Abstract
Microbial diversity has been well documented for the top 0–0.30 m of agricultural soils. However, spatio-temporal research into subsoil microbial diversity and the effects of agricultural management remains limited. Soil type may influence subsoil microbial diversity, particularly Vertosols. These soils lack distinct horizons and are known to facilitate the downward movement of organic matter, potentially supporting subsoil microbiota, removed from the crop root system (i.e., bulk soils). Our research used the MiSeq Illumina Platform to investigate microbial diversity down the profile of an agricultural Australian Vertosol to 1.0 m in bulk soils, as influenced by crop system (continuous cotton and cotton–maize) and sample time (pre- and in-crop samples collected over two seasons). Overall, both alpha- (Chao1, Gini–Simpson Diversity and Evenness indices) and beta-diversity (nMDS and Sørensen’s Index of Similarity) metrics indicated that both bacterial (16S) diversity and fungal (ITS) diversity decreased with increasing soil depth. The addition of a maize rotation did not significantly influence alpha-diversity metrics until 0.70–1.0 m depth in the soil, where bacterial diversity was significantly higher in this system, with beta-diversity measures indicating this is likely due to root system differences influencing dissolved organic carbon. Sample time did not significantly affect bacterial or fungal diversity over the two seasons, regardless of the crop type and status (i.e., crop in ground and post crop). The relatively stable subsoil fungal and overall microbial diversity in bulk soils over two crop seasons suggests that these microbiota have developed a tolerance to prolonged agricultural management.
Keywords:
Vertosol; Vertisol; Gossypium hirsutum; maize; community analysis; bacteria; fungi; bulk soil 1. Introduction
Soil microbiota mediate a range of functions within the soil environment, including the decomposition of organic matter (OM) and subsequent release and cycling of essential nutrients—carbon (C), nitrogen (N) and phosphorus (P)—into the terrestrial ecosystem [1,2,3,4,5,6]. These mechanisms are not only vital to a healthy soil, but are key in managing and maintaining sustainable agricultural practice [7,8,9]. As such, an understanding of the influence of disturbance, such as agricultural management and climate change on soil microbiota, is of utmost importance.
In cropping systems, microbiota are subject to disturbance from agricultural practice including minimum tillage, irrigation and crop rotations—all of which impose physical changes in the soil structure and nutrient availability [10]. These disturbances to the soil environment may have ramifications for microbial communities, exhibiting sensitivity (the extent to which a microbial community is altered in response to a disturbance), resistance (the extent to which a microbial community can resist change resulting from a disturbance) or resilience (the time taken for a microbial community to return to its original composition following disturbance) in their diversity and metabolic functionality [4,10,11]. Additionally, plant species are known to influence soil microbiota through variations in the composition of root exudates [12,13,14]. The response of microbial communities, therefore, contributes to the overall stability of the soil system and thus soil health and productivity.
Most research to date has focused on the effects of disturbance on microbial communities in the top 0–0.3 m of agricultural soils, principally in the northern hemisphere. Agricultural top soils have, therefore, been the primary focus for microbial related research due to their proximity to frequent OM (i.e., litter) and water inputs (i.e., rainfall and irrigation), and prolific crop root development [15]. However, some crop roots (i.e., cotton and maize) are known to grow deeper than 0.3 m, with the potential for root exudates to provide C sources, supporting subsoil microbiota in the deeper soil profile [16]. In addition, soils such as the Australian Vertosol (Australian Soil Classification [17]) or Vertisol (World Reference Base [18]) are known to exhibit vertical and lateral movements down the soil profile to at least 1.0 m, due to their high clay content (>35%) and vertic properties which include self-mulching, cracking down the soil profile, slickensides (slightly grooved, polished ped surfaces) and gilgai (continuous patterns of small mounds and depressions), resulting in soil horizons lacking distinction [17,18]. The dynamic nature of Vertosols facilitates the movement of soil organic matter (SOM) down the soil profile, with soils in close proximity to cracks and biopores having increased soil C, indicative of preferential flow pathways for nutrients and plant roots, with the potential to support subsoil microbiota [19]. The nature of Vertosol subsoils warrants further investigation into the stability of microbial communities. However, these soils are frequently used for agricultural purposes, adding further complexity in the study of these soils.
The majority of Australian cotton is grown in Vertosols, where minimum tillage and rotational crops are utilised to improve soil structure, promote and protect soil organic carbon (SOC), introduce and optimise nutrients in the system and suppress disease [20,21,22]. In soils that are considered low in OM and natural nutrient sources (compared to northern hemisphere soils), there is an increasing need to find innovative ways to maintain and enhance these depleted soil resources to meet the demands of a growing global population [23,24]. Therefore, research is beginning to move beyond the surface and into bulk and subsoils.
Our research was part of a larger project investigating the impact of agricultural management of cotton crop rotations on subsoil properties in Vertosols [25,26]. These experiments were undertaken on a long-term field site that has been the basis for studies into the impact of field management, crop rotation and rooting depth on soil physical and chemical properties for over 30 years [27,28,29]. Our study utilised high-throughput sequencing techniques to target prokaryotes (bacteria) and eukaryotes (fungi) to determine microbial diversity down to 1.0 m in the bulk soil profile of Vertosols growing continuous cotton (Gossypium hirsutum) and the influence of crop rotations on subsoil diversity, using a cotton–maize (Zea mays) system. We hypothesised that (1) microbial diversity would decrease with increasing depth in the soil profile [30,31,32,33,34], with (2) the addition of maize into the system increasing microbial diversity down the entire soil profile [35,36,37,38,39], in comparison to a cotton monoculture. We further hypothesised that (3) microbial diversity would converge (increase in similarity) when both systems were subject to the influences (i.e., root exudation composition) of a cotton rotation.
2. Materials and Methods
Crops were grown at the Australian Cotton Research Institute (ACRI) (Latitude 30°11′39.45″ S, Longitude 149°36′16.2″ E), located 26 km north west from the township of Narrabri, New South Wales (NSW). This area is subject to semi-arid climate, with mild winters (10 to 19 °C) and hot summers (20 to 32 °C), with a mean rainfall of 662 mm [40]. Crops were grown in a previously characterised, alkaline (pH 7.2 in 0.01M CaCl2 at 0–0.5 m, pH 7.4 from 0.5–1 m) black self-mulching Vertosol [15,17,41,42], with the mean particle size distribution to the depth of 1 m being 64% clay, 11% silt and 25% sand [43]. Both dissolved organic carbon (DOC) and soil organic carbon (SOC) decrease with depth from 76 mg/kg (DOC)/1.10% (SOC) at 0–0.15 m to 48 mg/kg (DOC)/0.7% (SOC) at 0.3–0.5 m and 32 mg/kg (DOC)/0.58% (SOC) at 0.7–1 m depth. Other soil characteristics (i.e., P, K, and Ca) for this site have been published and can be found in Polain, Guppy, Knox, Lisle, Wilson, Osanai and Siebers [26], Nachimuthu, Watkins, Hulugalle, Weaver, Finlay and McCorkell [29] and Hulugalle, Nachimuthu, Kirkby, Lonergan, Heimoana, Watkins and Finlay [27]. The field site is part of a long-term (approximately 30 years) study, where historical tillage and rotations constituted the main plot treatments, with maize being added as a rotation in 2011 as subplots. This site is managed by the NSW Department of Primary Industries (NSW DPI) and undergoes minimum tillage in the top 0.1 m of the soil profile, post picking, as part of an insect management regime to eliminate Helicoverpa spp. (Cotton bollworm or Native budworm) larvae in a process known as ‘pupae busting’ [44]. Crops are irrigated at a rate of 1 ML ha−1 (equivalent to 100 mm rain) at 7–20 day intervals with approximately 4–8 irrigations per season depending on the amount of seasonal rainfall. Both systems received 260 kg N ha−1 per season, with no P fertiliser applied due to critical Colwell P levels being <6 mg kg−1 for cotton [45]. Samples were collected at the midpoint between the head ditch and tail drainage ends (the site map can be viewed in Osanai, Knox, Nachimuthu and Wilson [25]) in continuous cotton (CC) and cotton–maize (CM) plots within the same field [27,29] over two seasons and at different crop status, these being post pupae busting and prior to planting in October 2015 and 2016 (pre-crop) and at flowering status (in-crop) January 2016 (CC—cotton; CM—maize) and 2017 (CC—cotton; CM—cotton). During season 1 (2015/2016), the minimum and maximum soil temperatures were 6.7 and 36.5 °C, respectively, with a total rainfall of 537 mm and irrigation volumes of 720 mm (CC) and 564 mm (CM) [28,46]. During season 2 (2016/2017), the minimum and maximum soil temperatures were 8.2 and 40.2 °C, respectively, with a total rainfall of 318 mm and irrigation volumes of 1008 mm for both systems [28,46].
