1. Introduction
Categorical predictors are widely used in healthcare data analysis as many key variables in medical research are naturally categorical, such as disease status, treatment groups, and demographic factors like gender, race, and socioeconomic status. These predictors allow researchers to assess differences across groups, identify risk factors, and tailor medical treatments to specific populations. Additionally, categorical variables often play a crucial role in clinical decision-making, where classifications like disease severity or diagnostic test results influence patient management. Properly incorporating categorical predictors in statistical models enables robust analysis, guiding evidence-based healthcare policies and personalized medicine. Different from numerical predictors, categorical predictors usually need to be transformed into a numeric format through an appropriate encoding method before being included in the statistical models [
1]. This transformation is necessary because statistical models typically require numerical inputs for computation. It must be decided how to encode categorical variables, with common methods including dummy coding, Helmert coding, and orthogonal contrasts, each emphasizing different aspects of category comparisons. The choice of encoding affects the interpretation of model coefficients, as different schemes highlight various contrasts among categories, such as comparisons to a reference group or overall mean differences. Consequently, a single categorical predictor can be represented in multiple ways within a model, influencing the insights drawn from the analysis and requiring careful selection to align with the study’s objectives.
The impact of coding strategy on variable selection and prediction with linear regression and least absolute shrinkage and selection operator, or LASSO [
2], has been investigated in [
3], which shows that although different coding schemes for categorical predictors do not affect the performance of linear regression, they do impact the performance of LASSO. Despite the success of their study, the results are based on using the point estimates produced from LASSO regression. Uncertainty quantification and statistical inference of LASSO regression using categorical predictors under different coding systems have not been considered. Such a technical gap has motivated us to tackle the problem from an Bayesian perspective, as it is well-acknowledged that fully Bayesian analysis can yield the entire posterior distributions of model parameters (including regression coefficients); therefore statistical inference with Bayesian credible intervals, posterior inclusion probabilities [
4], and hypothesis testing can be readily performed for a more comprehensive uncertainty assessment [
5]. Specifically, we propose to assess the performance of Bayesian LASSO [
6] in regression analysis with categorical predictors, which complements the analysis in [
3] by incorporating inference procedures. Furthermore, our limited literature search reveals that Bayesian analysis and inference with categorical predictors have rarely been reported [
7]. All these motivate us to evaluate how categorical coding strategies influence variable selection, predictive accuracy, and uncertainty quantification with the Bayesian LASSO framework.
Beyond its methodological contributions, our proposed analysis is also novel in terms of the data utilized. Although traditional methods like generalized linear models have been widely used to investigate healthcare datasets [
8,
9], Bayesian techniques have been seldom applied in analyzing chronic autoimmune diseases such as Multiple Sclerosis (MS) [
10,
11]. Multiple Sclerosis (MS) is a complex and chronic autoimmune neuroinflammatory disorder that affects the central nervous system and leads to a wide range of physical, cognitive, and emotional impairments. The disease typically manifests in early adulthood and progresses over time, contributing to long-term disability and substantially diminishing health-related quality of life (HRQoL). MS also places a significant economic burden on patients, families, and healthcare systems due to ongoing medical treatments, hospitalizations, and loss of productivity [
12,
13]. Understanding the predictors and risk factors associated with MS is essential for early detection, personalized management, and resource allocation. However, many of the relevant predictors-such as sex, race, region, educational attainment, and degree status-are categorical variables, requiring careful statistical handling. Exploring the performance and inference with Bayesian approaches will lead to more robust, interpretable, and clinically meaningful insights into the factors that influence MS onset, progression, and outcomes. By framing the Bayesian LASSO within a predictive modeling and probabilistic forecasting context, our approach emphasizes not only parameter estimation but also the generation of accurate and uncertainty quantification for health outcomes. This application provides an opportunity to evaluate how different categorical coding strategies influence predictive accuracy and uncertainty quantification in forecasting health-related outcomes among chronic autoimmune disease populations.
In this study, we investigate the impact of different coding strategies on categorical predictors in prediction, estimation, and inference procedures with Bayesian LASSO using the 2017–2022 Medical Expenditure Panel Survey (MEPS) from the Household Component (HC) and the corresponding Full-Year Consolidated data files. MEPS is a nationally representative survey of the U.S. civilian noninstitutionalized population, collecting detailed information on healthcare utilization, expenditures, insurance coverage, and sociodemographic characteristics. The study sample includes adult individuals aged 18 years and older, with and without a diagnosis of MS. Adults without MS are included in the non-MS group, which allows for the examination of sociodemographic characteristics, health conditions, and healthcare access between MS and non-MS adult populations in the US. Health-related quality of life (HRQoL) is measured using the Veterans RAND 12-Item Health Survey (VR-12) in the MEPS-HC, which yields two standardized scores: the Physical Component Summary (PCS) and the Mental Component Summary (MCS) [
9]. In this study, the response variable of interest is the PCS score, which reflects general physical health, activity limitations, role limitations due to physical health and pain. The dataset includes categorical predictors like sex, race, region, education degree, insurance level, age, and Number of Elixhauser comorbidity conditions [
14], among which there are categorical predictors such as sex and education, as well as continuous predictors such as age and exhauster Number of Elixhauser comorbidity conditions.
The structure of the article is as follows. First, we demonstrate four classic coding strategies, dummy coding, deviation coding, sequential coding, and Helmert coding, through fitting Bayesian linear regression with simulated data. We then provide a brief introduction to linear regression and LASSO, and use the inference procedure to motivate Bayesian LASSO. In a case study of MEPS data, Bayesian Lasso and alternative methods including linear regression have been to the data for estimation, prediction, and statistical inference. Bayesian analysis yields promising numeric results that have important practical implications, making it particularly powerful in uncertainty quantification, which provides insights for decision-making and scientific interpretation in complex disease analysis.
