Abstract
The first-hitting-time based model conceptualizes a random process for subjects’ latent health status. The time-to-event outcome is modeled as the first hitting time of the random process to a pre-specified threshold. Threshold regression with linear predictors has numerous benefits in causal survival analysis, such as the estimators’ collapsibility. We propose a neural network extension of the first-hitting-time based threshold regression model. With the flexibility of neural networks, the extended threshold regression model can efficiently capture complex relationships among predictors and underlying health processes while providing clinically meaningful interpretations, and also tackle the challenge of high-dimensional inputs. The proposed neural network extended threshold regression model can further be applied in causal survival analysis, such as performing as the Q-model in G-computation. More efficient causal estimations are expected given the algorithm’s robustness. Simulations were conducted to validate estimator collapsibility and threshold regression G-computation. The performance of the neural network extended threshold regression model is also illustrated by using simulated and real high-dimensional data from an observational study.
1. Introduction
Causal inference with survival outcomes has been a hot topic in recent years. When the research interest focused on the causal effect of intervention on the time until an individual’s first event of interest, extra care is needed in employing usual time-to-event models. The target population for a new drug often includes patients with diverse prognostic baseline factors. As a result, it is critical to consider confounders during the study design and data analysis. Furthermore, it is desirable to demonstrate the intervention efficacy by showing a significant marginal intervention effect. A well-designed randomized controlled trial has both the measured and unmeasured covariates balanced between intervention groups on average, thus even if the primary analysis does not consider any baseline covariates as confounders, the confounding bias can be eliminated efficiently [1]. However, collapsibility is a favorable model property that cannot be guaranteed by study design. Confounding and collapsibility are separate issues that have been emphasized in the literature [2,3,4]. In Section 3, we discuss the importance of confounding adjustment and collapsibility in causal survival analysis. It is known that the Cox Proportional Hazards (PH) model, the most prevalent survival model in medical research, is not a collapsible model and is not favored in causal survival analysis [5,6].
The first-hitting-time (FHT)-based threshold regression (abbreviated as TR) [7,8] is a well-accepted alternative method to analyze covariate effects on time-to-event endpoints while allowing for clinically meaningful interpretations [9,10,11]. We show in Section 4 that estimators from a TR Model using a Wiener process with linear predictors are collapsible and, hence, the TR model can be easily adapted for causal survival analysis. In particular, we would like to investigate the causal meaning of TR, as well as how to make a marginal causal conclusion from TR with the assistance of G-computation.
This paper is organized as follows: Section 2 presents the necessary notations and required causal assumptions for this manuscript. Section 3 reviews the definitions of confounding and collapsibility. Section 4 introduces the TR methodology as a useful survival model. Furthermore, this section discusses the estimator collapsibility property of TR that justifies its application in causal survival analysis. Section 5 provides simulation results that support this finding. Section 6 introduces TR G-computation, which can be used to estimate the marginal intervention causal effect. Next, Section 7 presents the highlight of this manuscript—a neural network extension of the TR model, TRNN. Section 8 provides further simulations which illustrate the accuracy of TRNN when the underlying model is correctly formulated and the potential bias when the model is misspecified. A real application is presented in Section 9. The manuscript is closed with discussions and limitations.
2. Notations and Causal Assumptions
The intervention is denoted by a dichotomous variable A that takes values 0 or 1. We defer the discussion of continuous treatment to future research. The outcome of interest is the subject’s time-to-first-event. Due to limited follow-up or competing risks failure, the true time-to-event random variable T may not be observed. Instead, the observed outcome variable , where C is the independent censoring random variable. We further use to denote whether an event is observed or not. The outcome Y is recorded once either during the study (event observed) or at the end of the study (right censored). The symbol Z denotes time-independent potential confounders. The observed data for individuals are, thus, drawn from the joint distribution of . In this paper, we only consider studies where the intervention was introduced and covariates were measured at baseline, without additional information being gathered before the occurrence of the first event.
There are three assumptions required for the discussions in this paper, consistency, exchangeability and positivity [12,13].
Consistency links the counterfactuals with the observables:
Exchangeability states that the conditional probability of receiving any value of treatment depends only on measured covariates Z:
The symbol denotes statistical independence. For example, means X is conditionally independent of Y given Z [14].
Positivity ensures that the probability of receiving any value of treatment conditional on Z is greater than zero, i.e., positive:
3. Confounding and Collapsibility
3.1. Time-Independent Confounding
A covariate Z is a confounder for the causal effect of the intervention A on the outcome Y if it has significant correlations with both A and Y, because Z is responsible for (at least part of) the apparent relationship between these two. However, the true causal effect of A on Y cannot be identified without a proper adjustment for the confounder Z. Another interesting complication of causal inference is “effect modification” which indicates that the effect of one variable on another varies across strata of a third [15]. Effect modification falls under the broader idea of “interaction” which is more familiar to statisticians. Traditional regression approaches are able to control confounding and deal with effect modification by producing conditional estimates, that is, by adding potential confounders to the regression models. Similar conditional estimation methods are prevalent in regression-based survival data analysis.
While it is important to take into account confounders that affect the outcome being studied and interactions between those factors, presenting the conditional estimates to policymakers is not always favorable. Conditional estimates are useful for understanding the specific effect of a variable, but they may not be the most relevant for policymakers who are looking for a more holistic understanding of the situation. On the other hand, the marginal effect provides a unified and simple way to quantify the causal relationship between the intervention and outcome, for the whole population. Thus, estimating the marginal intervention causal effect is often of research interest [16]. G-computation (also called standardization) [17] is an approach that allows one to estimate the marginal intervention effect. Even if the population contains different covariate groups, this method allows investigators to deduce the causality from observational studies as accurately as from perfectly randomized controlled trials, if a set of confounders is measured at each observation.
3.2. Model Collapsibility versus Estimator Collapsibility
When analyzing randomized clinical trial data, model collapsibility is a highly desirable model property. In order to introduce our findings and simulations presented in Section 4 and Section 5, in this subsection we review the distinction between collapsible models and collapsible estimators discussed in [18]. A collapsible model means that the marginal model after integrating out some covariates that are independent of the remaining will have the same regression coefficients for the remaining covariates. The most popular survival analysis approach, the Cox PH model, is not a collapsible model. The marginal regression parameter for a covariate in the Cox PH model does not equal the conditional parameter when other covariates are included in the model [19,20]. On the other hand, a collapsible estimator is an estimator which is consistent for the same value in the full and the marginal models, and so the estimate does not systematically change after removing independent covariates from the model [18].
In this paper, we focus on the collapsibility of parameter estimators for the TR model. This is of particular interest because causation conclusions are often derived based on parameter estimation results. If the model estimator is collapsible, the adjusted and unadjusted intervention-exposure association measures are the same, regardless of whether the control is made for covariates that are not associated with the intervention.
4. Parameter Estimators of TR Models with Linear Predictors Are Collapsible
The TR model conceptualizes a subject’s latent health as a random process that drifts down towards a threshold (which is usually set at zero) [7,8]. For a Wiener process where the health begins at and the boundary is the zero level, the distribution for the first-hitting-time (time-to-event) outcome T is given by an Inverse Gaussian (IG) distribution with p.d.f.
and survival function
where and denotes the c.d.f. of the standard normal distribution. In most situations, one expects the deterioration rate to be negative so that the process will hit a disease threshold. If the deterioration rate is positive, such that , the Wiener process may not hit the boundary threshold and the first-hitting-time T is not certain to be finite. This case can be used to model applications with a cure-rate, where the corresponding p.d.f. is improper with .
There are two parameters in the underlying latent health process, namely, the initial health status parameter , which describes the process starting point, and the deterioration rate parameter , which describes the rate at which the process drifts towards the threshold. To begin with, we considered TR with linear predictors in which the quantities and can be linked with observed regression covariate vectors and (with a leading unit to include an intercept term) and associated vectors of regression coefficients :
A logarithmic function is used to link the parameter with covariates in Equation (7) because the initial health status is set to be larger than 0. It is legitimate to use the same covariate vectors for both and , or to consider completely different sets of risk factors for these two linear predictors. In other words, covariates in x and z can completely differ, coincide, or partially agree. The most plausible functional form should come from expert knowledge of scientific research. The deterioration rate is not observable but can be connected with covariates, such as treatment regime A, by including the intervention A as a covariate in Equation (6). The effect size is then estimated by the coefficient of covariate A. Once the functional forms are determined by the investigator, the maximum likelihood estimation method is used to estimate coefficient vectors and . Note, though and incorporate randomness after including predictors, they can still be regarded as parameters when the covariates , are given.