Sampling was established as a split-plot design, based on the main plot treatments, where there were four replicate plots each for CC and CM, with one core extracted per replicate plot at each time of sampling in the plant line between cotton plants (in-crop), or from the reformed planting hill (pre-crop), which resulted in a total of 32 cores extracted over the two seasons. These soil cores were extracted using a powered soil corer, with a 42 mm inner diameter, without the use of lubricants [47] to the depth of 1 m and placed into polyvinyl chloride (PVC) half-pipes, wrapped with plastic wrap to store and support the core. Cores were transported to the laboratory in cooled, insulated containers, where subsamples were removed in triplicate from three depths (0–0.15, 0.3–0.5 and 0.7–1 m), avoiding any root material and stored at −20 °C until required.
DNA was extracted from each subsample using PowerSoil® DNA Isolation Kits (MO BIO Laboratories Inc. Carlsbad, CA, USA) according to the manufacturer’s instructions. Briefly, 0.25 g of soil was weighed and subjected to a series of steps to promote cell lysis, removal of inhibitors (i.e., non-DNA material), and washing of DNA, before being eluted in 65 µL elution buffer. The triplicate samples were pooled, with DNA quantity (1 ng/µL to 50 ng/µL required) and purity (A260/A280 ratio range 1.6 to 1.9) assessed using a NanoDrop™ 8000 Spectrophotometer (ThermoFisher Scientific, Waltham, MA, USA).
Three technical replicates of the pooled soil DNA soils were sent to the Australian Genome Research Facility (AGRF) in Brisbane (Queensland) for Diversity Profiling using the MiSeq Illumina Platform. This service identifies the relative proportion of organisms within mixed microbial communities [48]. AGRF-validated primers 341F and 806R were used to target 16S variable regions V3–V4 for bacteria and archaea, whilst primers 1F and 2R were used to target ITS regions for fungi (Table 1).

Table 1.
Forward and reverse primers used to target 16S and ITS regions [48].
Bioinformatics methods were completed by the AGRF as part of quality control [48]. Briefly, these methods involved the initial assembly of the paired ends of the forward and reverse reads using PEAR (version 0.9.5), followed by primer trimming [49]. The trimmed sequences were processed using Quantitative Insights into Microbial Ecology (QIIME 1.8) [50], USEARCH (version 8.0.1623) [51,52] and UPARSE [53] software. Sequences were quality filtered, with duplicate sequences, singletons and unique sequences removed and data sorted by abundance [48]. Sequences were clustered and chimera filtered using “rdp_gold” for 16S [52,54,55] and “Unite” for ITS [56] databases as references. The number of reads in each OTU were mapped back to OTUs with a minimum identity of 97%. QIIME was used to assign taxonomy using the Greengenes databases for 16S sequences [54] and the Unite database for ITS sequences [56]. Prior to the construction of mathematics functions (indices) to determine soil microbial diversity, the raw and quality control reads were statistically compared using IBM SPSS Statistics 25 program for statistical interrogation. Repeated-measures Analysis of Variance (ANOVA) was used to assess significant differences (p ≤ 0.05) between the experimental factors of time (season and crop status), depth, system, time by depth, time by system and depth by system.
Alpha-diversity (α-diversity), the diversity within a sample (i.e., the diversity within a sample obtained from a particular depth), was assessed using the Chao1 (SChao1) estimator for species richness [57,58], Gini–Simpson Diversity (DGS) and Gini–Simpson Evenness (EGS) indices (Table 2) [59,60,61]. Chao1 (Equation (1)) is a non-parametric method to estimate species richness, with the premise that rarer species infer more information about the number of species missing from a population [57,58].
where S is the number of species, F1 the number of singletons and F2 the number of doubletons in an assemblage [62]. The Gini–Simpson Diversity Index (Equation (2)) takes into consideration species richness and abundance, measuring the probability that two randomly selected individuals belong to two different species [58,60].
where S is the total number of species in a community and pi is the proportion of individuals of a particular species (OTUi), divided by the total number of species found (S) [60,63]. Ranging from 0 (low diversity) to 1 (high diversity), the DGS index favours abundant species, highlighting the dominant species in a microbial community [60]. The Gini–Simpson Evenness Index (Equation (3)) assesses the abundance (evenness) of the dominant species within an assemblage, with an increased evenness value indicating that the assemblage is more diverse [64].
where DGS is the Gini–Simpson Diversity value determined (Equation (2)) and S is the number of species observed [59]. The EGS value is considered meaningful when evaluating the microbial community’s ability to resist or recover from disturbances, where a value closer to 0 indicates a lower species abundance and, therefore, lower level of diversity [64].
These aforementioned indices were calculated and constructed in Microsoft Excel, then imported into IBM SPSS Statistics 25 program for statistical interrogation. Repeated-measures Analysis of Variance (ANOVA) was used to assess significant differences (p ≤ 0.05) between the experimental factors. Where data were not normally distributed, log 10 (Log10) or square root (√) transformations were performed prior to ANOVA.
Beta-diversity (β-diversity), the diversity observed between two samples (i.e., the diversity between samples recovered from CC and CM at 0–0.15 m depth) was determined using PRIMER-e v7 (Quest Research Limited, Albany, New Zealand) software, where system by depth absolute abundance OTU data at the phyla level were used for non-metric multidimensional scaling (nMDS) based on Bray–Curtis dissimilarity [65]. Permutational Multivariate Analysis of Variance (PERMANOVA) was used in the analysis of the Bray–Curtis distance data for depth, depth by system and time. Assessed and available soil environmental data (Table S1) were obtained and used with the β-diversity data to perform distance-based linear modelling (DISTLM) sequential tests, which were used to plot distance-based redundancy analysis (dbRDA), in an effort to identify some of the main drivers in our system (Figures S1 and S2). The results of these data interrogations have been used to support theories for system differences or similarities where relevant.
Finally, Sørensen’s Index of Similarity (IS) was used to observe microbial community similarities at the Phylum level [66,67]. Where
With C being the number of species two samples have in common, S1 is the total number of species in sample 1 and S2 is the total number of species in sample 2 [66].
3. Results
3.1. Quality Control and Microbial OTUs
The number of OTU reads significantly differed (p < 0.001) between the raw and quality control data, with a difference of 14,286 ± 358 for 16S and 39,879 ± 1302 for ITS data. Statistical analysis of the data set type (raw versus quality control) against depth (16S p = 0.965; ITS p = 0.320), crop system (16S p = 0.845; ITS p = 0.664) and sampling point (16S p = 0.942; ITS p = 743), revealed no significant differences. Statistical analysis of the data sets + depth + crop system (16S p = 1.000; ITS p = 0.742) and data sets + depth + sample point (16S p = 0.999; ITS p = 0.829) also revealed no significant differences.
The normalized sequence reads average bacterial (16S) OTUs were significantly greater (p < 0.001) than fungal (ITS) OTUs at 2614 ± 97 and 229 ± 15, respectively. The average number of OTUs decreased with increasing depth. However, bacterial OTU numbers were significantly greater (p = 0.001) with each depth (0–0.15 = 3203 ± 140; 0.30–0.50 = 2617 ± 101; 0.70–1.0 = 2023 ± 97) compared to the average number of fungal OTUs (0–0.15 = 358 ± 23; 0.30–0.50 = 198 ± 15; 0.70–1.0 = 131 ± 13). Both bacterial OTUs and fungal OTUs significantly differed (p = 0.004 and 0.020, respectively) with sampling time, with the average number of bacterial OTUs increasing from 2343 ± 187 (season 1 pre-crop) to 2974 ± 137 (season 2 in-crop), whilst fungal OTUs were significantly higher season 1 pre-crop (267 ± 34) and season 2 in-crop (252 ± 26), compared with season 1 in-crop (199 ± 33) and season 2 pre-crop (198 ± 25).
3.2. Alpha-Diversity
3.2.1. Bacteria (16S)
The average species richness (SChao1) estimate for bacteria (16S) was significantly influenced (p < 0.001) by depth with increased SChao1 values at 0.30–0.50 m (4101 ± 186) and 0.70–1.0 m (3380 ± 148), compared with SChao1 values at 0–0.15 m (2888 ± 191) values (Table 2). Average bacterial species richness estimates were not significantly affected by the remaining factors of crop system (p = 0.638) and sample time (p = 0.093). No significant difference was observed for the average species richness for bacteria at depth as influenced by crop system (p = 0.727) or for species richness by depth as influenced by crop system and sample time (p = 0.114) (Table 2). However, there was a significant difference (p = 0.01) in average bacterial species richness with depth as influenced by sample time, with season 2 in-crop SChao1 values at 0.30–0.50 m (4633 ± 146) being 2-fold higher than season 1 in-crop SChao1 values at 0–0.15 m (2367 ± 164).

Table 2.