4. Real Data Analysis
To explore the potential impacts of coding strategy on important predictors, we analyze the real-world healthcare data using Bayesian LASSO, LASSO and linear regression. This study utilized data from the MEPS, a population-based survey of U.S. adults that provides comprehensive information on healthcare utilization, expenditures, insurance coverage, and sociodemographic characteristics, to examine individuals with and without MS. The MEPS data set has a sample of 98,163 participants. In the analysis, the response of interest to evaluate the HRQoL is the PCS score. We consider six categorical predictors: MS (2 categories), Sex (2 categories), Race (4 categories), Region (4 categories), Education (5 categories), Insurance (3 categories), and two continuous variables: Age and ECI (Number of Elixhauser comorbidity conditions). The performance of three models under comparison has been examined under different coding strategies in terms of variable selection, prediction, and inference procedures. This study aims to assess how different coding strategies influence estimation, variable selection, prediction accuracy, and statistical inference with the Bayesian LASSO for factors associated with the PCS score.
4.1. Variable Selection
We investigate whether the choice of coding strategy influences the variable selection with Bayesian LASSO. The results are provided in
Table 6, which shows that selected variables vary under different coding strategies. For example, variable Race corresponds to three predictors after conversion. With dummy coding, all three race binary indicators are included in the model. With deviation coding, the predictor measuring the difference between non-Hispanic black and the average is excluded from the final model. However, the sequential coding strategy excludes the predictor indicating the difference between Hispanic and other, while the Helmert coding strategy excludes the predictor representing the difference between non-Hispanic black and other. From this example, we can find that a researcher may conclude that the PCS scores differ across the race of Hispanic and other with the dummy coding strategy, while using the sequential coding strategy, the opposite conclusions will be made. Variable selection with different coding strategies has also been assessed using LASSO and linear regression, as shown in
Table A3 and
Table A4 in the
Appendix A, respectively. For a direct comparison to the other two methods, under the linear regression model, we compute the 95% confidence interval for each regression coefficient, and exclude predictors if the corresponding confidence intervals cover zero. We can find that the important predictors identified through linear regression and LASSO vary depending on the coding strategy, as different coding schemes alter the model’s parameterization and thus the estimated regression coefficients.
A cross-comparison of results from
Table 6,
Table A3 and
Table A4 indicates that on MEPS data, Bayesian LASSO leads to exclusion of variables under all four types of coding. The number of excluded features under Dummy, Deviation, Sequential, and Helmert coding schemes are 2, 1, 1, and 2, respectively. In comparison, linear regression excludes 0, 1, 1, and 2 features under the same codings. The selection results between the two methods are more similar, as LASSO eliminates 0, 1, 0, and 1 variables, respectively. Although it may initially seem surprising that the variable selection results from Bayesian LASSO and linear regression are more similar to each other, a closer examination provides a clear explanation. Both Bayesian LASSO and linear regression rely on inference-based measures—such as confidence or credible intervals—for feature selection. In contrast, LASSO selects variables by applying regularization and eliminating features whose coefficient estimates are exactly zero. Overall, it can be concluded that the variable selection in both Bayesian LASSO, linear regression and LASSO is influenced by the choice of coding strategies for categorical variables.
4.2. Prediction Accuracy
Under different coding strategies, we have assessed the prediction performance for all methods under comparison in terms of (1) predicted category scores and (2) the least absolute deviation (LAD) error, which is defined as
. Choosing Education as an example, we examine whether the predicted score for each education level is the same under different coding strategies using Bayesian LASSO.
Table 7 shows predicted PCS scores using Bayesian LASSO corresponding to the five levels of Education, with the last column representing the actual category means. It can be observed that under different coding strategies, the predicted scores are of the same magnitude for the same category, with slight differences. Although the predicted scores are close to the actual category means, there are no models where the predicted scores are equal to the true category means. Similar patterns can be found with LASSO in
Table A2 in the
Appendix A. However, the predicted category scores are exactly the same when using linear regression, which are also equal to the actual category means, as shown in
Table A1 in the
Appendix A. Mathematically, any coding transformation corresponds to a change in the coefficient estimates but maintains the same fitted values in linear regression, as the full model space is explored without restriction.
Next, we assess predictive performance by computing the previously defined LAD error for all the three models by including all categorical and continuous variables under different coding strategies. The results are listed in
Table 8. In the case of linear regression, the LAD error remains unchanged across different coding strategies because the predicted values are invariant to how categorical variables are encoded. However, for Bayesian LASSO, shrinkage effects are not the same under different coding schemes, leading to different penalized estimates and, consequently, varying prediction errors in terms of LAD. Under the 4 parameterizations, the model performance is impacted under Bayesian LASSO and LASSO, while remaining the same with linear regression.
The differences observed across coding strategies in the LASSO and Bayesian LASSO can be understood as structural consequences of the penalization mechanism. In ordinary least squares regression, fitted values are invariant to full-rank reparameterizations of the design matrix because the estimation depends only on the column space of the predictors. By contrast, penalized estimators, such as the LASSO or Bayesian LASSO with independent Laplace priors on coefficients, are not invariant to reparameterization. The penalty acts directly on the coordinate system defined by the predictors. Hence, any change in the scaling or geometry of the design matrix alters the relative degree of shrinkage applied to each coefficient, leading to variations in performance across different coding strategies, even though the overall results remain comparable. This dependence indicates that such differences stem from the intrinsic structure of the regularization term, highlighting how the choice of coding can meaningfully influence model outcomes.
