Digital, Three-Dimensional Visualization of Root Systems in Peat
Abstract
1. Introduction
2. Materials and Methods
2.1. Sampling
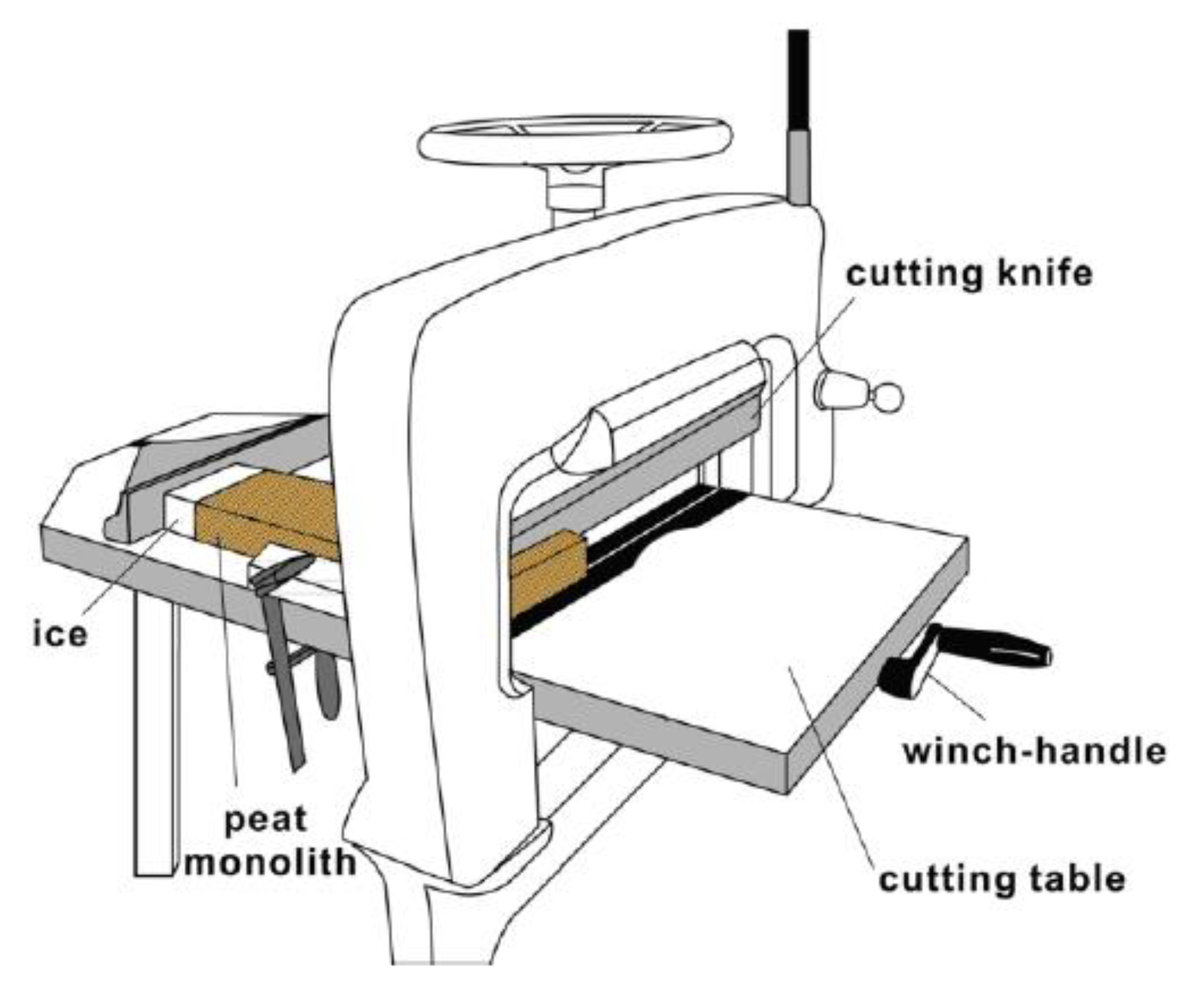
2.2. Sample Preparation
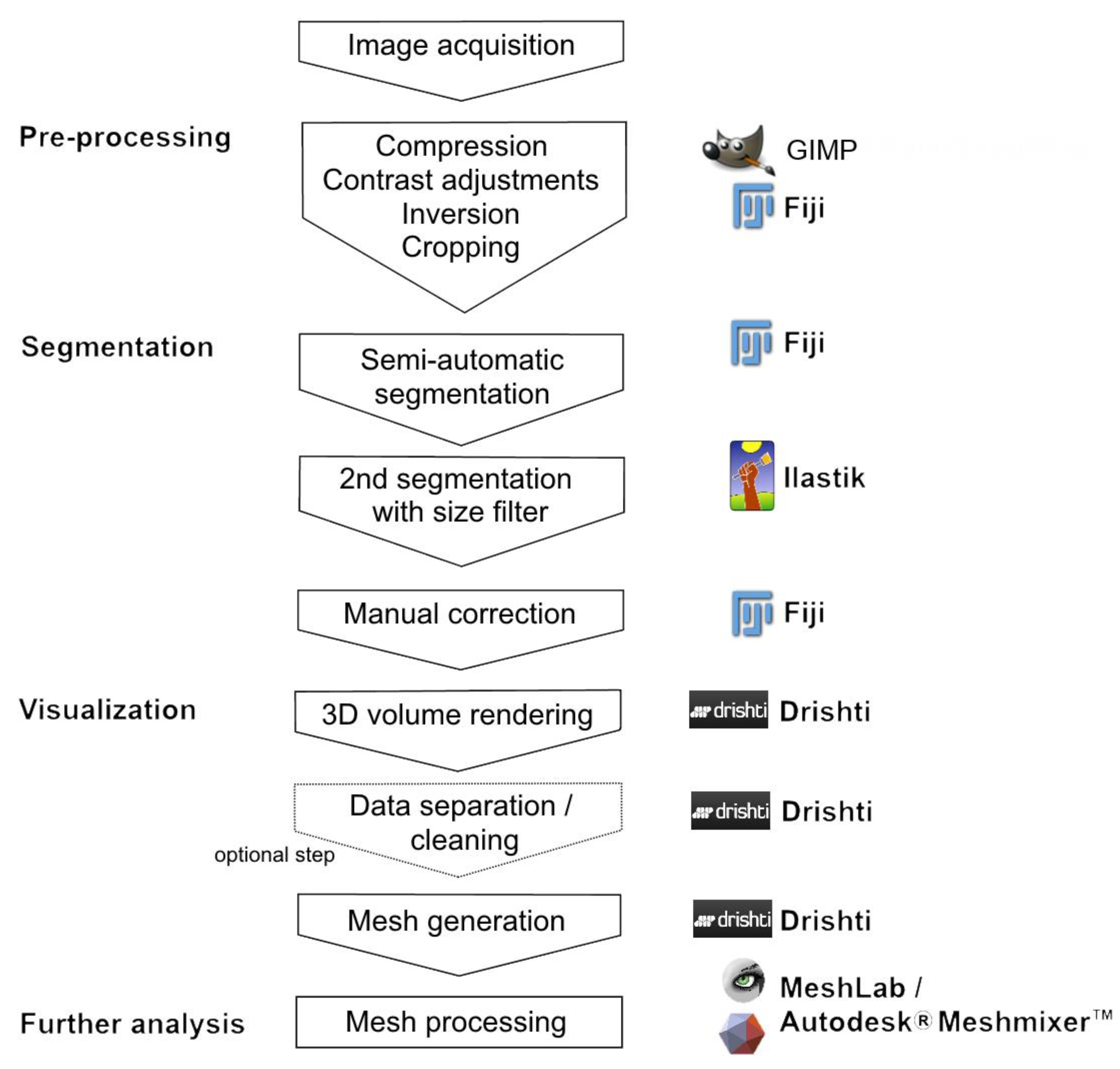