In most chronic disease (such as CVD or kidney disease) studies, the intervention aims at slowing the health deterioration rate. As a result, we focus on investigating the causal effect of the intervention on the deterioration rate . Figure 1 includes a binary intervention A and a potential confounder Z and illustrates the causal relationship among covariates, parameters, and the random variable of interest. The corresponding linear prediction function of Equation (6) includes predictors A and Z:
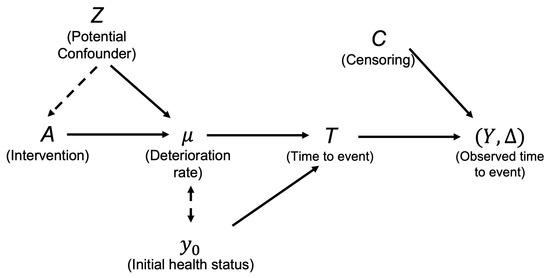
Figure 1.
Illustration of covariate effects on deterioration rate and first-hitting-time Y in TR.
The coefficient thus encodes the intervention effect on the deterioration rate .
We now claim that the MLE estimated treatment effect is collapsible; that is, the estimator does not systematically change after removing independent covariates (such as Z) from in Equation (8). See Appendix A for a proof. This property is especially pleasant because the removal of independent covariates does not change the parameters of remaining covariates when the initial health status is controlled correctly. On the other hand, the estimators from the Cox PH model do not have this benefit in most cases. It has been well established that the coefficient estimation before the intervention would be biased for the causal effect even when an uncorrelated covariate is omitted from the Cox PH model [21,22,23].
Since the Maximum Likelihood Estimates (MLEs) of are correlated, the initial health status has to be correctly adjusted to draw a valid causal conclusion regarding the deterioration rate in TR. Figure 1 includes a dashed arrow between and . This dashed arrow indicates may bias the estimation of the causal effect of A on . The only causal pathway we are interested in is the one from A to and, subsequently, flowing to T. To control the effect of the initial health status, in practice one may consider, (1) carefully recruiting subjects so that the participants do not dramatically vary in terms of the initial health status, or (2) correctly adjusting the initial health status during analysis. The former approach raises significant challenges because the initial health status is a latent variable that cannot be observed. The assumption may be true approximately if investigators carefully recruit subjects with similar demographics. Consequently, the generalizability of the study conclusion becomes infeasible. On the other hand, the schema of a complex disease process (which usually can be described by a Directed Acyclic Graph (DAG)) is not always available [24]. Thus, including the correct covariates is not an easy task. Considering the clinical meaning of each parameter in TR also helps us to select the correct set of covariates to be adjusted. For example, we recognized that a post-baseline effect should not be adjusted in the initial health status . Nonetheless, including unrelated predictors for will not influence the consistency and efficiency of the causal estimator due to the estimator collapsibility. As a result, it is encouraged to consider comprehensive predictor sets, for both and .
The hazard ratio (HR) has been criticized for decades for lacking causal meanings [5,6]. In contrast, the interpretation of the TR model is completely free from the hazard concept. For the causal problem investigated in this work, we would interpret the intervention’s causal effect as it causes the deterioration rate to increase or decrease by the estimated amount. With the same initial health level, a large deterioration rate means an earlier first-hitting-time, hence a shorter survival time. Furthermore, the causal inference community has been considering using other non-hazard-related models due to the interpretability issue of HR.
Not surprisingly, less model restrictive measures, such as differences between suitably adjusted and standardized survival curves or Restricted Mean Survival Time (RMST) [4,25], have drawn considerable attention. Some recent literature advocates the usage of RMST over a hazard ratio as the target causal measure [26]. In addition to focusing on interpreting the causal meaning of , RMST can also be estimated by TR. Given a fixed time horizon , RMST is defined with respect to the survival function as the area under the survival curves with a cut-off at the prespecified restrictive follow-up time (). We have also derived TR RMST formulas in a previous publication [27]. In general, the RMST for TR is calculated by
Theoretically, there are three formulas for TR RMST, depending on the sign of deterioration rate. When , a convenient and exact computational formula for evaluating is given by
Two other formulas when and can be easily obtained by variable transformation [27].
5. Simulation I—Estimator Collapsibility of TR with Linear Predictors
Now, we will use simulations to demonstrate the estimator collapsibility of the TR model with linear predictors. To investigate whether an intervention postpones an individual’s time to the first CVD event by slowing the deterioration rate, studies can be conducted to examine the relationship among intervention, potential confounder, and time-to-event outcome. In this simulation, we considered hypothesized studies with sample size . We simulated the dichotomous intervention A and two potential confounders . The intervention A was generated from a Bernoulli distribution, with a probability of receiving the intervention as . Confounder is a binary variable generated from a Bernoulli distribution, with . Some baseline characteristics, such as gender, follow this distribution. Confounder is a continuous variable, generated from a Normal distribution, with mean = 1 and standard deviation = 1. Following this setting, and are mutually independent. The time-to-event outcome T is simulated by an Inverse Gaussian (IG) distribution for TR as shown in (4) with and given later. The study is scheduled to end in 3 years. Thus, the endpoint is potentially right-censored due to the limited follow-up length. Later, we explore the case in which is a confounder for the intervention and outcome. By using to replace the aforementioned Bernoulli distribution which generates the intervention, we create an association between the intervention and confounder. The coefficient thus measures the magnitude of positive/negative association between A and .
Design 1: To start with, we consider Design 1 where the initial health status is the same for all individuals in the population. Intervention A and covariate both have causal effects on the deterioration rate . The linear predictors of and are simulated as:
Design 2: For a slightly more complicated design (Design 2), is related to the initial health status but not to by construction. Examples of such predictors include the genotype of an individual. If stands for gender as suggested earlier, the model is reasonable if the female has and females have a certain genetic makeup that protects them from CVD events. In conclusion, the simulation for deterioration rate remains unchanged whereas is added to the generation of :
As a result, the intervention causal effect is for both simulated studies. The effect can be interpreted as the intervention slowing the deterioration rate by . Now, we consider the fitting of and separately. For the deterioration rate, we consider a reduced model that includes A only versus a correct model that includes both A and . Then, for the initial health status, two models for were considered. One does not adjust for while the other does. In conclusion, for each design, with a specific association parameter , four TR models were explored. The simulation results of the intervention effect are summarized in Table 1; see the first six rows for Design 1 results and rows 7–12 for Design 2 results. The first two columns present results of the model with A only, and the next two columns present results from the model which includes both A and . The different models are summarized by rows, where “1” indicates the model with the intercept only and “1, ” indicates the model with intercept and . The standard error was computed based on 1000 repetitions. All models were fitted in R [28] with the threg package [29].

The simulation results of Design 1 first demonstrated that, when both the intervention A and the potential confounder are included in the deterioration model, the coefficient estimations are unbiased. Whether or not is included in the component, the estimation is not affected because the true initial health status is a constant. Now if the deterioration model only includes the intervention A in Design 1, the relationship between the intervention and the potential confounder needs to be further examined. Note that is not a confounder for the intervention and outcome if it is uncorrelated with A (i.e., ). By examining the first two rows of the table, the intervention causal effect is estimated consistently by the model, whether omitting or not. This is due to the independence between A and . Hence, the estimated coefficient for A provides the intervention causal meaning. The Cox PH model would fail to provide an unbiased estimation for the intervention when the uncorrelated covariate is omitted. The covariate has a beneficial effect on outcome by setting the coefficient to , the same as the intervention. Thus, if covariate is positively associated with the intervention (), exclusion of this confounder enlarges the intervention effect as the confounder beneficial effect has to be absorbed by the solely included intervention. If the association between the confounder and intervention is in a reverse direction (), the intervention effect is attenuated to account for the exclusion. For Design 2, the previous discoveries remain true, but now we have to correctly account for in the component. All reduced models failed to provide unbiased intervention estimation (serious underestimation), regardless of the format of . If both intervention and potential confounder are included in the deterioration rate model, we still obtain an unbiased estimation of the intervention effect. The unbiasedness is preserved if is not a confounder ().