4.3. Statistical Inference
Inference procedures play a crucial role in statistical analysis. However, frequentist high-dimensional methods, including LASSO, typically rely on complex asymptotic theory to develop inference procedures [
24]. This reliance poses significant challenges for practitioners seeking to understand and apply these methods to real-world data analysis. A more detailed discussion of the obstacles associated with implementing frequentist inference procedures in practical settings can be found in [
7]. On the other hand, Bayesian approaches overcome this difficulty by providing a principled and coherent framework of conducting statistical inference through posterior sampling regarding all model parameters [
5]. By building up Bayesian hierarchical models that leverage strength from prior information and observed data, fully Bayesian analysis can characterize the entire posterior distribution of model parameters via sampling based on Markov Chain Monte Carlo (MCMC) and techniques alike. Therefore, uncertainty quantification measures including standard summary statistics (such as median, mean and variance), credible intervals and posterior estimates with false discovery rates (FDR) can be readily obtained. Although estimation and prediction are inherently related, they represent conceptually distinct aspects of model evaluation. The inference procedure is essential for uncertainty quantification, providing a foundation for assessing the credibility of both parameter estimates and model predictions. In the Bayesian LASSO framework, predictions are derived directly from the posterior distribution, whereas in traditional LASSO and linear regression, they rely solely on point estimates.
We perform statistical inference in terms of marginal Bayesian credible intervals with Bayesian LASSO on the MEPS dataset under the four coding systems.
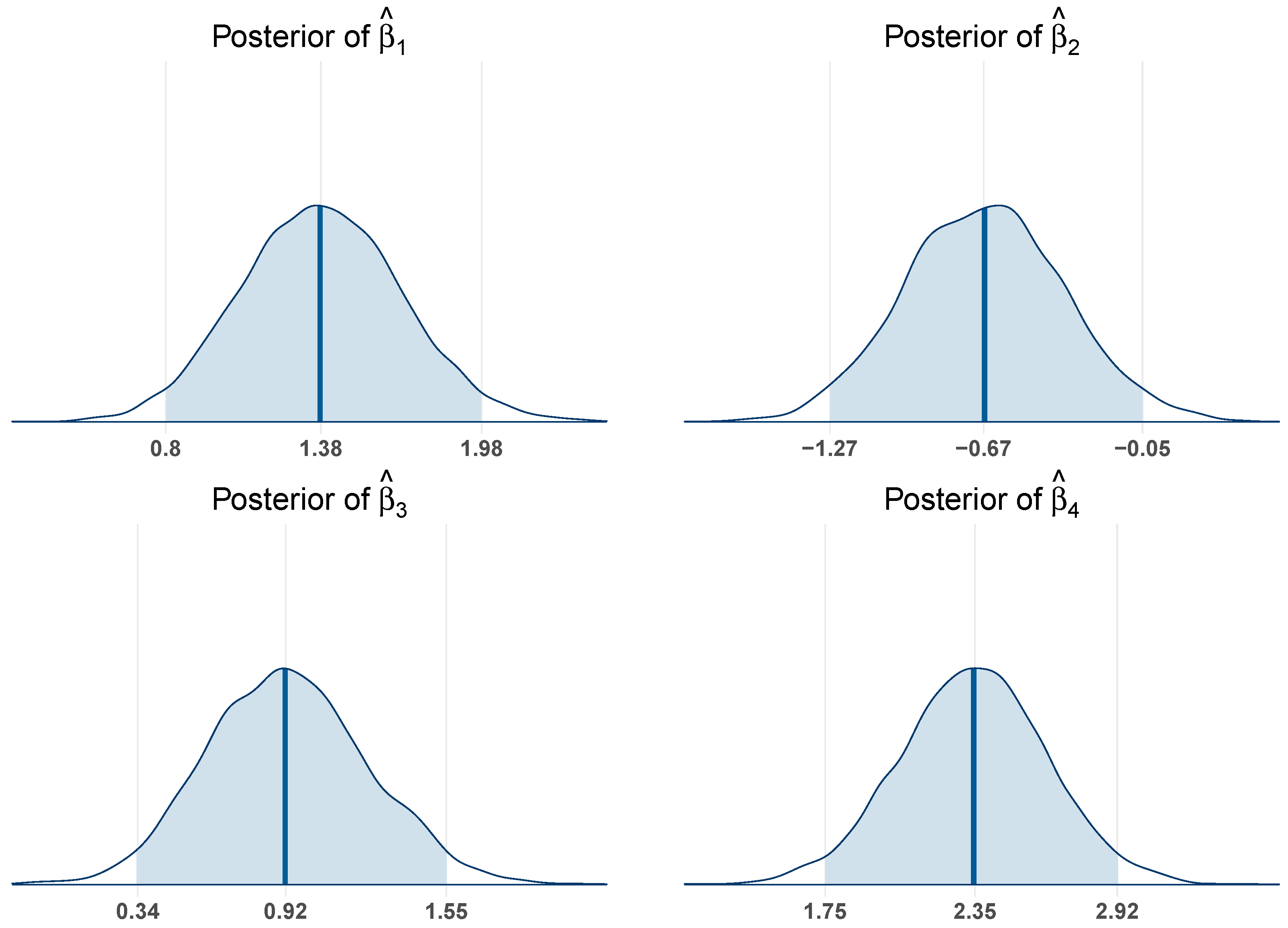
Figure 2 shows posterior distributions of three regression coefficients resulting from converting Race following dummy coding strategy. Using Hispanic individuals as the reference group, for Non-Hispanic white individuals, the posterior median is −0.45 with a 95% credible interval ranging from −0.58 to −0.31. Since this interval does not include zero, it suggests a statistically significant negative association compared to Hispanic individuals, indicating that Non-Hispanic whites have a significantly lower level of PCS score than the reference group. For Non-Hispanic black individuals, the posterior median is −0.73 with a 95% credible interval of −0.90 to −0.53. This interval also excludes zero, providing strong evidence of a significant negative difference in the outcome relative to the Hispanic group. Similarly, for individuals classified as other, the posterior median is −0.68, and the 95% credible interval extends from −0.90 to −0.46. This interval likewise does not include zero, indicating a statistically significant negative association with PCS score compared to Hispanic individuals. Taken together, these results demonstrate that all three racial groups exhibit significantly lower PCS scores compared to the Hispanic reference group. As the posterior credible intervals exclude zero, it suggests these predictors should be kept in the final model, which corresponds to the variable selection results in
Table 6.