Alpha-diversity of bacterial 16S OTUs down the soil profile as measured by the Chao1 species richness indicator (SChao1), the Gini–Simpson Diversity Index (DGS) and the Gini–Simpson Evenness Index (EGS) as influenced by system, sample time and sample time by depth. Lower case letters indicate significant differences based on least significant values (L.S.D).
Table 2.
Alpha-diversity of bacterial 16S OTUs down the soil profile as measured by the Chao1 species richness indicator (SChao1), the Gini–Simpson Diversity Index (DGS) and the Gini–Simpson Evenness Index (EGS) as influenced by system, sample time and sample time by depth. Lower case letters indicate significant differences based on least significant values (L.S.D).
| Factors | Chao1 (SChao1) | Gini–Simpson Diversity Index (DGS) | Gini–Simpson Evenness Index (EGS) | |||||||
|---|---|---|---|---|---|---|---|---|---|---|
| 0–0.15 m | 0.30–0.50 m | 0.0–1.0 m | 0–0.15 m | 0.30–0.50 m | 0.0–1.0 m | 0–0.15 m | 0.30–0.50 m | 0.0–1.0 m | ||
| Depth | 2888 a | 4104 b | 3380 c | 0.996 a | 0.993 b | 0.989 c | 4 × 10−4 | 4 × 10−4 | 5 × 10−4 | |
| (p < 0.001) | (L.S.D = 421) | (p < 0.001) | (L.S.D = 0.001) | (p = 0.391) | (L.S.D = 2 × 10−4) | |||||
| System | ||||||||||
| CC | 3014 | 4083 | 3388 | 0.996 a | 0.993 ab | 0.987 c | 3 × 10−4 | 4 × 10−4 | 5 × 10−4 | |
| CM | 2761 | 4125 | 3371 | 0.996 a | 0.994 ab | 0.991 b | 5 × 10−4 | 4 × 10−4 | 5 × 10−4 | |
| (p = 0.727) | (L.S.D = 595) | (p = 0.01) | (L.S.D = 0.002) | (p = 0.244) | (L.S.D = 3 × 10−4) | |||||
| Time | ||||||||||
| S1 Pre | 3630 ab | 3058 ab | 3028 ab | 0.996 | 0.993 | 0.986 | 8 × 10−4 | 4 × 10−4 | 6 × 10−4 | |
| S1 In | 2367 b | 4292 ab | 3168 ab | 0.995 | 0.992 | 0.988 | 3 × 10−4 | 4 × 10−4 | 6 × 10−4 | |
| S2 Pre | 2459 b | 4434 a | 3478 ab | 0.996 | 0.994 | 0.989 | 3 × 10−4 | 4 × 10−4 | 5 × 10−4 | |
| S2 In | 3094 ab | 4633 a | 3845 ab | 0.997 | 0.994 | 0.992 | 3 × 10−4 | 3 × 10−4 | 4 × 10−4 | |
| (p = 0.01) | (L.S.D = 903) | (p = 0.277) | (L.S.D = 0.003) | (p = 0.430) | (L.S.D = 5 × 10−4) | |||||
| Time x System | ||||||||||
| S1 Pre | CC | 4556 | 3108 | 2662 | 0.996 | 0.993 | 0.981 | 3 × 10−4 | 4 × 10−4 | 6 × 10−4 |
| CM | 2704 | 3009 | 3394 | 0.996 | 0.994 | 0.991 | 1 × 10−3 | 4 × 10−4 | 5 × 10−4 | |
| S1 In | CC | 2371 | 4367 | 3211 | 0.995 | 0.992 | 0.987 | 3 × 10−4 | 4 × 10−4 | 6 × 10−4 |
| CM | 2363 | 4217 | 3124 | 0.995 | 0.992 | 0.988 | 3 × 10−4 | 4 × 10−4 | 5 × 10−4 | |
| S2 Pre | CC | 2079 | 4151 | 3699 | 0.995 | 0.994 | 0.987 | 3 × 10−4 | 4 × 10−4 | 6 × 10−4 |
| CM | 2840 | 4718 | 3256 | 0.997 | 0.994 | 0.992 | 3 × 10−4 | 4 × 10−4 | 4 × 10−4 | |
| S2 In | CC | 3052 | 4709 | 3979 | 0.997 | 0.994 | 0.992 | 3 × 10−4 | 3 × 10−4 | 4 × 10−4 |
| CM | 3137 | 4558 | 3711 | 0.996 | 0.995 | 0.992 | 3 × 10−4 | 3 × 10−4 | 4 × 10−4 | |
| (p = 0.144) | (L.S.D = 1277) | (p = 0.247) | (L.S.D = 0.004) | (p =0.349) | (L.S.D = 7 × 10−4) | |||||
Depth significantly (p < 0.001) influenced the diversity of bacterial (16S) communities, with the average bacterial Gini–Simpson Diversity Index (DGS) decreasing from 1.00 ± 2 × 10−4 (0–0.15 m) to 0.99 ± 1 × 10−3 (0.70–1.0 m) (Table 2). Crop system significantly (p = 0.005) influenced DGS, with CM (0.99 ± 5 × 10−4) having increased diversity, in comparison to CC (0.99 ± 9 × 10−4). Crop system also significantly (p = 0.011) influenced bacterial diversity at depth (0.70–1.0 m), with a greater decrease in bacterial diversity in CC (average DGS = 0.99 ± 2 × 10−3) than CM (average DGS = 0.99 ± 9 × 10−4) (Table 2). No significant effect (p = 0.247) was detected for depth, as influenced by sample time and crop system on bacterial diversity, with DGS values averaging 0.99 ± 5 × 10−4. However, bacterial community diversity decreased with depth at each sample time over the two seasons, independent of crop system, except for the notable decreases in diversity in CC at 0.70–1.0 m, pre-crop for both seasons 1 and 2.
Depth did not significantly influence the evenness (EGS) of bacterial communities (p = 0.391) down the soil profile (Table 2). However, the average EGS value for bacterial communities increased between depths 0.30–0.50 m (4 × 10−4 ± 2 × 10−5) and 0.70–1.0 m (5 × 10−4 ± 2 × 10−5). Crop system did not significantly influence the evenness of bacterial (p = 0.388) communities down the soil profile, however, with the average bacterial EGS value for both CC and CM being 10−4 ± 6 × 10−5 (Table 2). The CM system trended in higher evenness of microbial communities at 0–0.15 and 0.30–0.50 m, whilst evenness increased in the CC system at 0.70–1.0 m. Sample time and crop system did not significantly influence the evenness of bacterial (p = 0.349) species down the soil profile (Table 2), with an average EGS of 4 × 10−4 ± 4 × 10−5. Whilst the evenness of bacterial communities was relatively consistent across systems, there was an increase in evenness during season 1 pre-crop at 0–0.15 m under CM, with an average of 1 × 10−3 ± 9 × 10−4, which was >2-fold greater than the remaining average values.
3.2.2. Fungi (ITS)
The average species richness (SChao1) estimate for fungi (ITS) was significantly influenced (p < 0.001) by depth (Table 3), with higher SChao1 values at 0–0.15 m (411 ± 25), compared to 0.30–0.50 m (238 ± 15) and 0.70–1.0 m (177 ± 13). Average species richness values for fungi were not significantly different for depth as influenced by crop system (p = 0.738) (or for species richness by depth as influenced by crop system and sample time (p = 0.057) (Table 3).

Table 3.
Alpha-diversity of fungal ITS OTUs down the soil profile as measured by the Chao1 species richness indicator (SChao1), the Gini–Simpson Diversity Index (DGS) and the Gini–Simpson Evenness Index (EGS) as influenced by system, sample time and sample time by depth. Lower case letters indicate significant differences based on least significant values (L.S.D).
There was no apparent influence of depth on fungal (ITS) community diversity (p = 0.637), with average fungal DGS differing by 0.01 from 0–0.15 to 0.70–1.0 m, with an increase of 0.04 at 0.30–0.50 m (Table 3). Fungal diversity down the soil profile was not significantly (p = 0.938) influenced by crop system. However, the average DGS was 0.03 ± 0.02 lower under the CM system (Table 3). No significant effect (p = 0.486) was detected for depth, as influenced by sample time and system on fungal diversity, with DGS values averaging 0.90 ± 1 × 10−2. Fungal community diversity did not demonstrate a clear change with soil depth over the two growing seasons although there was a notable, but not significant difference between crop system diversity at 0–0.15 m pre-crop, season 1, with a decrease in diversity in the CM system.