6. TR G-Computation Procedure with Time-Independent Covariates
G-computation [17] serves as a useful tool for the marginal causal effect estimation. Conducting G-computation for TR with linear predictors is straightforward. The first step of a G-computation is to fit a regression model of the outcome on the exposure and relevant covariates. The model fitted in this step is frequently called the “Q-model” [16]. In TR G-computation, the Q-model is nothing but a standard threshold regression of the time-to-event outcome with potential predictors, such as . A correct Q-model is required for G-computation to estimate the intervention causal effect unbiasedly.
To improve the chance of finding the correct model, the investigator may be able to apply classical model selection tools (for example, AIC or BIC) in picking the most plausible TR model. Classical model selection methods are valid for selecting among linear prediction models, such as those in the Q-model. AIC and BIC are useful for likelihood-based method. Model selection techniques for TR were discussed previously [30].
Once estimated, the Q-model is used to predict all counterfactual outcomes for each subject under all possible intervention regimes. If the intervention is dichotomous, this is accomplished by plugging a 1 and then subsequently a 0 as the intervention into the TR fit to obtain predicted outcomes. Depending on the research question, the counterfactual outcomes or other estimands of interest can be calculated. Specifically, corresponds to the potential deterioration rate that should be observed under continuing intervention and is the counterpart that would be observed without intervention. Predicting all potential outcomes for each subject provides researchers with a full dataset that is free of confounding under causal assumptions. In other words, comparison or contracts among different potential outcomes now can be estimated at the individual or population level.
Generally speaking, G-computation can be used to generate the full data under all possible intervention levels if an intervention with discrete levels is under investigation. If the research question is straightforward as to identifying the intervention’s causal effect on the deterioration rate, we can merely estimate the difference between and . Then, averaging the differences across the observed distribution of the confounders leads us to the marginal causal effect () of the intervention on the deterioration rate. For instance, the marginal intervention causal effect on for a population with size N can be written as
Alternatively, a Marginal Structural Model (MSM) [31] can be used to fit the counterfactual outcomes. The estimates from the MSM are also equipped with causal meanings under certain regularization conditions. Under the binary intervention case, these two approaches yield the same estimation. Nevertheless, if the researcher plans to use a more clinically related measure, such as RMST, estimates of and can be plugged into Equation (9) to obtain the counterfactual individual RMST. The marginal intervention causal effect on RMST with a specific can be calculated similarly to what has been implemented for the deterioration rate:
7. Machine Learning-Assisted TR—TRNN
TR models with linear predictors discussed in previous sections enjoy benefits such as easy interpretation and estimator collapsibility. Nevertheless, the linear predictors of and (i.e., Equations (6) and (7)) can be improved. To mitigate the risk of model misspecification, one may propose adding all relevant covariates along with all possible interactions and higher-order terms into the model. Yet, a model with linear predictors is likely to fail when the input dimension is close to or even higher than the number of observations [32]. A recent research work [11] introduced a novel approach to fit the TR model with linear predictors in a high-dimensional framework. Using their suggested boosting algorithm, estimated coefficients can be obtained. In causal survival analysis, one may not only have to deal with cases with high-dimensional inputs but also have to consider possible non-linearity among predictors. To address these challenges, TR with non-linear predictors is considered using machine learning methods.
Deep learning techniques, such as neural networks, are useful tools for survival data analysis [33]. In this article, we propose a neural network extension for the TR model—TRNN. TRNN includes two separate neural networks to predict and in a TR model. It is reasonable to assume that the deterioration rate and the initial health may be influenced by different biological factors that are specific to them, in varied manners. Thus, different inputs and structures of the neural networks for and can reflect potential distinct trajectories. Nevertheless, the neural networks for and are connected by a log-likelihood-loss for right censored data and are optimized together. Trainable parameters in these two networks are updated simultaneously while the model is trained with respect to the objective function. This is the first time that neural networks are employed to fit a TR.
To start, the linear Predictors (6) and (7) can be replaced by neural networks without a hidden layer:
and
In above equations, and are activation functions. The quantities and are weight matrices, and and are bias vectors, all of which must be estimated. To echo the parameter range for TR, one can choose an activation function with output on the real line (such as a linear activation function) for and an activation function with a strictly positive output range (such as an exponential activation function) for . When the activation is the identity and the logarithm function, respectively, Equations (15) and (16) simply reduce to Equations (6) and (7), which have linear formats. More generally, hidden layers can be added between the input and output layers with user-specified numbers of neurons. With increased complexity, neural networks can approximate any functions [34]. The number of neurons in the output layer decides the output dimension. Regardless of the number of hidden layers and the number of neurons that each layer contains, the output layer of the and networks should always have one neuron. As a result, each network outputs a one-dimensional value for or in TRNN. Equations (15) and (16) can be generalized to and , in which and are some neural networks. An example of TRNN which takes two three-dimensional inputs is displayed in Figure 2. This TRNN has one hidden layer with 5 units for both and .
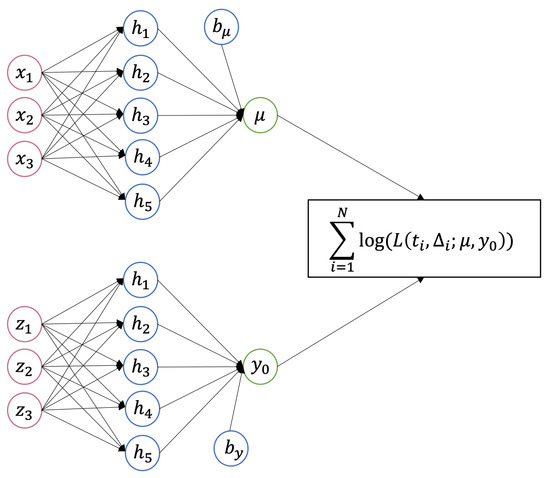
Figure 2.
A TRNN with a 3-dimensional input and 5 units in the hidden layer for both and .
The loss function for TRNN is based on the log-likelihood function for TR. With the right censored data of N individuals, the total log-likelihood is the sum of the log density or log survival functions:
The neural network solves the optimization problem by minimizing the loss. Consequently, the negative log-likelihood is the final loss function for TRNN. The MLE of Inverse Gaussian with censored observations is unique [35] and, thus, creates no challenge for the optimization problem. The well-accepted optimization algorithms, such as Adam [36], can be used to train the model. To facilitate model convergence, we can first fit an Inverse Gaussian with intercept only (for both and ) using observed data, then use the estimated intercept terms at the initial bias for each network. Or, we can simply set the initial bias for at and for at 1. The usual neural network training techniques are all applicable to train a TRNN. For instance, the validation loss can be monitored during training and the algorithm will be stopped early if the validation loss decrements between iterations do not exceed a prespecified value (delta). The weights that return the best validation loss will be reported as the final weights. Employing early stopping can effectively avoid over-fitting. See Section 9 for a real example of fitting a TRNN.
The benefits of using TRNN are multi-fold, either when applying TR for a conventional survival analysis or a causal survival analysis. TRNN is capable of handling high-dimensional data such as gene expression. In Section 6, we have claimed that a correct Q-model is the prerequisite for the unbiased marginal effect estimation using G-computation. A TRNN can replace a regular TR model for the Q-model for TR G-computation. Once the TRNN is optimized, it can be used to predict and under all possible regimes. Once the Q-model provides the complete data set with and under different regimes for each subject, the marginal causal effect can be evaluated as before, depending on the causal estimands of interest. All ML models and experiments were implemented in the R package keras [37]. Keras [38] is a high-level neural networks Application Programming Interface (API) of either TensorFlow or Theano. The kerasR interfaces with Python 3.8.8 and Tensorflow 2.9.1 [39]. Sample code running a TRNN using R is included in Appendix B.