The posterior median, 95% credible interval and the corresponding interval length of regression coefficients associated with all predictors under the four coding strategies using Bayesian LASSO have been shown in
Table A5 in the
Appendix A. The covariates with the credible interval excluding zero are considered to be significantly associated with the outcome and are included in the final model, which is listed in
Table 6. We can find that while the posterior medians and credible intervals of regression coefficients vary across different coding strategies, their overall magnitudes are close. Although the underlying relationships between the predictors and outcome remain the same across coding strategies, the parameterization changes. Thus, while the parameter estimates and their associated uncertainty intervals differ, the substantive conclusions about the strength and direction of associations remain relatively stable. This explains why the posterior medians and credible intervals vary numerically but reflect comparable effect sizes across coding strategies. This suggests that although the numerical representations of the categorical variables influence the specific parameter estimates and their associated uncertainty ranges, the underlying effect sizes are consistently captured across coding strategies. These differences in posterior summaries highlight the sensitivity of Bayesian LASSO inference to the choice of coding strategies, yet the comparable magnitude of estimates indicates that the substantive conclusions about variable importance remain relatively stable. As a comparison, we present estimation results along with the 95% confidence interval for uncertainty quantification by fitting linear regression in
Table A6 in the
Appendix A.
Table A7 in the
Appendix A shows the estimation results using LASSO, which is similar to the analysis conducted in [