Depth did not significantly influence the evenness of fungal communities (p = 0.715) down the soil profile (Table 3), with average EGS increasing from 0–0.15 and 0.30–0.50 m (3 × 10−3 ± 7 × 10−4) were lower in comparison to 0.70–1.0 m (8 × 10−3 ± 7 × 10−4). Crop system did not significantly influence the evenness of fungal (p = 0.556) communities down the soil profile, with the average EGS values differing by less than 0.001 between each system, at each depth (Table 3). The CM system trended in higher evenness of fungal communities at 0–0.15 and 0.70–1.0 m, whilst evenness was increased in the CC system at 0.30–0.50 and 0.70–1.0 m. Sample time and crop system did not significantly influence the evenness fungal (p = 0.744) species down the soil profile (Table 3), with an average EGS of 6 × 10−3 ± 4 × 10−4. Overall, the evenness of fungal communities fluctuated over the two growing seasons, with the EGS index particularly variable at 0.70–1.0 m.
3.3. Beta-Diversity
3.3.1. Dissimilarities between Microbial Communities
PERMANOVA analysis of absolute abundance OTUs for bacterial (16S) found a significant difference between samples collected at different depths (p = 0.001) and samples collected at depth under the different crop rotations (p = 0.001). However, there were no significant differences between the diversity of samples collected from different crop systems (p = 0.139) or sample time (p = 0.087). Non-metric multidimensional scaling (nMDS) was used to visualise the differences in bacterial communities down the soil profile and between the two systems. Clear separation of clusters with depth and to a lesser extent, separation by system, was evident particularly between CC and CM at 0–0.15 m and 0.70–1.0 m (Figure 1).
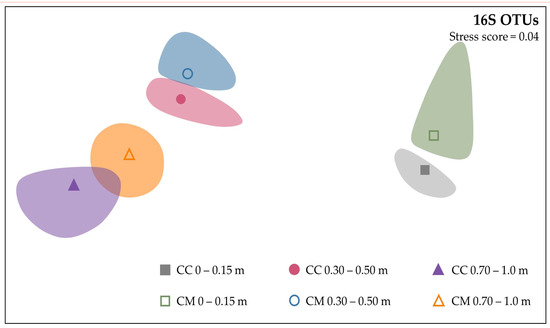
Figure 1.
Dissimilarities between bacterial (16S) communities down the soil profile, as influenced by continuous cotton (CC) and cotton–maize (CM) crop systems using non-metric multidimensional scaling (nMDS) of absolute abundance OTUs based on the square root transformation of Bray–Curtis dissimilarity, with a stress score of 0.04.
PERMANOVA analysis of absolute abundance OTUs for fungal (ITS) found a significant difference between samples collected at different depths (p = 0.001) and samples collected at depth under the different crop rotations (p = 0.003). However, there were no significant differences between the diversity of samples collected from different crop systems (p = 0.451) or sample time (p = 0.446). Non-metric multidimensional scaling (nMDS) was used to visualise the differences in fungal communities down the soil profile and between the two systems. A depth difference was evident in nMDS, with 0–0.15 m communities clustering separately from communities at 0.30–0.50 m and 0.70–1.0 m (Figure 2). A crop system by depth influence was evident at 0.30–0.50 m, with separation between clusters for CC and CM.
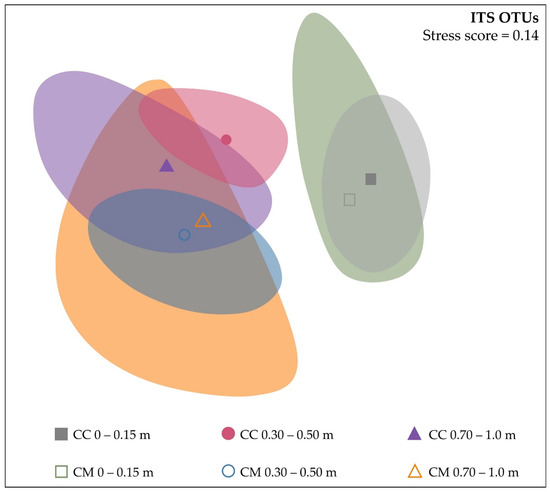
Figure 2.
Dissimilarities between fungal (ITS) communities down the soil profile, as influenced by continuous cotton (CC) and cotton–maize (CM) crop systems, using non-metric multidimensional scaling (nMDS) of absolute abundance OTUs based on the square root transformation of Bray–Curtis dissimilarity, with a stress score of 0.14.
3.3.2. Similarities between Microbial Communities
Similarities between bacteria and fungi community profiles significantly decreased (p < 0.001) between the depths of 0–0.15 cm and 0.70–1.0 m, with Sørensen’s Index of Similarity (IS) values of 0.77 and 0.53 for bacterial and fungal communities, respectively (Figure 3). At each depth, Acidobacteria (14–19%), Actinobacteria (29–49%) and Proteobacteria (14–27%) were the prevalent bacterial phyla (Figure 3a). Both Actinobacteria and Nitrospira increased with each depth increment sampled, being more prevalent in the 0.30–0.50 and 0.70–1.0 m samples than they were in the 0–0.15 m samples. Ascomycota (47–61%) and Basidiomycota (19–35%) were amongst the most prevalent phyla for fungi (Figure 3b). Under the CC system, the fungal groups were relatively constant, with the exception of Zygomycota becoming less abundant at 0.30–0.50 cm, before returning to a higher abundance at 0.70–1.0 m. In contrast, under the CM system, the relative abundance of Agaricomycetes, belonging to the Basidiomycota phylum, increased with depth, whilst Ascomycota, namely the Sordariomycetes, decreased in relative abundance. Whilst crop system did not significantly influence bacteria and fungi profiles with depth, IS values between CC and CM systems decreased by 0.08 (bacteria) and 0.16 (fungi) from 0–0.15 to 0.70–1.0 m (Figure 3). The decrease in similarity with depth appears to be characterised by increases in Actinobacteria at 0.70–1.0 m under the CC system (Figure 3a) and increase in Basidiomycota at 0.70–1.0 m under the CM system (Figure 3b). However, this was not a significant difference.
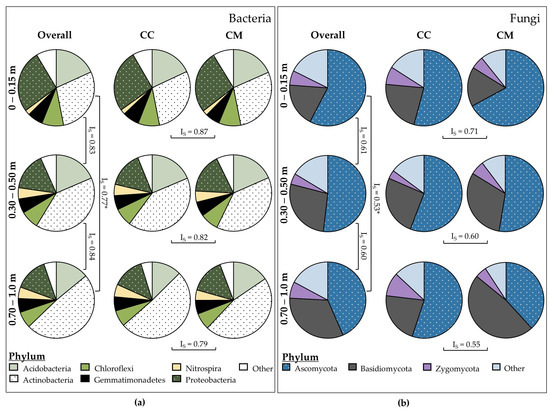
Figure 3.
Similarities between microbial diversity profiles as determined by Sørensen’s Index of Similarity (IS), with an asterisk (*) indicating a significant difference of p < 0.001 when α = 0.05. (a) Bacterial community profiles down the soil profile (overall) and as influenced by continuous cotton (CC) and cotton–maize (CM). (b) Fungal community profiles down the soil profile (overall) and as influenced by continuous cotton (CC) and cotton–maize (CM).
4. Discussion
To our knowledge, this is one of the first studies investigating subsoil and bulk soil microbial diversity using high-throughput sequencing in an agriculturally managed Vertosol, in the southern hemisphere [31,68,69]. Whilst our data have followed trends observed in studies from the northern hemisphere (i.e., decline in diversity with increasing depth), there have been a few surprises [30,70]. The first is that depth appears to have a lower level of influence on fungal (ITS) diversity for both alpha- and beta-diversity [33,34,71]. The second is that the imposition of maize into cotton rotations had a limited effect on bacterial α-diversity (significantly noticeable at 0.70–1.0 m) and no effect on fungal α-diversity. However, significant differences between systems was evident in β-diversity measures for bacteria (0–0.15 and 0.30–0.50 m) and fungi (0.30–0.50 m). Finally, sample time did not significantly influence microbial alpha- or beta-diversity, with the exception of bacterial α-diversity with depth, with increased Chao1 values during season 2 at 0.30–0.50 m.
