New insights into bacterial chemoreceptor array structure and assembly from electron cryotomography. Biochemistry 2014, 53, 1575–1585. [Google Scholar] [CrossRef] [PubMed]
- Webb, B.A.; Hildreth, S.; Helm, R.F.; Scharf, B.E. Sinorhizobium meliloti chemoreceptor McpU mediates chemotaxis toward host plant exudates through direct proline sensing. Appl. Environ. Microbiol. 2014, 80, 3404–3415. [Google Scholar] [CrossRef] [PubMed]
- Webb, B.A.; Compton, K.K.; Del Campo, J.S.M.; Taylor, D.; Sobrado, P.; Scharf, B.E. Sinorhizobium meliloti chemotaxis to multiple amino acids is mediated by the chemoreceptor McpU. Mol. Plant-Microbe Interact. 2017, 30, 770–777. [Google Scholar] [CrossRef] [PubMed]
- He, K.; Bauer, C.E. Chemosensory signaling systems that control bacterial survival. Trends Microbiol. 2014, 22, 389–398. [Google Scholar] [CrossRef] [PubMed]
- Bladergroen, M.R.; Badelt, K.; Spaink, H.P. Infection-blocking genes of a symbiotic Rhizobium leguminosarum strain that are involved in temperature-dependent protein secretion. Mol. Plant-Microbe Interact. 2003, 16, 53–64. [Google Scholar] [CrossRef] [PubMed]
- Chai, Y.; Zhu, J.; Winans, S.C. TrlR, a defective TraR-like protein of Agrobacterium tumefaciens, blocks TraR function in vitro by forming inactive TrlR:TraR dimers. Mol. Microbiol. 2001, 40, 414–421. [Google Scholar] [CrossRef] [PubMed]
- Gawronski, J.D.; Wong, S.M.; Giannoukos, G.; Ward, D.V.; Akerley, B.J. Tracking insertion mutants within libraries by deep sequencing and a genome-wide screen for Haemophilus genes required in the lung. Proc. Natl. Acad. Sci. USA 2009, 106, 16422–16427. [Google Scholar] [CrossRef] [PubMed]
- Goodman, A.L.; Wu, M.; Gordon, J.I. Identifying microbial fitness determinants by insertion sequencing using genome-wide transposon mutant libraries. Nat. Protoc. 2011, 6, 1969–1980. [Google Scholar] [CrossRef] [PubMed]
- Langridge, G.C.; Phan, M.D.; Turner, D.J.; Perkins, T.T.; Parts, L.; Haase, J.; Charles, I.; Maskell, D.J.; Peters, S.E.; Dougan, G.; et al. Simultaneous assay of every Salmonella typhi gene using one million transposon mutants. Genome Res. 2009, 19, 2308–2316. [Google Scholar] [CrossRef] [PubMed]
- Van Opijnen, T.; Bodi, K.L.; Camilli, A. Tn-Seq: High-throughput parallel sequencing for fitness and genetic interaction studies in microorganisms. Nat. Methods 2009, 6, 767–772. [Google Scholar] [CrossRef] [PubMed]