3] where only point estimates of LASSO have been considered, while Bayesian LASSO assess the uncertainty by providing the posterior distribution, yielding a more interpretable and comprehensive result. In the linear regression analysis, the estimated coefficients remain consistent across different coding strategies. In contrast, the LASSO approach exhibits greater sensitivity to coding choices and certain coefficients are shrunk exactly to zero under some strategies, while the same coefficients are retained as nonzero under others. Unlike Bayesian methods, both linear regression and LASSO provide only point estimates of the coefficients, without a direct framework for quantifying uncertainty.
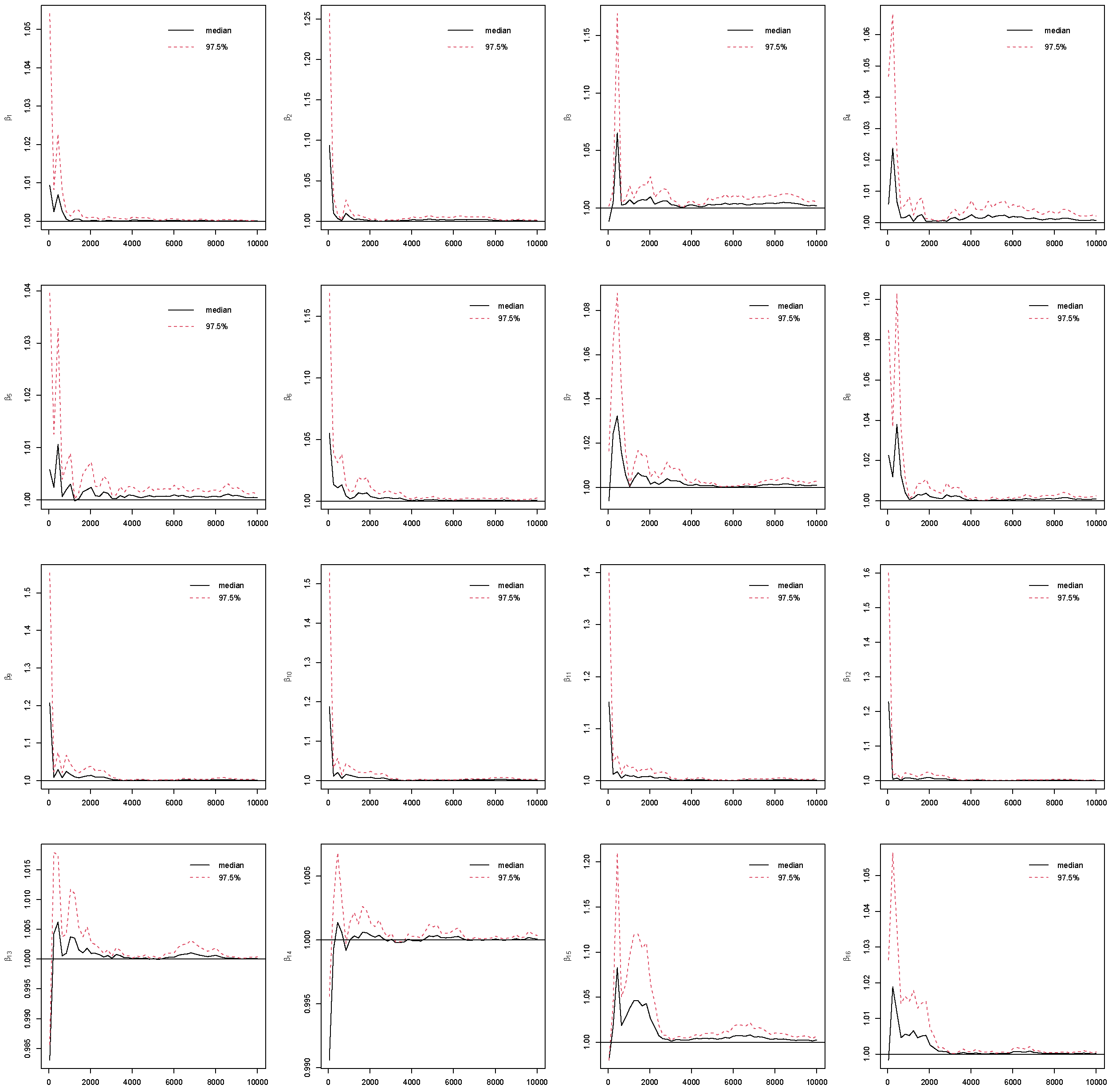
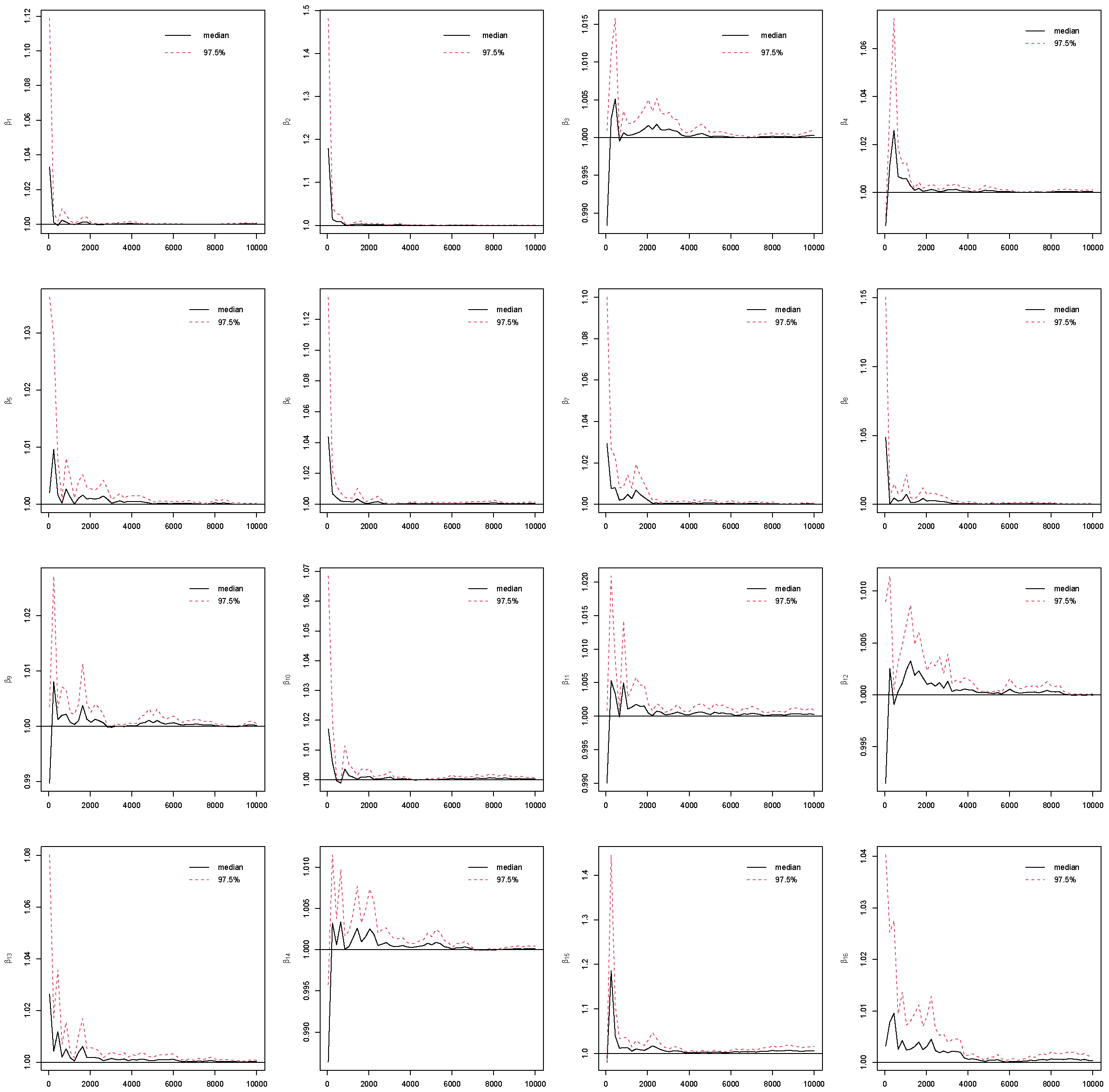
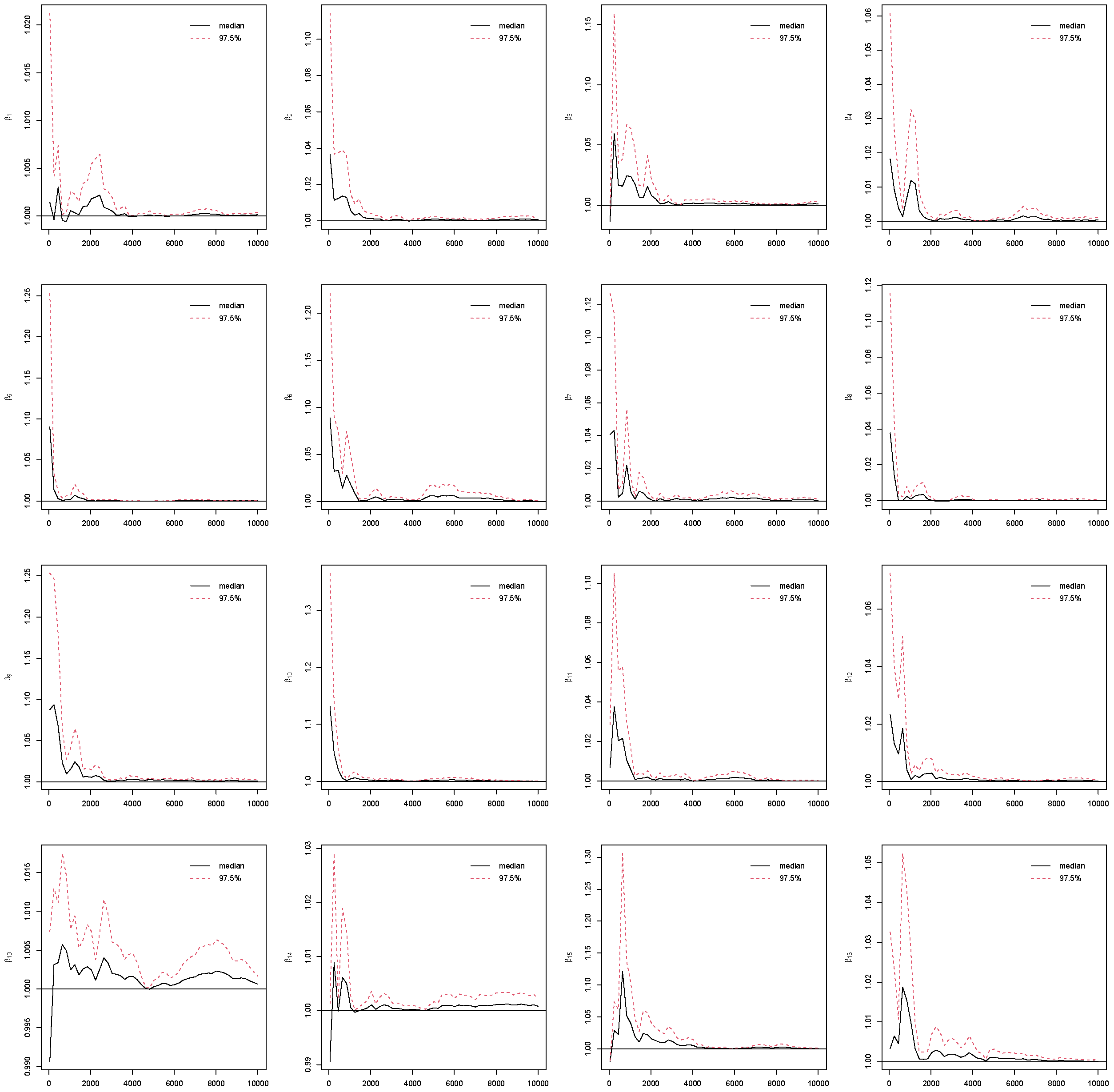
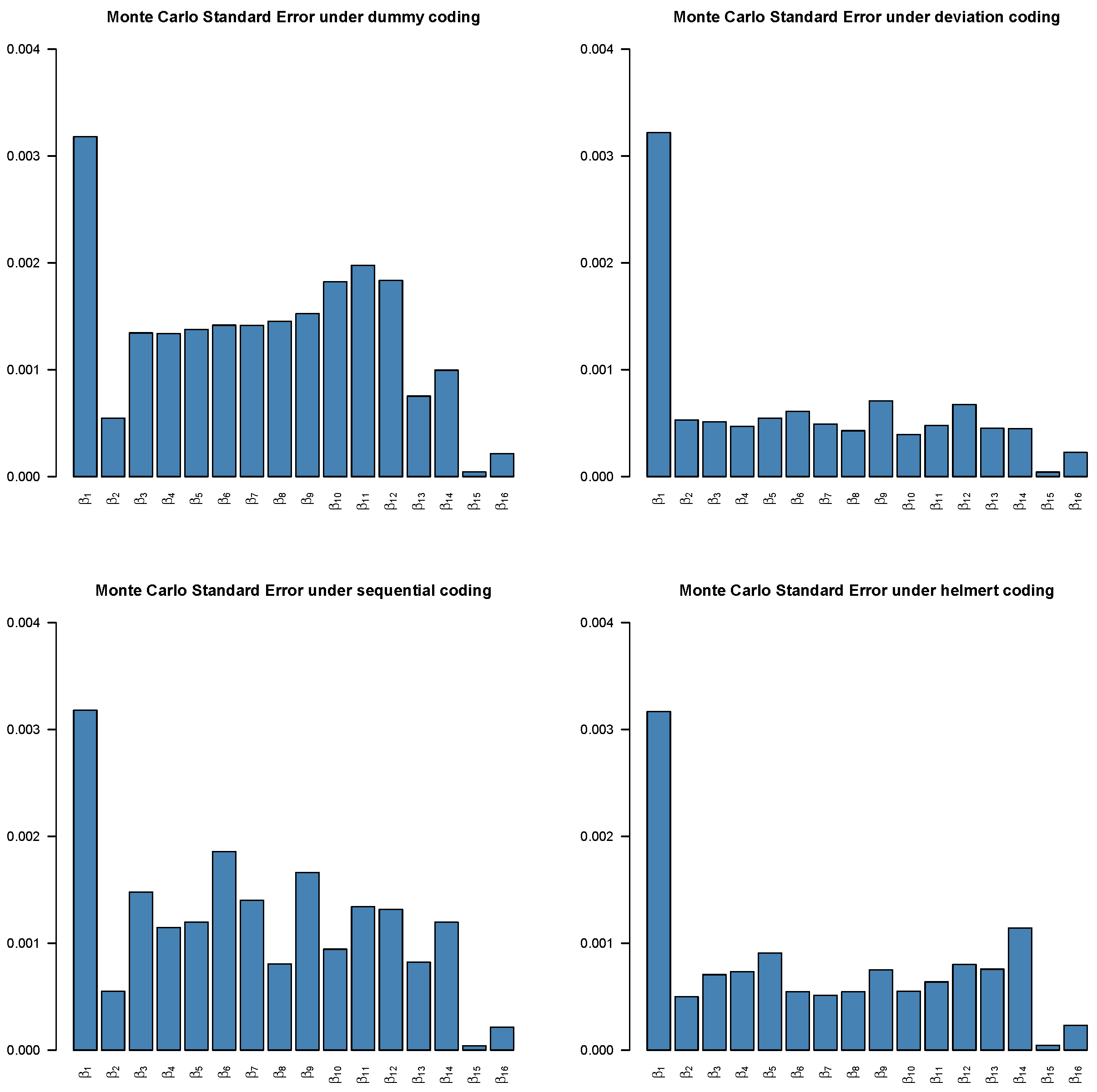
4.4. Convergence