Depth was a significant factor influencing the diversity of bacteria, both within a sample (α-diversity) and between samples (β-diversity), whilst depth was only a significant factor for fungal β-diversity. Despite a significantly higher species richness (SChao1) at 0.30–0.50 m, bacterial α-diversity (DGS) was overall lower as depth increased, with the low evenness values (EGS) indicating low abundance of the different bacterial species present with increasing depth. A decrease in bacterial diversity with increasing depth is not a surprising result, as availability and accessibility to C, N and P sources becomes increasingly difficult with depth (Table S1) in both natural and managed ecosystems [30,70,71,72,73]. The trend of decreasing bacterial diversity with increasing depth was also apparent in β-diversity analyses, with nMDS showing pronounced separation between each depth assessed. We performed distance-based linear modelling (DISTLM) sequential tests and plotted distance-based redundancy analysis (dbRDA) on a subset of diversity data against environmental factors, and both DOC and gravimetric water content (GWC) were equally proportional (29.9% each) in explaining 73.6% of total variation (Figure S1) in bacterial diversity. It appears that bacterial communities in the top 0–0.15 m of the soil profile are more positively correlated to DOC, whilst subsoil bacterial diversity (i.e., 0.30–0.50 and 0.70–1.0 m) was negatively correlated to DOC, despite subsoils having a higher GWC. Sørensen’s Index of Similarity (IS) indicated that both Proteobacteria and Actinobacteria dominated the top 0–0.15 m of the soil profile, an observation consistent with these copiotrophs preferences for nutrient-rich environments [74,75,76]. Interestingly, the relative abundance of Actinobacteria increased with depth, whilst the relative abundance of the oligotrophic (adaptable to low-nutrient habitats) Acidobacteria [75,76] decreased with depth at 0.7–1.0 m, in an apparent negative correlation. There have been reports that water-extractable organic carbon (i.e., DOC) has decreased availability in soils with increased clay content, and therefore the DOC content in moist soil conditions does not indicate the availability of this C to microbiota [77,78]. It is, therefore, plausible that the decrease in DOC accessibility down the soil profile resulted in overall lower bacterial diversity in subsoils, which was monopolized by the highly competitive Actinobacteria [72,79].
The lack of significant differences in fungal α-diversity with depth (with the exception of SChao1) is surprising, as fungal diversity has been reported as being limited to surface soils, where oxygen and OM are abundant [70,71,80]. Granted, SChao1 indicates a significant decrease in species richness below 0.15 m but, overall, the abundance of these fungal species is low. It is plausible that the high clay content and subsequent cracking in the Vertosol profile facilitates the movement of O2, OM and H2O down the soil profile, providing micropockets of nutrients to be accessed by fungal hyphae networks [19,81,82]. β-diversity analyses using nMDS highlight the effect of depth on fungal diversity, with communities in the top 0–0.15 m separate from the remaining depths, which were clustered together. In contrast to our observation with bacterial diversity, DISTLM sequential analyses indicated that GWC (48.9%) was the highest contributor to the 75.8% total variation in diversity data on the dbRDA plot (Figure S2), when environmental factors were taken into consideration. In this context, GWC was negatively correlated with DOC, which in turn was driving the separation of fungal diversity in the top 0–0.15 m of the soil profile. Sørensen’s Index of Similarity (IS) indicated the dominance of Ascomycota in these top soils, largely comprised of members belonging to Sordariomycetes, which are known to prefer warm, moist and well-oxygenated environments [83]. As depth increased, the prevalence of Basidiomycota, (predominantly members of Agaricomycetes) increased, possibly reflecting differences in responses to water and carbon between these two classes of fungi [83].
Crop system influence on α-diversity down the soil profile was only apparent in subsoil (0.70–1.0 m), with a decrease in bacterial diversity (DGS) under CC prior to planting (pre-crop) in both seasons. However, time was not a significant factor. The decrease in bacterial diversity in the CC system at 0.70–1.0 m depth is mostly likely attributed to the prevalence of maize crop roots in the CM system after crop removal, as observed by Rasse and Smucker [84]. Although not measured as part of these experiments, the presence and distribution of roots have been examined in this rotation and field previously [15,85]. The prevalence of maize roots would create diverse biopore networks, with OM accumulating and being flushed through the soil profile upon irrigation [19,43,86], thus becoming available to the subsoil microorganisms in this system. Another possible contributor to consider is the increased P in the 0.70–1.0 m soils under CM (Table S1). While these soil measurements were taken during season 1, the major P pool in these soils is calcium phosphate, which has a long turnover time and is thus unlikely to have changed [26]. Therefore, increased accessibility of C and increased P could both be co-contributors to increased diversity in CM at 0.70–1.0 m
Crop system influence down the soil profile was apparent in β-diversity measures for both bacteria and fungi. The crop system differences were more pronounced for bacteria at 0–0.15 m, but a clear dispersion between system mean values at 0.30–0.50 m was also observed. In contrast, system differences occurred only at 0.30–0.50 m for fungi. The drivers behind microbial diversity patterns between the two investigated systems remain unclear. The DISTLM analysis and dbRDA plots demonstrated that DOC and GWC were key players in microbial diversity, but there was little separation between systems. However, it seems that fungi diversity at 0.30–0.50 m in CC was more positively correlated with DOC, compared to fungal diversity in CM at the same depth (Figure S2). A further observation that is worth mentioning is the distinct increase in Basidiomycota at 0.70–1.0 m under CM, as indicated by Sørensen’s Index of Similarity. Whilst not a significant increase, it is highly possible that the Basidiomycota have proliferated in this region due to the persistence of lignin containing maize roots in subsoils, acting as a C source for members of this phyla with lignin degrading metabolic pathways [15,84,85,87,88].
Sample time (i.e., prior to planting, mid-crop development, post crop removal, and rotational phase) has been linked to belowground changes in soil microbiota diversity [14,89,90,91,92]. However, this was not evident down the soil profile in the systems we studied, with the exception of bacterial α-diversity at 0.30–0.50 m during season 2. Season 2 was characterised by temperatures falling outside the ideal temperature range (11–36 °C) for cotton growth and drought setting into the region [40]. This resulted in poor crop establishment, which, in turn, would have had a knock on effect for root growth and soil microbiota, thus disrupting the structure of microbial communities [14,89]. The increased SChao1 values at 0.30–0.50 m could be reflective of previously suppressed bacterial species proliferating in favourable conditions, which would contribute to the increased DGS values, although these were not significant increases [70,93]. Whilst we cannot definitively state that microbial communities are resistant to the influence of agricultural disturbance, without post crop diversity comparisons, there does appear to be a level of resilience in the microbial communities studied [10,94,95].
This study represents an initial exploration into Australian subsoil microbial diversity in cotton systems, establishing a baseline for understanding these vital biota and their resilience in highly dynamic soils. This work provides a framework against which to explore the functional diversity of subsoil microbiota in future research, which may assist in defining how the addition of maize has influenced changes in microbial diversity, which remains unclear [39,96,97]. Our research investigating both long- and short-term microbial activity in these soils suggest that the differences between systems is likely a result of how microbiota respond and allocate C and other nutrients for metabolic processes [26,71,98].
5. Conclusions
The majority of agricultural soil microbial diversity studies have focused on the top 0–0.30 m, with exploration into subsoil (>0.3 m) limited and microbial diversity in rotational cotton systems cultivated in Vertosols non-existent. Vertosols are highly dynamic, with both vertical and lateral soil movements facilitating the downward movement of organic matter, oxygen and water, with the potential to support microbial life beyond the top 0.3 m. Despite fungal diversity patterns deviating from published research, our first hypothesis, relating to decreasing microbial diversity with increasing depth, was upheld. Our second hypothesis that microbial diversity would increase down the soil profile under the cotton–maize system was partially refuted in relation to α-diversity, with significant differences only occurring at 0.70–1.0 m for bacteria. Significant differences in β-diversity between systems was evident. However, the exact mechanisms behind diversity differences were masked by the strong depth effect. Our final hypothesis, that microbial diversity would increase in similarity when both systems were under the same crop rotation (cotton), was neither refuted nor supported, as there were no significant differences between the systems at any given time point, indicating a level of resilience in microbial communities to agricultural management.
The dynamic nature of these soils is likely to be responsible for the general lack of difference observed in the α-diversity of microbial communities between the continuous cotton and cotton–maize rotations. Differences with depth, within the bacterial and fungal β-diversity, appear to be attributable to changes in DOC and available water, both resulting from changes in plant rooting patterns within this dynamic soil. The distinct lack of diversity between sample times as influenced by season and crop status is reflective of the resilience in the microbial communities studied. The data presented in this paper are, to our knowledge, the first investigation into Vertosol subsoils in agricultural crops, using high-throughput sequencing. As such, we present a baseline for future investigations into subsoil diversity of Australian agricultural soils and indeed Vertosols.
Supplementary Materials
The following are available online at https://www.mdpi.com/2571-8789/4/3/44/s1, Figure S1: Distance-based redundancy analysis (dbRDA) plot of 16S absolute abundance OTU data against environmental factors (Table S1) where, based on a distance-based linear model (DISTLM) sequential tests [1,2], Figure S2: Distance-based redundancy analysis (dbRDA) plot of ITS absolute abundance OTU data against environmental factors (Table S1), based on a distance-based linear model (DISTLM) sequential tests [1,2], Table S1: Compilation of available soil characteristics and environmental parameters over the two growing seasons. Underlined superscript numbers correspond to footnote numbers.