8. Simulation II—TR G-Computation and RMST
This section uses simulated data to demonstrate the application of G-computation with TR, either with the deterioration rate or RMST as the target causal estimand wherein the sample size . The outcome of interest is the time until the subject’s first CVD event. The observed outcome is potentially right-censored at a prespecified study end time, at 10 years. We further simulated two potential confounders. Similar to Simulation I, the binary variable gender () was generated from a Bernoulli distribution, with a probability of female being , and is a continuous baseline variable that was generated from a Normal (1, 1). An example of such a predictor is population-standardized blood pressure. The covariates and are independent of each other. The binary intervention variable A was assigned by:
This assignment mechanism indicates that subjects with higher blood pressure and/or male are more likely to receive the intervention. This is just a sample intervention assignment mechanism. Nevertheless, the estimation procedure described in this section follows even if other parametrizations that create associations between covariates and intervention are considered. In observational studies, assuming intervention is given at random may not be reasonable. Instead, it is safer to consider the intervention as randomly assigned, but the randomization probability depends on some baseline covariates. The time-to-event outcome T is simulated by an Inverse Gaussian (IG) distribution as shown in (4). Linear predictors of and are formulated as:
Figure 3 illustrates covariate effects on the outcome of interest for the simulated study.
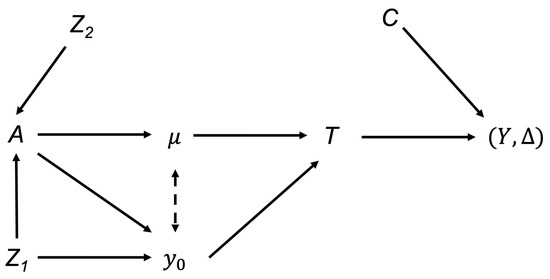
Figure 3.
Illustrations of covariates effects on the deterioration rate for the simulated study.
Under this simple simulation setting, two covariates ( and ) must be considered when analyzing the effects of intervention A on the outcome Y. Gender () is a confounder for the intervention and outcome because it affects both A and the initial health status (subsequently Y). Without any adjustments, one may incorrectly conclude that the intervention A has a causal effect on Y even when there is no such causal pathway from A to Y. The association appears only due to the unadjusted confounder, Gender. In contrast, blood pressure () affects the intervention, yet has no independent effect on the deterioration rate (subsequently Y). Thus, is just an effect modifier for the intervention effect on , evident by the interaction term between and A and lacking the causal pathway from to . This data generation schema implies that the intervention has a positive effect on slowing the deterioration rate and the effect is further modified by while the intervention benefit increases as blood pressure increases. In terms of the initial health status, subjects who received the intervention and/or were female have larger initial health values.
We explored TR G-computation of four TR models (with different linear predictors for and/or ), as well as a TRNN as the Q-model. Model 1 assumes that the intervention influences both the initial health status and the deterioration rate. Thus, one only adjusts the intervention in both and .
Model 1 (linear predictors with intervention only):
Model 1 is incorrect for as it ignores the interaction effect and is also incorrect for by omitting the gender main effect. Model 2 is another incorrect model with a similar assumption for the covariate .
Model 2 (linear predictors with only):
This model includes the main effect of when estimating but this causal pathway is lacking, and only influences by interacting with intervention A. Further, wrongly replaces for . Thus, the model fails to model both the deterioration rate and initial health status flawlessly. Next, Model 3 is the correct model.
Model 3 (true simulated model as specified in Equation (19)):
Given the fact that picking the correct TR model may not be an easy task for the user, it is more realistic to consider a full model which includes all potential confounders, as well as the interaction term.
Model 4 (full model with intervention, , interaction):
Table 2 summarizes the intervention effect estimated by TR, Table 3 summarizes the marginal intervention causal effect estimation provided by the G-computation. The estimation standard deviation was calculated by non-parametric bootstrapping with 1000 times bootstrap samples.

Table 2.
Model estimation of TR Models 1–4. TR Model 3 is the true model, TR Model 4 is the full model.

Table 3.
G-computation results of marginal intervention causal effect by using TR Models 1–4 and TRNN as the Q-model, respectively. TR Model 3 is the true model, TR Model 4 is the full model.
RMST may be more interpretable for clinicians and patients. With the same simulated data, using , Table 4 summarized the RMST estimation results for each group and the group-wise difference. The estimations were obtained by using TR Model 1–4, TRNN (same parameter settings as used in previous G-computation), Kaplan–Meier curves, and the Cox model. The Cox model used in the simulation is similar to the TR full model which includes and an interaction between intervention A and as covariates. The baseline hazard was estimated by the Breslow method [40].

Table 4.
Group specific and group-wise difference RMST estimations from TR Models 1–4, TRNN, Kaplan–Meier (KM), and Cox model. The RMST difference is calculated by the intervention group minus the control group.
The simulated true marginal effect on deterioration rate is per Equation (19). Specifically, the true marginal intervention causal effect on the deterioration rate is the summation of the main effect and the interaction effect then averaged over the population distribution of the effect modifier. We observed that the intervention causal effects estimated by TR are the same as by TR with G-computation, for TR Models 1 and 2. The reason is that the TR with G-computation is estimating the same marginal effect as the TR models if no interaction term was captured. Nonetheless, these TR models are biased due to model misspecifications. Both TR Model 1 and 2 have the intervention effect slightly overestimated since part of the intervention effect modified by BP is absorbed by the only included main intervention effect. Though TR is estimator collapsible, these TR models failed to provide consistent estimation as the excluded interaction term is correlated with the included main effect. Further, intervention A correlates with both and under this simulation setting. This emphasizes again the importance of confounder adjustment. Next, when the TR model was correctly specified (Model 3), TR with G-computation returned a consistent estimation of the marginal intervention causal effect. This marginal causal effect has not only accounted for the main intervention effect but also the effect modified by . After adjusting for gender effect in the initial health level and the effect modifier for intervention in the deterioration rate, we were able to conclude that receiving the intervention will slow the deterioration rate by marginally, for this population. The TR full model (Model 4) with G-computation also provided an unbiased estimation of the marginal intervention effect. Though the standard error for the TR model 4 estimations increased due to unnecessary terms that have been added to the model, the marginal causal intervention effect estimation has a similar standard error to the correct model.
Model 5 (TRNN): Furthermore, as suggested in Section 7, we used a TRNN to fit and . The algorithm took as input, for both and networks. We chose a network with one hidden layer (10 units, activation is tanh) for the network and a network without a hidden layer for the network. This TRNN can be conceptualized as TR Model 5 in which there were no specific forms of and assumed. Each simulated dataset was first split into a training set (90%) and a testing set (10%). Then 25% of the training data were randomly selected as validation data and the model was trained with the rest of the training set. The optimization process would be potentially stopped early if the validation loss no longer decreases. Once the and networks were trained and we obtained the predictions of and , the G-computation for the marginal intervention effect followed as demonstrated in Section 6. However, the estimation will only be evaluated using the test data. The marginal intervention causal effect on the deterioration rate we obtained by using TRNN as the Q-model is , and on the logarithm of initial health status is (Table 3). Though only three main effects were fed into the model, using neural networks allows us to capture the non-linear relationship among covariates. The estimations were fairly close given the fact that no specific model is assumed. One should expect the correct parametric models to provide more efficient estimates as the estimates are derived based on maximal likelihood. The standard deviations of marginal effect estimation were even smaller than those obtained from TR Models 3 and 4.
We would like to further quantify the marginal causal intervention effect in terms of RMST. Table 4 provides the RMST estimation results of TR Models 1-4, TRNN, Kaplan–Meier (KM), and the Cox Model, with G-computation. First, the results obtained by KM were from group-specific non-parametric Kaplan–Meier curves. They serve as the true values and were the average RMST in each intervention group with diverse gender and BP values.