- Perry, B.J.; Yost, C.K. Construction of a mariner-based transposon vector for use in insertion sequence mutagenesis in selected members of the rhizobiaceae. BMC Microbiol. 2014, 14, 298. [Google Scholar] [CrossRef] [PubMed]
- Perry, B.J.; Akter, M.S.; Yost, C.K. The use of transposon insertion sequencing to interrogate the core functional genome of the legume symbiont Rhizobium leguminosarum. Front. Microbiol. 2016, 7, 1873. [Google Scholar] [CrossRef] [PubMed]
- Wheatley, R.M.; Ramachandran, V.K.; Geddes, B.A.; Perry, B.J.; Yost, C.K.; Poole, P.S. Role of O2 in the growth of Rhizobium leguminosarum bv. viciae 3841 on glucose and succinate. J. Bacteriol. 2017, 199, e00572-16. [Google Scholar] [CrossRef] [PubMed]
- Hynes, M.F.; Quandt, J.; O’Connell, M.P.; Puhler, A. Direct selection for curing and deletion of Rhizobium plasmids using transposons carrying the Bacillus subtilis sacB gene. Gene 1989, 78, 111–120. [Google Scholar] [CrossRef]
- Sanchez-Canizares, C.; Palacios, J. Construction of a marker system for the evaluation of competitiveness for legume nodulation in Rhizobium strains. J. Microbiol. Methods 2013, 92, 246–249. [Google Scholar] [CrossRef] [PubMed]
| Rlv UPM791 | R. etli CFN42T | Rlv 3841 | Rlt CB782 | Rlt WSM1325 | Rlt WSM1689 | Rlt WSM2304 | |
|---|---|---|---|---|---|---|---|
| Rlv UPM791 | |||||||
| R. etli CFN42T | 87.89 | ||||||
| Rlv 3841 | 93.05 | 87.89 | |||||
| Rlt CB782 | 89.16 | 88.15 | 89.14 | ||||
| Rlt WSM1325 | 93.06 | 87.82 | 94.16 | 89.07 | |||
| Rlt WSM1689 | 92.50 | 87.85 | 94.38 | 89.06 | 93.56 | ||
| Rlt WSM2304 | 89.36 | 88.35 | 89.33 | 90.82 | 89.28 | 89.21 |
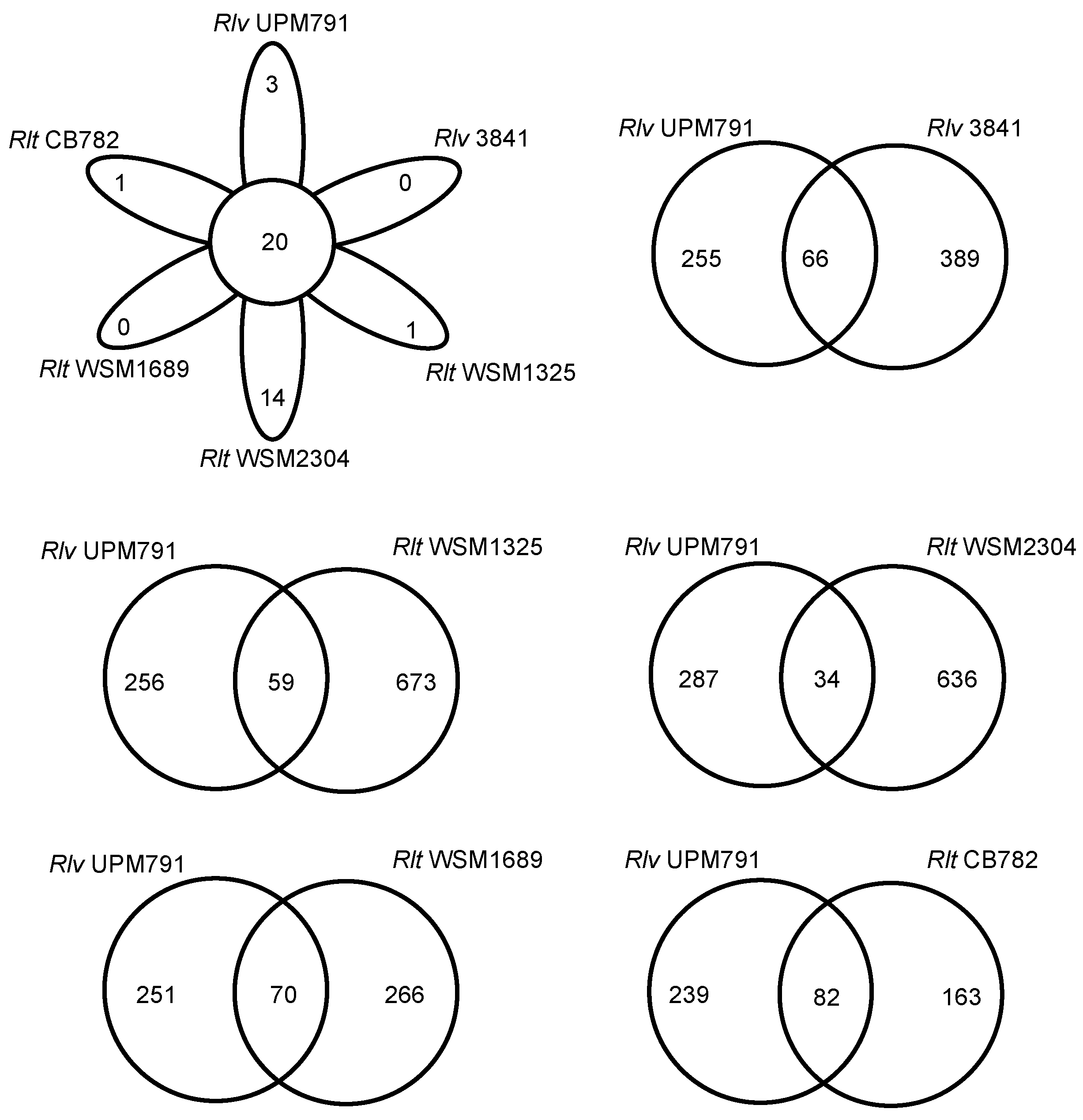
| Rl viciae | Rl viciae | Rl trifolii | Rl trifolii | Rl trifolii | |
|---|---|---|---|---|---|
| UPM791 | 3841 | WSM1325 | WSM2304 | WSM1689 | |
| Host plant | pea, lentil, vetch, Lathyrus | pea, lentil, vetch, Lathyrus | clover | clover | clover |
| Genome size (Mb) | 7.84 | 7.75 | 7.42 | 6.87 | 6.90 |
| GC content (%) | 60.51 | 60.86 | 60.77 | 61.18 | 60.94 |
| Ribosomal RNA operons | 3 | 3 | 3 | 3 | 3 |
| Transfer RNAs | 57 | 52 | 51 | 53 | 51 |
| Total protein-coding genes | 7318 | 7276 | 7232 | 6581 | 6709 |
| Plasmid no. | 5 | 6 | 5 | 4 | 5 |
| % genome in plasmids | 39 | 34.8 | 35.7 | 34 | 42.2 |
| Assembly No. | PRJNA417467 * | ASM926v1 | ASM2318v1 | ASM2134v1 | ASM51760v1 |