Author Contributions
Conceptualization, K.P., O.K. and L.P.; methodology, K.P., O.K. and L.P.; formal analysis, K.P. and O.K.; investigation, K.P.; data curation, K.P.; writing—original draft preparation, K.P.; writing—review and editing, O.K. and B.W.; visualisation, K.P.; supervision, O.K., L.P. and B.W.; funding acquisition, O.K. and B.W. All authors have read and agreed to the published version of the manuscript.
Funding
This research was funded by Cotton Research and Development Corporation (CRDC), grant number UNE1601.
Acknowledgments
Gunasekhar Nachimuthu from the New South Wales Department of Primary Industries, for managing the study and assisting with sample collection; Yui Osanai for advice on data analyses; Australian Genome Research Facility (AGRF) for use of the service and facilities.
Conflicts of Interest
The authors declare no conflict of interest.
References
- Gupta, V.V. Beneficial microorganisms for sustainable agriculture. Microbiol. Aust. 2012, 33, 113–115. [Google Scholar] [CrossRef]
- Gupta, V.; Neate, S.; Leonard, E. Life in the Soil-The Relationship between Agriculture and Soil Organisms; Cooperative Research Centre for Soil & Land Management: Glen Osmond, Adelaide, Australia, 2007. [Google Scholar]
- Bender, S.F.; Wagg, C.; van der Heijden, M.G. An underground revolution: Biodiversity and soil ecological engineering for agricultural sustainability. Trends Ecol. Evol. 2016, 31, 440–452. [Google Scholar] [CrossRef] [PubMed]
- Lehman, R.M.; Acosta-Martinez, V.; Buyer, J.S.; Cambardella, C.A.; Collins, H.P.; Ducey, T.F.; Halvorson, J.J.; Jin, V.L.; Johnson, J.M.; Kremer, R.J. Soil biology for resilient, healthy soil. J. Soil Water Conserv. 2015, 70, 12A–18A. [Google Scholar] [CrossRef]
- Torsvik, V.; Øvreås, L. Microbial diversity and function in soil: From genes to ecosystems. Curr. Opin. Microbiol. 2002, 5, 240–245. [Google Scholar] [CrossRef]
- Tecon, R.; Or, D. Biophysical processes supporting the diversity of microbial life in soil. FEMS Microbiol. Rev. 2017, 41, 599–623. [Google Scholar] [CrossRef] [PubMed]
- Koch, A.; McBratney, A.; Adams, M.; Field, D.; Hill, R.; Crawford, J.; Minasny, B.; Lal, R.; Abbott, L.; O’Donnell, A. Soil security: Solving the global soil crisis. Glob. Policy 2013, 4, 434–441. [Google Scholar] [CrossRef]
- Gupta, V. Intensive Cropping Starts with the Soil. Available online: https://grdc.com.au/resources-and-publications/groundcover/ground-cover-issue-48/intensive-cropping-starts-with-the-soil (accessed on 16 November 2016).
- Lal, R. Restoring soil quality to mitigate soil degradation. Sustainability 2015, 7, 5875–5895. [Google Scholar] [CrossRef]
- Shade, A.; Peter, H.; Allison, S.D.; Baho, D.; Berga, M.; Bürgmann, H.; Huber, D.H.; Langenheder, S.; Lennon, J.T.; Martiny, J.B. Fundamentals of microbial community resistance and resilience. Front. Microbiol. 2012, 3, 417. [Google Scholar] [CrossRef]
- Allison, S.D.; Martiny, J.B. Resistance, resilience, and redundancy in microbial communities. Proc. Natl. Acad. Sci. USA 2008, 105, 11512–11519. [Google Scholar] [CrossRef]
- Berg, G.; Smalla, K. Plant species and soil type cooperatively shape the structure and function of microbial communities in the rhizosphere. FEMS Microbiol. Ecol. 2009, 68, 1–13. [Google Scholar] [CrossRef]
- Doornbos, R.F.; van Loon, L.C.; Bakker, P.A. Impact of root exudates and plant defense signaling on bacterial communities in the rhizosphere. A review. Agron. Sustain. Dev. 2012, 32, 227–243. [Google Scholar] [CrossRef]
- Chaparro, J.M.; Badri, D.V.; Vivanco, J.M. Rhizosphere microbiome assemblage is affected by plant development. ISME J. 2014, 8, 790–803. [Google Scholar] [CrossRef] [PubMed]
- Hulugalle, N.R.; Broughton, K.J.; Tan, D.K.Y. Root growth of irrigated summer crops in cotton-based farming systems sown in Vertosols of northern New South Wales. Crop Pasture Sci. 2015, 66, 158–167. [Google Scholar] [CrossRef]
- Gunina, A.; Kuzyakov, Y. Sugars in soil and sweets for microorganisms: Review of origin, content, composition and fate. Soil Biol. Biochem. 2015, 90, 87–100. [Google Scholar] [CrossRef]
- Isbell, R. The Australian Soil Classification; CSIRO publishing: Collingwood, Australia; Clayton, Australia, 2016. [Google Scholar]
- IUSS Working Group WRB. World Reference Base for Soil Resources 2014, Update 2015: International Soil Classification System for Naming Soils and Creating Legends for Soil Maps, 106; FAO Rome: Roma, Italy, 2015; p. 192. [Google Scholar]
- Hullugalle, N.; Weaver, T.; Finlay, L.; Entwistle, P. Physical and chemical properties of soil near cracks in irrigated vertisols sown with cotton-wheat rotations. Arid Land Res. Manag. 2001, 15, 13–22. [Google Scholar] [CrossRef]
- Cattle, S.R.; Field, D.J. A review of the soil science research legacy of the triumvirate of cotton CRC. Crop Pasture Sci. 2014, 64, 1076–1094. [Google Scholar] [CrossRef]
- Dias, T.; Dukes, A.; Antunes, P.M. Accounting for soil biotic effects on soil health and crop productivity in the design of crop rotations. J. Sci. Food Agric. 2015, 95, 447–454. [Google Scholar] [CrossRef]
- Dalal, R.C.; Allen, D.E.; Chan, K.Y.; Singh, B.P. Soil organic matter, soil health and climate change. In Soil Health and Climate Change; Springer: Berlin, Germany, 2011; pp. 87–106. [Google Scholar]
- Lal, R. Soil health and carbon management. Food Energy Secur. 2016, 5, 212–222. [Google Scholar] [CrossRef]
- Koch, A.; Chappell, A.; Eyres, M.; Scott, E. Monitor soil degradation or triage for soil security? An Australian challenge. Sustainability 2015, 7, 4870–4892. [Google Scholar] [CrossRef]
- Osanai, Y.; Knox, O.; Nachimuthu, G.; Wilson, B. Increasing soil organic carbon with maize in cotton-based cropping systems: Mechanisms and potential. Agric. Ecosyst. Environ. 2020, 299, 106985. [Google Scholar] [CrossRef]
- Polain, K.; Guppy, C.; Knox, O.; Lisle, L.; Wilson, B.; Osanai, Y.; Siebers, N. Determination of Agricultural Impact on Soil Microbial Activity Using δ18OP HCl and Respiration Experiments. ACS Earth Space Chem. 2018, 2, 683–691. [Google Scholar] [CrossRef]
- Hulugalle, N.; Nachimuthu, G.; Kirkby, K.; Lonergan, P.; Heimoana, V.; Watkins, M.; Finlay, L. Sowing maize as a rotation crop in irrigated cotton cropping systems in a Vertosol: Effects on soil properties, greenhouse gas emissions, black root rot incidence, cotton lint yield and fibre quality. Soil Res. 2020, 58, 137–150. [Google Scholar] [CrossRef]
- Nachimuthu, G.; Hulugalle, N.R.; Watkins, M.D.; Finlay, L.A.; McCorkell, B. Irrigation induced surface carbon flow in a Vertisol under furrow irrigated cotton cropping systems. Soil Tillage Res. 2018, 183, 8–18. [Google Scholar] [CrossRef]
- Nachimuthu, G.; Watkins, M.D.; Hulugalle, N.R.; Weaver, T.B.; Finlay, L.A.; McCorkell, B.E. Leaching of dissolved organic carbon and nitrogen under cotton farming systems in a Vertisol. Soil Use Manag. 2019, 35, 443–452. [Google Scholar] [CrossRef]
- Eilers, K.G.; Debenport, S.; Anderson, S.; Fierer, N. Digging deeper to find unique microbial communities: The strong effect of depth on the structure of bacterial and archaeal communities in soil. Soil Biol. Biochem. 2012, 50, 58–65. [Google Scholar] [CrossRef]
- Hsiao, C.-J.; Sassenrath, G.F.; Zeglin, L.H.; Hettiarachchi, G.M.; Rice, C.W. Vertical changes of soil microbial properties in claypan soils. Soil Biol. Biochem. 2018, 121, 154–164. [Google Scholar] [CrossRef]
- Kramer, S.; Marhan, S.; Haslwimmer, H.; Ruess, L.; Kandeler, E. Temporal variation in surface and subsoil abundance and function of the soil microbial community in an arable soil. Soil Biol. Biochem. 2013, 61, 76–85. [Google Scholar] [CrossRef]
- Moll, J.; Hoppe, B.; König, S.; Wubet, T.; Buscot, F.; Krüger, D. Spatial distribution of fungal communities in an arable soil. PLoS ONE 2016, 11, e0148130. [Google Scholar] [CrossRef]
- Schlatter, D.C.; Kahl, K.; Carlson, B.; Huggins, D.R.; Paulitz, T. Fungal community composition and diversity vary with soil depth and landscape position in a no-till wheat-based cropping system. FEMS Microbiol. Ecol. 2018, 94, fiy098. [Google Scholar] [CrossRef]
- Ashworth, A.; DeBruyn, J.; Allen, F.; Radosevich, M.; Owens, P. Microbial community structure is affected by cropping sequences and poultry litter under long-term no-tillage. Soil Biol. Biochem. 2017, 114, 210–219. [Google Scholar] [CrossRef]
- Dorr de Quadros, P.; Zhalnina, K.; Davis-Richardson, A.; Fagen, J.R.; Drew, J.; Bayer, C.; Camargo, F.A.; Triplett, E.W. The effect of tillage system and crop rotation on soil microbial diversity and composition in a subtropical acrisol. Diversity 2012, 4, 375–395. [Google Scholar] [CrossRef]
- Mbuthia, L.W.; Acosta-Martinez, V.; DeBryun, J.; Schaeffer, S.; Tyler, D.; Odoi, E.; Mpheshea, M.; Walker, F.; Eash, N. Long term tillage, cover crop, and fertilization effects on microbial community structure, activity: Implications for soil quality. Soil Biol. Biochem. 2015, 89, 24–34. [Google Scholar] [CrossRef]
- Silvestro, L.B.; Biganzoli, F.; Stenglein, S.A.; Forjan, H.; Manso, L.; Moreno, M.V. Mixed cropping regimes promote the soil fungal community under zero tillage. Antonie van Leeuwenhoek 2018, 111, 1055–1064. [Google Scholar] [CrossRef]
- Tiemann, L.; Grandy, A.; Atkinson, E.; Marin-Spiotta, E.; McDaniel, M. Crop rotational diversity enhances belowground communities and functions in an agroecosystem. Ecol. Lett. 2015, 18, 761–771. [Google Scholar] [CrossRef] [PubMed]
- Meterology, B.O. Climate Statistics for Australian Locations, 14 March 2019 ed.; Australian Government: Canberra, Australia, 2019.