The RMST difference was simply calculated as the difference between the areas under group-specific survival curves. Next, for TR/Cox models, the group-specific estimations came directly from the estimated survival probabilities while the between-group difference requires an extra step of G-computation. The TR Models 3 and 4 provide nearly identical results for both group-specific estimations, as well as the between-group difference. Moreover, these two TR models provided estimations that are close to the non-parametric estimation. Thus, these two models with G-computation both provided unbiased marginal intervention causal effect on RMST as the intervention effect modified by has been correctly accounted for. Yet, TR Models 1 and 2 with G-computation fail this purpose because their underlying Q-models were incorrect. The intervention effect on RMST was underestimated. It can be seen from the TR Models 3 and 4 estimation result that the intervention was beneficial as the RMST increased by years when the follow-up horizon was set to 10 years. In other words, the population-level RMST will be almost doubled if everyone switches to the intervention.
Finally, TRNN outperforms all other TR models as it provides the closest results to the non-parametric method, though the standard deviations of estimation increase significantly as well. The Cox model estimated the group-specific RMST inaccurately; however, the estimated RMST between-group difference is almost the same as TR estimated. Note that the standard error of Cox model estimation is much higher than that of TR models 3 and 4.
9. Application—CARDIA
The Coronary Artery Risk Development in Young Adults (CARDIA) study [41,42] examined the development and determinants of clinical and subclinical cardiovascular disease and their risk factors. This study is a population-based observational study of 5115 participants aged around 18–30 years recruited during 1985–1986, from four centers in the United States (Birmingham, AL; Chicago, IL; Minneapolis, MN; and Oakland, CA). The sample was designed to achieve approximately balanced subgroups of race, gender, education (high school or less, and more than high school), and age (18–24 and 25–30 years old). After the enrollment, follow-up in-person examinations were conducted in 1987–1988 (Year 2), 1990–1991 (Year 5), 1992–1993 (Year 7), 1995–1996 (Year 10), 2000–2001 (Year 15), 2005–2006 (Year 20), 2010–2011 (Year 25), and 2015–2016 (Year 30), with retention of 90.5%, 85.6%, 80.6%, 78.3%, 73.6%, 72.0%, 72.2%, and 71.0% of living participants, respectively. Because this study started to follow subjects during their young adulthood, it has provided evidence that supports intervening during young adulthood to prevent CVD mortality in later decades [43].
To examine the smoking causal effect on subjects’ time to first CVD, we consider the baseline measurement and the individual’s first CVD event time measured until the end of the study. We implemented gentle data cleaning techniques, such as removing observations with missing covariates, and combining categories for extremely unbalanced categorical data. The cleaned data ( obs) are used to demonstrate the TRNN application and the following G-computation. We are interested in the marginal causal effect of smoking on the heart health deterioration rate . Since the competing risk censoring (e.g., death, lost-to-follow-up) is likely to be informative, we first use Inverse Probability of Censoring Weighting (IPCW) to adjust for the informative censoring bias. A subject who is competing risk censored will receive weights of 0, while the remaining subjects will be inversely weighted by the probabilities of not competing risk censored. The likelihood-based loss function used in TRNN is updated accordingly to incorporate these weights. Figure 4 shows the Kaplan–Meier survival curves of CVD events of smokers vs. non-smokers. The outcome is the subject’s time to his/her first CVD event. We truncate the follow-up time at month 400.
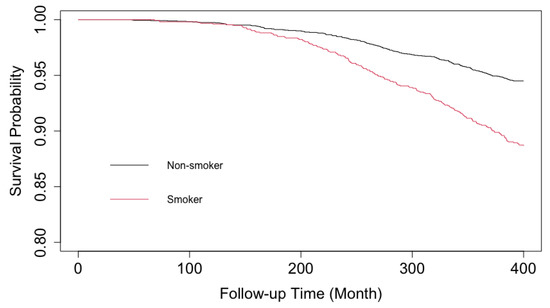
Figure 4.
Kaplan–Meier Curves of Time-to-First CVD Event, Smokers (red) vs Non-smokers (black), CARDIA. Note the y-axis starts from .
As can be seen from the survival curves, there is an obvious trend of delayed exposure effect. The trend is expected as participants’ likelihood of experiencing their first CVD event grows as they age.
To identify the optimal structure of the TRNN, we started with determining the hyperparameters to be used in the network. Five-fold cross-validation [44] was used for the tuning process. From the simulation, we observed that one hidden layer is sufficient to capture the non-linear nature of the covariates. In this application, we would like to consider sigmoid and tanh activation functions for the hidden layer. Additional hyperparameters we tuned included hidden unit number for network , hidden unit number for network , learning rate , and delta for early stopping . There are many other hyperparameters, such as batch size, to be tuned. The truth is that it is impossible to tune all hyperparameters exhaustively as the set of hyperparameters must be large. Therefore, we only include a working example here with a limited set of hyperparameters. The choice of network structure and hyperparameter set are mostly experience-based. For instance, in this dataset the event rate is low. We do not encourage using a small batch size—the algorithm may not perform well if a batch of data does not include a single event. Thus, we set the batch size to 256 and refrain from tuning this parameter. The number of hidden units also depends on the input dimension. Referring to the proposed network structure in Figure 2, one can select different covariates for the and networks. In this application, for the network, we considered baseline covariates, including the smoking status (smoker vs non-smoker), gender (female vs male), race (white vs non-white), age at enrollment (younger than 18, 19–25, 25–29, and older than 30), marital status (married or living as married, divorced or separated or widowed, never married), baseline BMI, and baseline SBP. For the network, we considered gender (female vs male), race (white vs non-white), age at enrollment (younger than 19, 19–25, 25–29, and older than 29), marital status (married or living as married, divorced or separated or widowed, never married), baseline BMI, baseline SBP, exercise intensity score, employment status (full-time/part-time/unemployed), education level (less than high school or GED/some college or college/college above), self-reported diabetic, hypertension, high cholesterol, whether father/mother had a heart attack, and whether father/mother had a stroke. Nevertheless, TRNN is capable of handling high-dimensional input. Thus, many other potentially important covariates can be included. Using domain knowledge, we narrowed down the range of covariates, as well as the hyperparameter set, for demonstration purposes. The five-fold cross-validation selects neuron numbers 15 and 2, activation function sigmoid and tanh for and network, respectively. We set the learning rate to and the delta for early stopping to . With selected hyperparameters, the data were further split into the training set (70%), the validation set (20%), and the testing set (10%). Once the model is trained, the G-computation is performed with 10% testing data that the model never used previously. Note without setting a random seed, the resultant neural networks parameter estimations are different due to the stochastic optimization. With 100 refits of the neural networks, smoking changes the deterioration rate by in this population.
In terms of RMST ( months) of the first CVD event free time, the difference between smokers and non-smokers on average is months. The confidence interval was computed using the bootstrapped standard deviation. The marginal smoking causal effect we detected using G-computation and TRNN is significant.
10. Discussion
The threshold regression (TR) model with inverse Gaussian first hitting time distribution provides profound clinical interpretations because it models the initial health status , as well as the deterioration rate . These two parameters jointly describe the first hitting time of a latent health process. Predictors linked to these two parameters influence the health process and can be interpreted with different biological meanings. We further argue that the estimators from the TR model with linear predictors for the deterioration rate are collapsible if is correctly controlled. Collapsibility of the estimator is a desirable property for a causal model because one often feels uncomfortable interpreting the conditional effect estimation. Unlike the Cox PH model, the TR model ensures an unbiased estimation of the intervention’s causal effect on the outcome, if an uncorrelated covariate is omitted from the model.
When analyzing the intervention effect, any proposed model will generally not be exactly correct, and results can be difficult to interpret if the model is misspecified and treatment effects substantially differ across subgroups [1]. The lack of subgroup information may make things more complicated. As a result, the marginal intervention effect is of research interest as it provides population summaries with a simple interpretation. A typical randomized controlled trial attempts to estimate the marginal effect of the investigated intervention, which is the intervention effect on the population level regardless of the diverse characteristics. Estimator collapsibility is a particularly desirable model property because it provides a model with a much higher likelihood to estimate the marginal effect unbiasedly. Furthermore, estimator collapsibility allows for other more advanced causal inference techniques, such as the instrumental variable (IV) analysis [45]. It has been shown that TR offers an appealing opportunity to apply IV analysis in a survival context [9].