| Reference | This work | [6] | [7] | [1] | [8] |
| Rlv UPM791 Plasmids | ORFs Included | Strain and Plasmid | ANIb % | % Sequence Coverage |
|---|---|---|---|---|
| pRlvA | RLV_0001-RLV_1156 | Rlv 3841 pRL12 | 90.44 | 43.03 |
| Rlt WSM1325 pRL132501 | 90.36 | 39.59 | ||
| Rlt WSM2304 pRLG201 | 83.02 | 33.75 | ||
| pRlvB | RLV_1157-RLV_1685 | Rlv 3841 pRL11 | 92.76 | 67.36 |
| Rlt WSM1325 pRL132502 | 91.89 | 63.57 | ||
| Rlt WSM2304 pRLG202 | 88.44 | 56.04 | ||
| pRlvC | RLV_1686-RLV_2021 | Rlv 3841 pRL10 | 88.59 | 20.41 |
| pRlvD | RLV_2022-RLV_2227 | Rlv 3841 pRL7 | 91.81 | 34.15 |
| pRlvE | RLV_2228-RLV_2741 | Rlv 3841 pRL10 | 91.62 | 40.42 |
| Rlv 3841 pRL9 | 88.75 | 37.44 | ||
| Rlt WSM1325 pRL132505 | 90.97 | 35.39 | ||
| Rlt WSM1325 pRL132504 | 88.80 | 37.39 | ||
| Rlt WSM2304 pRLG201 | 84.22 | 36.78 | ||
| Rlt WSM2304 pRLG204 | 86.98 | 33.77 | ||
| Chr | RLV_2742- RLV_7380 | Rlv 3841 chr | 93.08 | 81.83 |
| Rlt WSM1325 chr | 92.79 | 82.72 | ||
| Rlt WSM2304 chr | 88.85 | 73.06 |
© 2018 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Sánchez-Cañizares, C.; Jorrín, B.; Durán, D.; Nadendla, S.; Albareda, M.; Rubio-Sanz, L.; Lanza, M.; González-Guerrero, M.; Prieto, R.I.; Brito, B.; et al. Genomic Diversity in the Endosymbiotic Bacterium Rhizobium leguminosarum. Genes 2018, 9, 60. https://doi.org/10.3390/genes9020060
Sánchez-Cañizares C, Jorrín B, Durán D, Nadendla S, Albareda M, Rubio-Sanz L, Lanza M, González-Guerrero M, Prieto RI, Brito B, et al. Genomic Diversity in the Endosymbiotic Bacterium Rhizobium leguminosarum. Genes. 2018; 9(2):60. https://doi.org/10.3390/genes9020060
Chicago/Turabian StyleSánchez-Cañizares, Carmen, Beatriz Jorrín, David Durán, Suvarna Nadendla, Marta Albareda, Laura Rubio-Sanz, Mónica Lanza, Manuel González-Guerrero, Rosa Isabel Prieto, Belén Brito, and et al. 2018. "Genomic Diversity in the Endosymbiotic Bacterium Rhizobium leguminosarum" Genes 9, no. 2: 60. https://doi.org/10.3390/genes9020060
APA StyleSánchez-Cañizares, C., Jorrín, B., Durán, D., Nadendla, S., Albareda, M., Rubio-Sanz, L., Lanza, M., González-Guerrero, M., Prieto, R. I., Brito, B., Giglio, M. G., Rey, L., Ruiz-Argüeso, T., Palacios, J. M., & Imperial, J. (2018). Genomic Diversity in the Endosymbiotic Bacterium Rhizobium leguminosarum. Genes, 9(2), 60. https://doi.org/10.3390/genes9020060