- Rochester, I.J. Nutrient uptake and export from an Australian cotton field. Nutr. Cycl. Agroecosyst. 2007, 77, 213–223. [Google Scholar] [CrossRef]
- Staff, S.S. Keys to Soil Taxonomy; Department of Agriculture, Natural Resources Conservation Service: Washington, DC, USA, 2003.
- Hulugalle, N.; Weaver, T.; Finlay, L.; Luelf, N.; Tan, D. Potential contribution by cotton roots to soil carbon stocks in irrigated Vertosols. Soil Res. 2009, 47, 243–252. [Google Scholar] [CrossRef]
- Ceeney, S.; Williams, S.; Maas, S. Integrated Pest Management & Resistance Management; Cotton Research and Development Corporation: Queensland, Australia, 2016; pp. 54–62. [Google Scholar]
- Dorahy, C.G.; Rochester, I.J.; Blair, G.J. Response of field-grown cotton (Gossypium hirsutum L.) to phosphorus fertilisation on alkaline soils in eastern Australia. Soil Res. 2004, 42, 913–920. [Google Scholar] [CrossRef]
- CSIRO. CottASSIST. Available online: https://www.cottassist.com.au/ (accessed on 9 July 2019).
- Polain, K.; Joice, G.; Jones, D.; Pereg, L.; Nachimuthu, G.; Knox, O.G. Coring lubricants can increase soil microbial activity in Vertisols. J. Microbiol. Methods 2019. [Google Scholar] [CrossRef]
- AGRF. Australian Genomic Research Facility: Next Generation Sequencing Resources. Available online: http://www.agrf.org.au/resources/applications/-next-gen-sequencing#Diversity (accessed on 28 February 2017).
- Zhang, J.; Kobert, K.; Flouri, T.; Stamatakis, A. PEAR: A fast and accurate Illumina Paired-End reAd mergeR. Bioinformatics 2014, 30, 614–620. [Google Scholar] [CrossRef]
- Caporaso, J.G.; Kuczynski, J.; Stombaugh, J.; Bittinger, K.; Bushman, F.D.; Costello, E.K.; Fierer, N.; Pena, A.G.; Goodrich, J.K.; Gordon, J.I. QIIME allows analysis of high-throughput community sequencing data. Nat. Methods 2010, 7, 335–336. [Google Scholar] [CrossRef]
- Edgar, R.C. Search and clustering orders of magnitude faster than BLAST. Bioinformatics 2010, 26, 2460–2461. [Google Scholar] [CrossRef] [PubMed]
- Edgar, R.C.; Haas, B.J.; Clemente, J.C.; Quince, C.; Knight, R. UCHIME improves sensitivity and speed of chimera detection. Bioinformatics 2011, 27, 2194–2200. [Google Scholar] [CrossRef] [PubMed]
- Edgar, R.C. UPARSE: Highly accurate OTU sequences from microbial amplicon reads. Nat. Methods 2013, 10, 996–998. [Google Scholar] [CrossRef] [PubMed]
- DeSantis, T.Z.; Hugenholtz, P.; Larsen, N.; Rojas, M.; Brodie, E.L.; Keller, K.; Huber, T.; Dalevi, D.; Hu, P.; Andersen, G.L. Greengenes, a chimera-checked 16S rRNA gene database and workbench compatible with ARB. Appl. Environ. Microbiol. 2006, 72, 5069–5072. [Google Scholar] [CrossRef]
- Cole, J.R.; Wang, Q.; Fish, J.A.; Chai, B.; McGarrell, D.M.; Sun, Y.; Brown, C.T.; Porras-Alfaro, A.; Kuske, C.R.; Tiedje, J.M. Ribosomal Database Project: Data and tools for high throughput rRNA analysis. Nucleic Acids Res. 2014, 42, D633–D642. [Google Scholar] [CrossRef]
- Kõljalg, U.; Larsson, K.H.; Abarenkov, K.; Nilsson, R.H.; Alexander, I.J.; Eberhardt, U.; Erland, S.; Høiland, K.; Kjøller, R.; Larsson, E. UNITE: A database providing web-based methods for the molecular identification of ectomycorrhizal fungi. New Phytol. 2005, 166, 1063–1068. [Google Scholar] [CrossRef]
- Chao, A.; Chiu, C.H. Species richness: Estimation and comparison. Wiley StatsRef Stat. Ref. Online 2014. [Google Scholar] [CrossRef]
- Gotelli, N.J.; Chai, A. Measuring and Estimating Species Richness, Species Diversity, and Biotic Similarity from Sampling Data. In Encyclopedia of Biodiversity, 2nd ed.; Levin, S.A., Ed.; Academic Press: Waltham, MA, USA, 2013; Volume 5, pp. 195–211. [Google Scholar]
- Maurer, B.A.; McGill, B.J. Measurement of species diversity. In Biological Diversity: Frontiers in Measurement and Assessment; Maurer, B.A., Ed.; Oxfold University Press: Oxfold, UK, 2011; pp. 55–56. [Google Scholar]
- Kim, B.-R.; Shin, J.; Guevarra, R.B.; Lee, J.H.; Kim, D.W.; Seol, K.-H.; Lee, J.-H.; Kim, H.B.; Isaacson, R.E. Deciphering diversity indices for better understanding of the microbial communities. J. Microbiol. Biotechnol. 2017, 27, 2089–2093. [Google Scholar] [CrossRef]
- Tuomisto, H. An updated consumer’s guide to evenness and related indices. Oikos 2012, 121, 1203–1218. [Google Scholar] [CrossRef]
- Chao, A. Nonparametric estimation of the number of classes in a population. Scand. J. Stat. 1984, 11, 265–270. [Google Scholar]
- Simpson, E.H. Measurement of diversity. Nature 1949, 163, 688. [Google Scholar] [CrossRef]
- Magurran, A.E. Biological diversity. Curr. Biol. 2005, 15, R116–R118. [Google Scholar] [CrossRef] [PubMed]
- Anderson, M.J.; Gorley, R.N.; Clarke, K.R. PERMANOVA+ for PRIMER: Guide to Software and Statistical Methods, 1st ed.; PRIMER-E: Plymouth, UK, 2008. [Google Scholar]
- Benizri, E.; Amiaud, B. Relationship between plants and soil microbial communities in fertilized grasslands. Soil Biol. Biochem. 2005, 37, 2055–2064. [Google Scholar] [CrossRef]
- Hugerth, L.W.; Andersson, A.F. Analysing microbial community composition through amplicon sequencing: From sampling to hypothesis testing. Front. Microbiol. 2017, 8, 1561. [Google Scholar] [CrossRef] [PubMed]
- Shrestha, K.; Stevens, S.; Shrestha, P.; Adetutu, E.M.; Walsh, K.B.; Ball, A.S.; Midmore, D.J. Characterisation of the soil microbial community of cultivated and uncultivated vertisol in Australia under several management regimes. Agric. Ecosyst. Environ. 2015, 199, 418–427. [Google Scholar] [CrossRef]
- Song, X.; Tao, B.; Guo, J.; Li, J.; Chen, G. Changes in the microbial community structure and soil chemical properties of vertisols under different cropping systems in Northern China. Front. Environ. Sci. 2018, 6, 132. [Google Scholar] [CrossRef]
- Jiao, S.; Chen, W.; Wang, J.; Du, N.; Li, Q.; Wei, G. Soil microbiomes with distinct assemblies through vertical soil profiles drive the cycling of multiple nutrients in reforested ecosystems. Microbiome 2018, 6, 1–13. [Google Scholar] [CrossRef]