The causal aspect of TR is underrated because of the availability of TR causal inference [9]. Section 4 and Simulation 5 have demonstrated the collapsibility of estimators from TR with linear predictors when one would like to make causal conclusions for the deterioration rate, condition on the initial health status is correctly controlled. The part in TR allows one to control for the population baseline heterogeneity. Thus, we shall not treat the necessity of controlling in TR deterioration rate analysis as a disadvantage. Previous research has shown that the intervention effect may interact with the baseline genotypes [46]. This is a common cause for the crossing survival curves observed in medical research. Subjects in the intervention group seem to experience the event sooner than the control group in the early stage of follow-up due to subgroup initial health status heterogeneity. However, the intervention benefit emerges later because the intervention has significant effects on the deterioration rate. The Cox model may lose power significantly when the PH assumption is violated, not to mention the lack of collapsibility and interpretation issues. Alternatively, TR is capable of addressing distinct hazard types [10]. As we have shown in Section 5, the genotype or other baseline characteristics can be adjusted as part of the initial health status. Once the baseline heterogeneity has been effectively controlled, the marginal intervention causal effect can be viewed as slowing the health process deterioration, providing a solid causal interpretation of the intervention on the deterioration rate .
Though the non-parametric marginal causal estimation is feasible under limited cases, such as when all covariates are discrete and there are data collected for each sub-category, parametric models are more realistic choices in practice. One may have to seek help from the G-computation method when confounders for both the intervention and outcome or an effect modifier (interaction) are present. G-computation is a statistically reliable method of covariate adjustment for an unconditional treatment effect that produces a resulting estimator [1]. We have demonstrated that, with correct model specifications, TR is capable of estimating the intervention effect marginally besides the usual conditional estimation with G-computation. G-computation with TR can also provide a valid marginal causal estimation if other estimands, such as RMST, are chosen as the target causal estimand. Simulation in Section 8 advocates using a comprehensive model as the Q-model to mitigate the risk of model misspecification. Plus, a doubly robust estimation that combines the outcome model (TR in our case) and IPW is feasible. The combined strategies are generally superior to either strategy alone [47].
TRNN is the neural network extension of the TR model in which the linear predictor structure is replaced by nonlinear neural networks. TRNN can not only handle high-dimensional input easily, but also capture more sophisticated relationships between covariates and deterioration rate and/or initial health status. From our observations, TRNN serves as a competitive Q-model for the G-computation. As we have shown with simulations in Table 3 and Table 4, using TRNN can free practitioners from selecting the “true” model. Yet, as in other ML methods, the network structures and hyperparameters have to be tuned before implementation. With an increasing pool of ML toolkits, we only expect this method to become more accessible in the future. Last, the convergence of TRNN is fairly quick. Under the simulated setting, 100 training epochs are sufficient, thus no significant increased computation time was observed compared to a non-NN-based TR model.
As a flexible parametric survival model, surrogate estimands, such as RMST, can be readily obtained for TR. Using this estimand also solves the dilemma that an investigated intervention may provide reverse effects for the initial health status and the deterioration rate. Given a fixed end of follow-up horizon (), slowing the degradation rate and/or improving the initial health status directly improves the survival rate, subsequently the RMST. We can also consider other causal estimands, such as causal survival probability difference.
There exist other aspects of TR, which make it more valuable in practice. For instance, when , TR with a cure rate can handle the case where a substantial proportion of subjects are believed “cured” or immune from the endpoint events [48].
11. Limitations and Future Work
The discovery of the correct DAG falls beyond the scope of this paper. Yet, the correctness of the DAG is often the prerequisite of any valid causal inference, as well as an unbiased G-computation. Though G-computation enables one to draw marginal causal effect conclusions, the generalization of the results requires extra attention because the confounder distribution may vary significantly across multiple populations. Even within the same population, making inferences for unobserved subgroups requires extrapolations that are often dangerous. Applying G-computation to TR in this paper requires the assumption that the underlying survival distribution is Inverse Gaussian. In practice, the assumption is not necessarily true. The advantage of the model is its rich parameterization which provides good opportunities for modeling the underlying health process. TR models equipped with other processes are outside the scope of this paper, and, hence, omitted from discussion. Interested readers can refer to other literature [7].
In order to consider longitudinal TR with time-varying covariates [49], the deterioration rate at an earlier interval is a causal factor of initial health status at the beginning of the next interval. It, thus, requires a rethink of the model from the random process perspective. Moreover, time-varying confounding may be a common complication when the exposure and covariates are both measured multiple times along the study. The structural nested AFT model [50] has been shown as an effective method to handle time-varying confounding. How to do a Structural Nested Threshold Regression Model (SNTRM) is an open question and reserved for further research.
12. Conclusions
Threshold regression is a model that treats an individual’s latent health status as a random process. The TR model can address a wide variety of applications by employing different random processes. Notably, unlike the Cox PH model that only deals with the constant hazard case, TR can model a time-varying hazard function and, therefore, can handle more complicated cases, such as delayed or crossover treatment effects. The TR model is straightforward to implement for time-to-event outcomes by modeling the health process’s first-hitting-time of a boundary. The model provides clinically meaningful interpretations. Various causal questions can be posed for the deterioration rate and/or the initial health status . In this work, we focus on using TR to model the time to the occurrence of the subject’s first event of interest, without considering the longitudinal case. The research interest centers on whether an intervention causes a change in the deterioration rate when is correctly controlled. It is shown that a TR model with linear predictors for has collapsible estimators, if is correctly controlled. The estimator collapsibility makes this model a useful tool in causal survival analysis. Furthermore, TR G-computation is easy to implement. This method can provide the marginal causal effect estimation on deterioration rate.
In conclusion, TR has proved its effectiveness in causal survival analysis. The neural network extension of TR, namely TRNN, expands the model’s capabilities of handling high-dimensional and interconnected data, thus providing a new working direction for TR in the machine learning era.
Author Contributions
Conceptualization, Y.C. and M.-L.T.L.; methodology, Y.C.; simulation and validation, Y.C.; writing—original draft preparation, Y.C.; writing—review and editing, Y.C., M.-L.T.L. and P.J.S.; supervision, M.-L.T.L. All authors have read and agreed to the published version of the manuscript.
Funding
Research of M.-L.T.L. was partially supported by NIH grant R01EY022445.
Institutional Review Board Statement
Ethical review and approval were waived for this study because this is a secondary data analysis. The authors did not involved with the conduct of the trial. The received data have been de-identified.
Informed Consent Statement
Not applicable.
Data Availability Statement
The CARDIA datasets generated during and/or analyzed during the current study are available in the Biologic Specimen and Data Repository Information Coordinating Center (BioLINCC) repository. Available online: https://biolincc.nhlbi.nih.gov/home (accessed on 24 April 2023).
Acknowledgments
The authors thank Yen-Tsung Huang for his review and helpful comments on an earlier draft. The authors thank the BioLINCC for granting access to the CARDIA data.
Conflicts of Interest
The authors declare no conflict of interest.
Abbreviations
The following abbreviations are used in this manuscript:
| AIC | Akaike Information Criterion |
| BIC | Bayesian Information Criterion |
| BP | Blood Pressure |
| CVD | Cardiovascular Disease |
| DAG | Directed Acyclic Graph |
| FHT | First Hitting Time |
| HR | Hazards Ratio |
| IG | Inverse Gaussian |
| IV | Instrumental Variable |
| IPW | Inverse Probability Weighting |
| KM | Kaplan–Meier |
| ML | Machine Learning |
| MLE | Maximum Likelihood Estimates |
| MSM | Marginal Structural Model |
| NN | Neural Network |
| RMST | Restricted Mean Survival Time |
| TR | Threshold Regression |
Appendix A. Proof of the Collapsibility of TR Estimators
We focus on the discussion of the simplest case in which there are two covariates and for the deterioration rate . The resultant linear format of reads as:
The extension to the more covariates case is straightforward hence omitted. Following discussion in Section 4, covariates and are assumed to be independent and correctly controlled. First, it can be seen from the p.d.f of the TR distribution
that the model is no longer a TR model if we integrate the random variable out. Hence, we cannot claim TR as a collapsible model.