Assessing convergence of Markov Chain Monte Carlo (MCMC) is critical in Bayesian analysis, as the validity of all Bayesian posterior estimates and inferences depends on that MCMC has converged and reached the corresponding stationary distribution [
5]. If the chains do not converge properly, the posterior samples drawn from may not accurately characterize correct underlying posterior distributions, leading to biased estimates and inferences, as well as misleading conclusions [
25]. To assess the reliability of posterior estimates, we evaluated standard MCMC convergence diagnostics for all model parameters with the potential scale reduction factor (PSRF) [
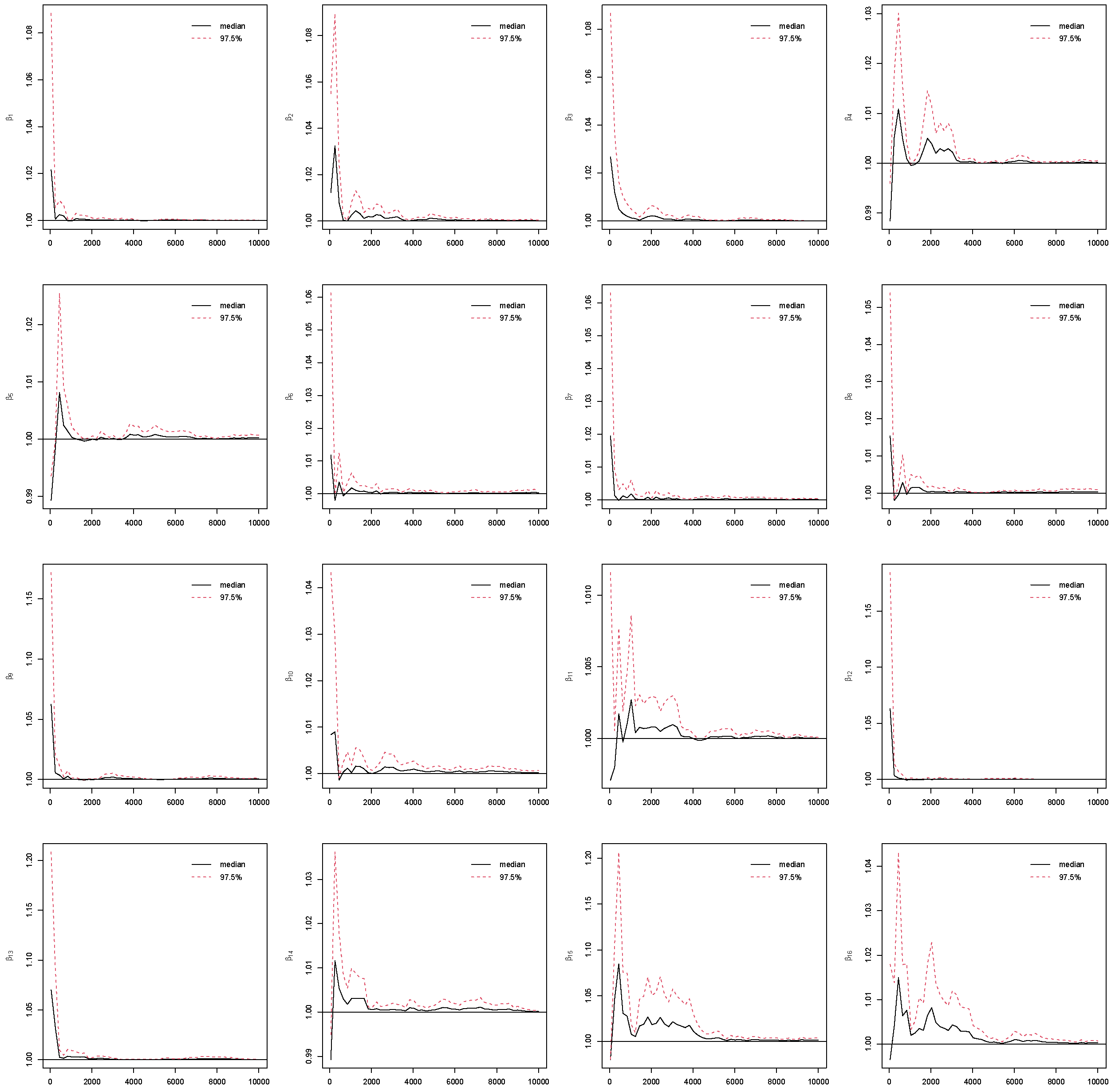
26]. By running multiple MCMC chains on the same dataset, PSRF compares the variance within chains to the variance between chains. If all chains have converged to the target posterior distribution, these variances should be similar, leading to a value close to 1. Otherwise, the chains do not mix well if PSRF value is much larger than 1.