- Fierer, N.; Schimel, J.P.; Holden, P.A. Variations in microbial community composition through two soil depth profiles. Soil Biol. Biochem. 2003, 35, 167–176. [Google Scholar] [CrossRef]
- Hartmann, M.; Lee, S.; Hallam, S.J.; Mohn, W.W. Bacterial, archaeal and eukaryal community structures throughout soil horizons of harvested and naturally disturbed forest stands. Environ. Microbiol. 2009, 11, 3045–3062. [Google Scholar] [CrossRef]
- Gordon, H.; Haygarth, P.M.; Bardgett, R.D. Drying and rewetting effects on soil microbial community composition and nutrient leaching. Soil Biol. Biochem. 2008, 40, 302–311. [Google Scholar] [CrossRef]
- Goldfarb, K.C.; Karaoz, U.; Hanson, C.A.; Santee, C.A.; Bradford, M.A.; Treseder, K.K.; Wallenstein, M.D.; Brodie, E.L. Differential growth responses of soil bacterial taxa to carbon substrates of varying chemical recalcitrance. Front. Microbiol. 2011, 2, 94. [Google Scholar] [CrossRef] [PubMed]
- Fierer, N.; Lauber, C.L.; Ramirez, K.S.; Zaneveld, J.; Bradford, M.A.; Knight, R. Comparative metagenomic, phylogenetic and physiological analyses of soil microbial communities across nitrogen gradients. ISME J. 2012, 6, 1007–1017. [Google Scholar] [CrossRef] [PubMed]
- Finn, D.; Kopittke, P.M.; Dennis, P.G.; Dalal, R.C. Microbial energy and matter transformation in agricultural soils. Soil Biol. Biochem. 2017, 111, 176–192. [Google Scholar] [CrossRef]
- Nelson, P.; Dictor, M.; Soulas, G. Availability of organic carbon in soluble and particle-size fractions from a soil profile. Soil Biol. Biochem. 1994, 26, 1549–1555. [Google Scholar] [CrossRef]
- Lundquist, E.J.; Jackson, L.; Scow, K. Wet–dry cycles affect dissolved organic carbon in two California agricultural soils. Soil Biol. Biochem. 1999, 31, 1031–1038. [Google Scholar] [CrossRef]
- McCarthy, A.J.; Williams, S.T. Actinomycetes as agents of biodegradation in the environment—A review. Gene 1992, 115, 189–192. [Google Scholar] [CrossRef]
- Li, X.; Sun, J.; Wang, H.; Li, X.; Wang, J.; Zhang, H. Changes in the soil microbial phospholipid fatty acid profile with depth in three soil types of paddy fields in China. Geoderma 2017, 290, 69–74. [Google Scholar] [CrossRef]
- Michéli, E.; Schad, P.; Spaargaren, O. World Reference Base for Soil Resources 2006: A Framework for International Classification, Correlation and Communication; Food and agriculture organization of the United nations (FAO): Rome, Italy, 2006. [Google Scholar]
- Strickland, M.S.; Rousk, J. Considering fungal: Bacterial dominance in soils–methods, controls, and ecosystem implications. Soil Biol. Biochem. 2010, 42, 1385–1395. [Google Scholar] [CrossRef]
- Taylor, D.L.; Sinsabaugh, R.L. The soil Fungi: Occurrence, phylogeny, and ecology. Soil Microbiol. Ecol. Biochem. 2015, 4, 77–109. [Google Scholar]
- Rasse, D.P.; Smucker, A.J. Root recolonization of previous root channels in corn and alfalfa rotations. Plant Soil 1998, 204, 203–212. [Google Scholar] [CrossRef]
- Hulugalle, N.R.; Broughton, K.J.; Tan, D.K.Y. Fine root production and mortality in irrigated cotton, maize and sorghum sown in vertisols of northern New South Wales, Australia. Soil Tillage Res. 2015, 146 Pt B, 313–322. [Google Scholar] [CrossRef]
- Hulugalle, N.R.; Weaver, T.B.; Finlay, L.A. Carbon inputs by irrigated corn roots to a Vertisol. Plant Root 2010, 4, 18–21. [Google Scholar] [CrossRef]
- Boer, W.d.; Folman, L.B.; Summerbell, R.C.; Boddy, L. Living in a fungal world: Impact of fungi on soil bacterial niche development. FEMS Microbiol. Rev. 2005, 29, 795–811. [Google Scholar] [CrossRef] [PubMed]
- Costa, C.A.E.; Coleman, W.; Dube, M.; Rodrigues, A.E.; Pinto, P.C.R. Assessment of key features of lignin from lignocellulosic crops: Stalks and roots of corn, cotton, sugarcane, and tobacco. Ind. Crops Prod. 2016, 92, 136–148. [Google Scholar] [CrossRef]
- Houlden, A.; Timms-Wilson, T.M.; Day, M.J.; Bailey, M.J. Influence of plant developmental stage on microbial community structure and activity in the rhizosphere of three field crops. FEMS Microbiol. Ecol. 2008, 65, 193–201. [Google Scholar] [CrossRef] [PubMed]
- Qiao, Q.; Wang, F.; Zhang, J.; Chen, Y.; Zhang, C.; Liu, G.; Zhang, H.; Ma, C.; Zhang, J. The variation in the rhizosphere microbiome of cotton with soil type, genotype and developmental stage. Sci. Rep. 2017, 7, 1–10. [Google Scholar] [CrossRef]
- Ai, C.; Zhang, S.; Zhang, X.; Guo, D.; Zhou, W.; Huang, S. Distinct responses of soil bacterial and fungal communities to changes in fertilization regime and crop rotation. Geoderma 2018, 319, 156–166. [Google Scholar] [CrossRef]
- Hilton, S.; Bennett, A.J.; Chandler, D.; Mills, P.; Bending, G.D. Preceding crop and seasonal effects influence fungal, bacterial and nematode diversity in wheat and oilseed rape rhizosphere and soil. Appl. Soil Ecol. 2018, 126, 34–46. [Google Scholar] [CrossRef]
- Kuzyakov, Y.; Blagodatskaya, E. Microbial hotspots and hot moments in soil: Concept & review. Soil Biol. Biochem. 2015, 83, 184–199. [Google Scholar]
- Sikorski, J. The prokaryotic biology of soil. Soil Org. 2015, 87, 1–28. [Google Scholar]
- Chen, X.; Henriksen, T.M.; Svensson, K.; Korsaeth, A. Long-term effects of agricultural production systems on structure and function of the soil microbial community. Appl. Soil Ecol. 2019. [Google Scholar] [CrossRef]
- Fierer, N. Embracing the unknown: Disentangling the complexities of the soil microbiome. Nat. Rev. Microbiol. 2017, 15, 579–590. [Google Scholar] [CrossRef] [PubMed]
- Delgado-Baquerizo, M.; Maestre, F.T.; Reich, P.B.; Jeffries, T.C.; Gaitan, J.J.; Encinar, D.; Berdugo, M.; Campbell, C.D.; Singh, B.K. Microbial diversity drives multifunctionality in terrestrial ecosystems. Nat. Commun. 2016, 7, 1–8. [Google Scholar] [CrossRef] [PubMed]
- Polain, K.; Knox, O.; Wilson, B.; Guppy, C.; Lisle, L.; Nachimuthu, G.; Osanai, Y.; Siebers, N. Distribution of subsoil microbial activity and biomass under Australian rotational cotton as influenced by system, crop status and season. Soil Res. 2020. accepted publication. [Google Scholar]
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).