We instead focus on showing that the estimator from the full model which includes does not systemically differ from the which is estimated from the reduced model with only. This is of research interest because using TR, one would interpret the estimated coefficients for deterioration rate or and further translate the coefficients’ biological meanings.
If one formulate the deterioration rate as in Equation (A1) and does not consider any covariate for , the parameters can be estimated by maximum likelihood. The likelihood of the full model with sample size N is:
After taking the log of the likelihood, the system of score equations we are going to solve is (with irrelevant terms omitted):
To ease the notations, we let
Given , the last three equations of (A5) are a linear system with constant terms depending on . Their solutions are linear functions of . These solutions can be plugged into the first equation in (A5) to obtain an updated value of . This forms the basis of an iterative scheme to estimate all four parameters in (A5). After some algebra, the formula for as a function of is derived as
Similarly, the solved from score equations of the reduced model as functions of is
when , given the fact that covariates are independent, some important linear terms in Equation (A6), such as , , and are zero, if consider their expectation values. As a result, there is no asymptotic difference between Equations (A6) and (A7).
The above derivation only discusses the non-censored case. When censored observations present, we need to take another round of integration of censored observations on their outcome space. Yet, the main MLE idea stays the same [35]. The estimated coefficients of the deterioration rate would still be collapsible if irrelevant covariates are omitted from the model when is successfully controlled (mainly for correct model specification purposes). Intuitively, the collapsibility mainly comes from the fact that covariates are added to the deterioration rate model linearly. Furthermore, the set of score functions to be solved has these parameters in additive format.
Appendix B. TRNN R Code
- library(tensorflow)
- library(keras)
- tfp <- tf_probability()
- tfp <- import("tensorflow_probability")
- tfd <- tfp$distributions
- Normal<- tfd$Normal(loc = 0, scale = 1)
- ##loss function
- TR_loglik_continuous=function(y_true, y_pred){
- t = y_true[, 1]
- c = y_true[, 2]
- mu = y_pred[, 1]
- y0 = y_pred[, 2]
- s1=(mu*t+y0)/k_sqrt(t)
- s2=(mu*t-y0)/k_sqrt(t)
- f=k_log(y0+ k_epsilon())-1.5*k_log(t)-k_square(y0+mu*t)/(2*t)
- S=k_log(Normal$cdf(s1)-
- k_exp(-2*y0*mu)*Normal$cdf(s2)+ k_epsilon())
- return (k_mean(-1 * (c*f+(1-c)*S)))
- }
- ##network
- k_clear_session()
- sigmoid1<- function(x) {
- return(-10*(k_sigmoid(x)))
- }
- log1<- function(x) {
- return(-k_exp(x))
- }
- act_exp<- function(x) {
- return(k_exp(x))
- }
- #len_mu - length of predictors for mu
- #len_y0 - length of predictors for y0
- input_mu <- layer_input(shape = c(len_mu),
- dtype = ’float32’,
- name = ’input_mu’)
- input_y0 <- layer_input(shape = c(len_y0),
- dtype = ’float32’,
- name = ’input_y0’)
- ##many parameters in below network are subject to tuning
- output_mu=input_mu%>%
- layer_dense(units = 10,activation = "tanh")%>%
- layer_dense(units = 1,activation = "linear",name = ’mu’,
- kernel_initializer =
- initializer_random_uniform(minval = -0.05,
- maxval = 0.05),
- bias_initializer =
- initializer_constant(0))
- output_y0=input_y0%>%
- layer_dense(units = 1,activation = act_exp,name = ’y0’,
- kernel_initializer =
- initializer_random_uniform(minval = -0.05,
- maxval = 0.05),
- bias_initializer =initializer_constant(1))
- output_tr = layer_concatenate(list(output_mu,output_y0))
- tr_model <- keras_model(
- inputs = c(input_mu,input_y0),
- outputs = c(output_tr))
- #summary(tr_model)
- early_stop <-
- callback_early_stopping(monitor = "val_loss",
- min_delta = 0.01,
- patience = 10,
- restore_best_weights = TRUE,
- verbose = 0)
- tr_model %>%
- compile(loss = c(TR_loglik_continuous),
- optimizer=optimizer_nadam(learning_rate=0.002))
References
- U.S. Food and Drug Administration. Adjusting for Covariates in Randomized Clinical Trials for Drugs and Biological Products Guidance for Industry. 2021. Available online: https://www.fda.gov/regulatory-information/search-fda-guidance-documents/adjusting-covariates-randomized-clinical-trials-drugs-and-biological-products (accessed on 11 November 2022).
- Greenland, S. Absence of Confounding Does Not Correspond to Collapsibility of the Rate Ratio or Rate Difference. Epidemiology 1996, 7, 498–501. [Google Scholar] [CrossRef] [PubMed]
- Greenland, S.; Pearl, J. Adjustments and their Consequences-Collapsibility Analysis using Graphical Models. Int. Stat. Rev. 2011, 79, 401–426. [Google Scholar] [CrossRef]
- Didelez, V.; Stensrud, M.J. On the logic of collapsibility for causal effect measures. Biom. J. 2022, 64, 235–242. [Google Scholar] [CrossRef] [PubMed]
- Hernán, M.A. The Hazards of Hazard Ratios. Epidemiology 2010, 21, 13–15. [Google Scholar] [CrossRef] [PubMed]
- Aalen, O.O.; Cook, R.J.; Røysland, K. Does Cox analysis of a randomized survival study yield a causal treatment effect? Lifetime Data Anal. 2015, 21, 579–593. [Google Scholar] [CrossRef]
- Lee, M.L.T.; Whitmore, G.A. Threshold Regression for Survival Analysis: Modeling Event Times by a Stochastic Process Reaching a Boundary. Statist. Sci. 2006, 21, 501–513. [Google Scholar] [CrossRef]
- Lee, M.L.T.; Whitmore, G.A. Proportional hazards and threshold regression: Their theoretical and practical connections. Lifetime Data Anal. 2010, 16, 196–214. [Google Scholar] [CrossRef]
- Hellier, J.; Emsley, R.; Pickles, A. Estimating dose-response for time to remission with instrumental variable adjustment: The obscuring effects of drug titration in Genome Based Therapeutic Drugs for Depression Trial (GENDEP): Clinical trial data. Trials 2020, 21, 10. [Google Scholar] [CrossRef]
- Chen, Y.; Lawrence, J.; Lee, M.L.T. Group sequential design for randomized trials using “first hitting time” model. Stat. Med. 2022, 41, 2375–2402. [Google Scholar] [CrossRef]
- Bin, R.D.; Stikbakke, V.G. A boosting first-hitting-time model for survival analysis in high-dimensional settings. Lifetime Data Anal. 2023, 29, 420–440. [Google Scholar] [CrossRef]
- Rosenbaum, P.R.; Rubin, D.B. The central role of the propensity score in observational studies for causal effects. Biometrika 1983, 70, 41–55. [Google Scholar] [CrossRef]
- Hernán, M.A.; Robins, J.M. Causal Inference: What If; Chapman and Hall/CRC: Boca Raton, FL, USA, 2020. [Google Scholar]
- Dawid, A.P. Conditional Independence in Statistical Theory. J. R. Stat. Soc. Ser. Methodol. 1979, 41, 1–15. [Google Scholar] [CrossRef]