In this study, we use PSRF ≤ 1.1 [
27] as the cut-off point which indicates that chains converge to a stationary distribution. In MCMC, the initial iterations are often affected by the choice of starting values and may not adequately represent draws from the stationary posterior distribution. To mitigate this influence and reduce bias in posterior estimation, these early samples are commonly discarded as burn-in iterations [
5]. In this study, the Gibbs sampling is implemented with 10,000 iterations with the first 5000 as burn-ins. The convergence of the MCMC chains after burn-ins has been checked for all predictors under the four coding strategies, which can be found in
Figure A1,
Figure A2,
Figure A3,
Figure A4 in the
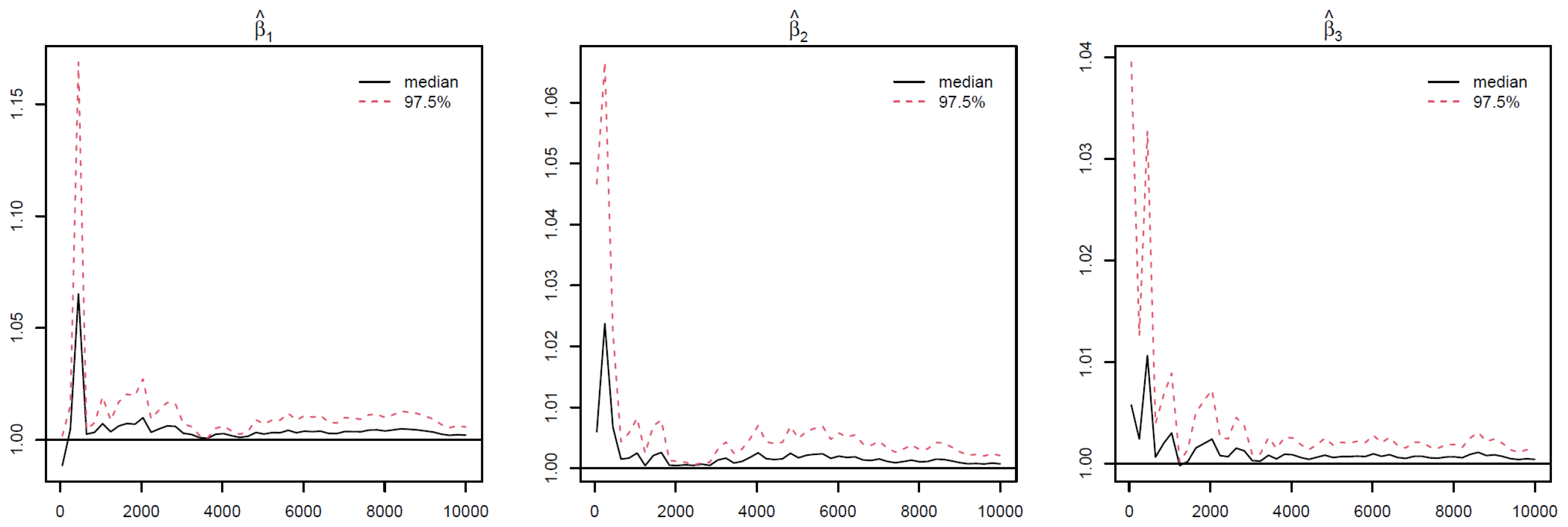
Appendix B. Take Race from dummy coding as an example in
Figure 3. The PSRF trajectories for the three Race dummy variables corresponding to Non-Hispanic white, Non-Hispanic black, and other race categories, respectively. For each parameter, the median PSRF (solid black line) rapidly approaches and stabilizes near 1.00, and the upper 97.5% quantile (dashed red line) remains well below the commonly used threshold of 1.1 after early iterations. This pattern indicates good mixing and convergence of the Markov chains for all three race coefficients. The initial variability observed in the early iterations diminishes quickly, further supporting the conclusion that the chains have converged. These results suggest that posterior inference for the race-related coefficients is reliable and based on well-converged MCMC samples. The convergence assessment is further supported by the effective sample size (ESS) and Monte Carlo standard error (MCSE) analyses. As shown in
Figure A5 and
Figure A6 in the
Appendix B, all coefficients across the four coding strategies achieved ESS values well above the recommended threshold of 400, indicating highly efficient sampling and low autocorrelation within chains [
28]. Correspondingly, the value of MCSE remains small for all coefficients, suggesting that the Monte Carlo uncertainty is negligible relative to posterior variability [
28,
29]. Taken together, the PSRF, ESS, and MCSE diagnostics consistently demonstrate that the Gibbs sampling algorithm converged satisfactorily and produced stable, reliable posterior estimates for all model parameters.
5. Discussion
In this study, we investigate the impact of coding strategies on categorical predictors in variable selection, prediction, and statistical inference with Bayesian LASSO in MEPS data. Comparisons against frequentist approaches such as linear regression and LASSO have also been performed. Our study complements existing work, such as [
3], by adopting a Bayesian framework and incorporating formal statistical inference procedures into the analysis. Moreover, by applying this Bayesian framework to the nationally representative MEPS dataset, our study systematically evaluates how different categorical coding strategies influence estimation, prediction, and uncertainty quantification in real-world healthcare data, thereby highlighting their practical implications for modeling population-level health outcomes, which has been rarely examined in prior studies. Huang et al. [
3] have also evaluated group LASSO as an alternative to LASSO on regression with categorical predictors, and concluded that group LASSO leads to overfitting few instead of all dummy predictors within the same group are needed. By applying the penalty to each group, group LASSO ensures that all predictions within a group are selected or excluded simultaneously, making the selection process invariant to the specific coding strategy. However, in our application using the MEPS data, there is no inherent group structure among covariates. The predictors include sociodemographic and clinical factors that do not naturally form distinct, hierarchical groups. Therefore, our modeling objective is to achieve sparsity at the individual coefficient level, identifying the most influential predictors rather than entire variable groups. Under these conditions, the standard and Bayesian LASSO are more appropriate, as they promote coefficient-level sparsity directly.While we agree with the conclusion in [
3] regarding the application of group LASSO, we want to point out that sparse group LASSO [
30,
31] is a promising regularization method that seeks to achieve sparsity on both group level and within group level. Therefore, sparse group LASSO type of methods are potentially promising in the scenario when selection of important categorical predictors within groups are of interest.
Due to heterogeneity of complex diseases such as multiple sclerosis and cancers, disease phenotypes of interest usually follow heavy-tailed distributions and have outlying observation. Therefore, robust statistical methods, especially robust regularization and variable selection methods that can safeguard against outliers and skewed distributions are demanded [
19]. Recently, the advantages of robust Bayesian variable selection methods over their frequentist counterparts, particularly in the context of statistical inference, have been investigated in [
7]. It will be interesting to explore how robust Bayesian analysis can facilitate modeling with categorical predictors. For example, the robust Bayesian sparse group LASSO model proposed in [
32] offers uncertainty quantification, which is typically unavailable in corresponding frequentist approaches [
30,
31]. In future work, we also plan to extend the current methodology to other types of phenotypic traits, such as survival and longitudinal outcomes.