- VanderWeele, T.J.; Robins, J.M. Four Types of Effect Modification: A Classification Based on Directed Acyclic Graphs. Epidemiology 2007, 18, 561–568. [Google Scholar] [CrossRef] [PubMed]
- Snowden, J.M.; Rose, S.; Mortimer, K.M. Implementation of G-Computation on a Simulated Data Set: Demonstration of a Causal Inference Technique. Am. J. Epidemiol. 2011, 173, 731–738. [Google Scholar] [CrossRef] [PubMed]
- Robins, J.M. A new approach to causal inference in mortality studies with a sustained exposure period—Application to control of the healthy worker survivor effect. Math. Model. 1986, 7, 1393–1512. [Google Scholar] [CrossRef]
- Samuelsen, S.O. Cox regression can be collapsible and Aalen regression can be non-collapsible. Lifetime Data Anal. 2023, 29, 403–419. [Google Scholar] [CrossRef]
- Ford, I.; Norrie, J.; Ahmadi, S. Model inconsistency, illustrated by the cox proportional hazards model. Stat. Med. 1995, 14, 735–746. [Google Scholar] [CrossRef]
- Burgess, S. Commentary. Epidemiology 2015, 26, 411–413. [Google Scholar] [CrossRef]
- Martinussen, T.; Vansteelandt, S. On collapsibility and confounding bias in Cox and Aalen regression models. Lifetime Data Anal. 2013, 19, 279–296. [Google Scholar] [CrossRef]
- Sjölander, A.; Dahlqwist, E.; Zetterqvist, J. A Note on the Noncollapsibility of Rate Differences and Rate Ratios. Epidemiology 2016, 27, 356–359. [Google Scholar] [CrossRef]
- Crowther, M.J.; Royston, P.; Clements, M. A flexible parametric accelerated failure time model and the extension to time-dependent acceleration factors. Biostatistics 2022, 5, kxac009. [Google Scholar] [CrossRef] [PubMed]
- VanderWeele, T.J. Principles of confounder selection. Eur. J. Epidemiol. 2019, 34, 211–219. [Google Scholar] [CrossRef]
- Uno, H.; Claggett, B.; Tian, L.; Inoue, E.; Gallo, P.; Miyata, T.; Schrag, D.; Takeuchi, M.; Uyama, Y.; Zhao, L.; et al. Moving beyond the hazard ratio in quantifying the between-group difference in survival analysis. J. Clin. Oncol. Off. J. Am. Soc. Clin. Oncol. 2014, 32, 2380–2385. [Google Scholar] [CrossRef]
- Stensrud, M.J.; Aalen, J.M.; Aalen, O.O.; Valberg, M. Limitations of hazard ratios in clinical trials. Eur. Heart J. 2018, 40, 1378–1383. [Google Scholar] [CrossRef] [PubMed]
- Lee, M.L.T.; Lawrence, J.; Chen, Y.; Whitmore, G.A. Accounting for delayed entry into observational studies and clinical trials: Length-biased sampling and restricted mean survival time. Lifetime Data Anal. 2022, 28, 637–658. [Google Scholar] [CrossRef]
- R Core Team. R: A Language and Environment for Statistical Computing; R Foundation for Statistical Computing: Vienna, Austria, 2021. [Google Scholar]
- Xiao, T.; Whitmore, G.A.; He, X.; Lee, M.L.T. The R Package to Implement Threshold Regression Models. J. Stat. Softw. 2015, 66, 1–16. [Google Scholar] [CrossRef]
- Saegusa, T.; Ma, T.; Li, G.; Chen, Y.Q.; Lee, M.L.T. Variable Selection in Threshold Regression Model with Applications to HIV Drug Adherence Data. Stat. Biosci. J. Int. Chin. Stat. Assoc. 2020, 12, 376–398. [Google Scholar] [CrossRef]
- Robins, J.M.; Hernán, M.Á.; Brumback, B. Marginal Structural Models and Causal Inference in Epidemiology. Epidemiology 2000, 11, 550–560. [Google Scholar] [CrossRef]
- Johnstone, I.M.; Titterington, D.M. Statistical challenges of high-dimensional data. Philos. Trans. R. Soc. Math. Phys. Eng. Sci. 2009, 367, 4237–4253. [Google Scholar] [CrossRef]
- Faraggi, D.; Simon, R. A neural network model for survival data. Stat. Med. 1995, 14, 73–82. [Google Scholar] [CrossRef]
- Pinkus, A. Approximation theory of the MLP model in neural networks. Acta Numer. 1999, 8, 143–195. [Google Scholar] [CrossRef]
- Whitmore, G.A. A regression method for censored inverse-Gaussian data. Can. J. Stat. 1983, 11, 305–315. [Google Scholar] [CrossRef]
- Kingma, D.P.; Ba, J. Adam: A Method for Stochastic Optimization. arXiv 2015, arXiv:1412.6980. [Google Scholar]
- Chollet, F.; Allaire, J. keras: R Interface to ’Keras’. 2023. Available online: https://CRAN.R-project.org/package=keras (accessed on 24 April 2023).
- Chollet, F. Keras. 2015. Available online: https://keras.io (accessed on 26 April 2023).
- Abadi, M.; Agarwal, A.; Barham, P.; Brevdo, E.; Chen, Z.; Citro, C.; Corrado, G.S.; Davis, A.; Dean, J.; Devin, M.; et al. TensorFlow: Large-Scale Machine Learning on Heterogeneous Systems. 2015. Available online: tensorflow.org (accessed on 26 April 2023).
- Breslow, N.E. Discussion on Professor Cox’s Paper. J. R. Stat. Soc. Ser. Methodol. 1972, 34, 202–220. [Google Scholar] [CrossRef]
- Friedman, G.D.; Cutter, G.R.; Donahue, R.P.; Hughes, G.H.; Hulley, S.B.; Jacobs, D.R.; Liu, K.; Savage, P.J. Cardia: Study design, recruitment, and some characteristics of the examined subjects. J. Clin. Epidemiol. 1988, 41, 1105–1116. [Google Scholar] [CrossRef]
- Loria, C.M.; Liu, K.; Lewis, C.E.; Hulley, S.B.; Sidney, S.; Schreiner, P.J.; Williams, O.D.; Bild, D.E.; Detrano, R. Early Adult Risk Factor Levels and Subsequent Coronary Artery Calcification. J. Am. Coll. Cardiol. 2007, 49, 2013–2020. [Google Scholar] [CrossRef]
- Liu, K.; Daviglus, M.L.; Loria, C.M.; Colangelo, L.A.; Spring, B.; Moller, A.C.; Lloyd-Jones, D.M. Healthy Lifestyle Through Young Adulthood and the Presence of Low Cardiovascular Disease Risk Profile in Middle Age. Circulation 2012, 125, 996–1004. [Google Scholar] [CrossRef]
- Hastie, T.; Tibshirani, R.; Friedman, J. The Elements of Statistical Learning: Data Mining, Inference and Prediction, 2nd ed.; Springer: Berlin/Heidelberg, Germany, 2009. [Google Scholar]
- Tchetgen, E.J.T.; Walter, S.; Vansteelandt, S.; Martinussen, T.; Glymour, M. Instrumental Variable Estimation in a Survival Context. Epidemiology 2015, 26, 402–410. [Google Scholar] [CrossRef]
- Mok, T.S.; Wu, Y.L.; Thongprasert, S.; Yang, C.H.; Chu, D.T.; Saijo, N.; Sunpaweravong, P.; Han, B.; Margono, B.; Ichinose, Y.; et al. Gefitinib or carboplatin-paclitaxel in pulmonary adenocarcinoma. N. Engl. J. Med. 2009, 361, 947–957. [Google Scholar] [CrossRef]
- Little, R.J.; Rubin, D.B. Causal Effects in Clinical and Epidemiological Studies Via Potential Outcomes: Concepts and Analytical Approaches. Annu. Rev. Public Health 2000, 21, 121–145. [Google Scholar] [CrossRef] [PubMed]
- Lee, M.L.T.; Whitmore, G.A. Multivariate Threshold Regression Models with Cure Rates: Identification and Estimation in the Presence of the Esscher Property. Stats 2022, 5, 172–189. [Google Scholar] [CrossRef]
- Lee, M.L.T.; Whitmore, G.A.; Rosner, B.A. Threshold regression for survival data with time-varying covariates. Stat. Med. 2010, 29, 896–905. [Google Scholar] [CrossRef] [PubMed]
- Robins, J.M. Estimation of the time-dependent accelerated failure time model in the presence of confounding factors. Biometrika 1992, 79, 321–334. [Google Scholar] [CrossRef]
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).