2.3. Digital Image Analysis
2.4. Volume Measurements
3. Results
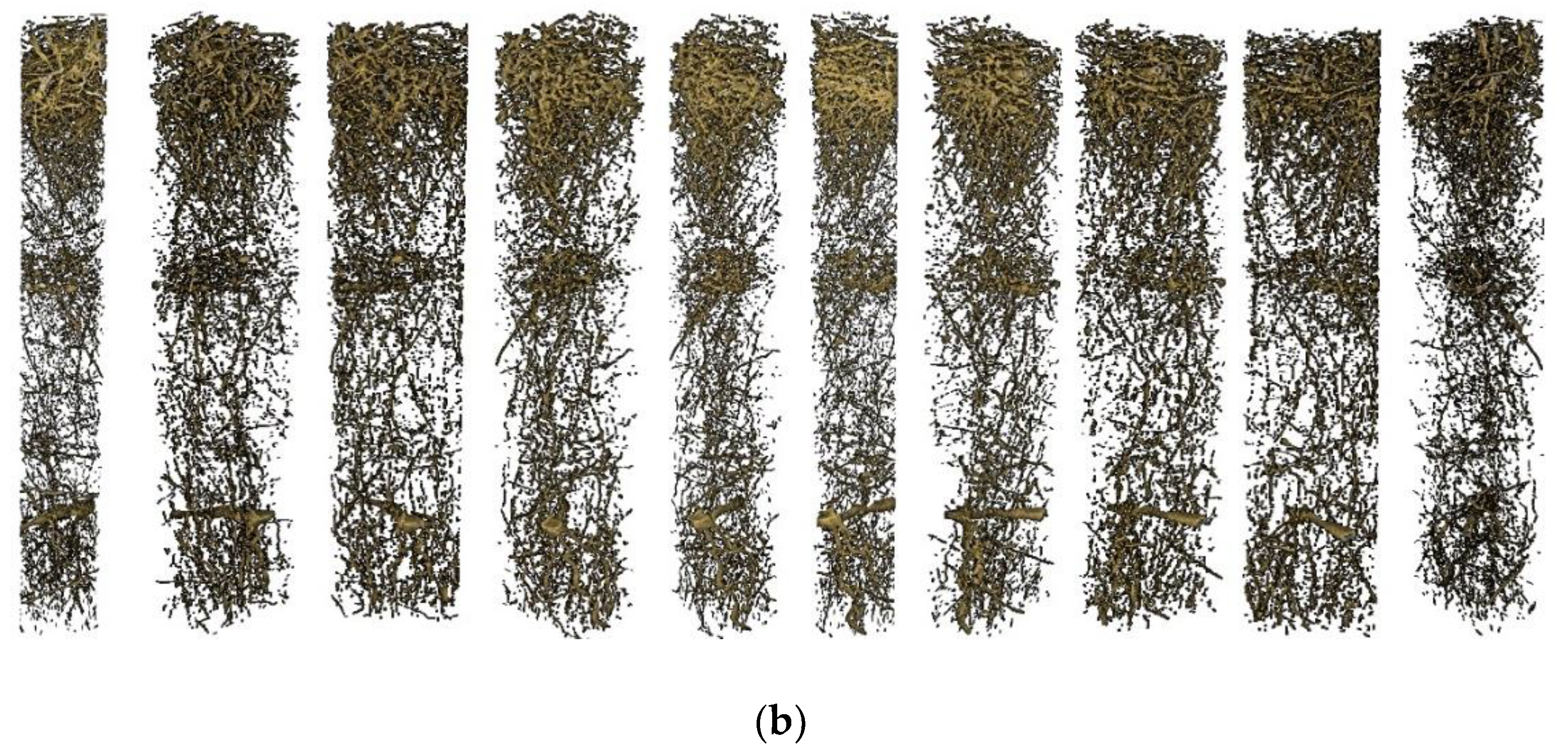
3.1. 3D Model Visualization
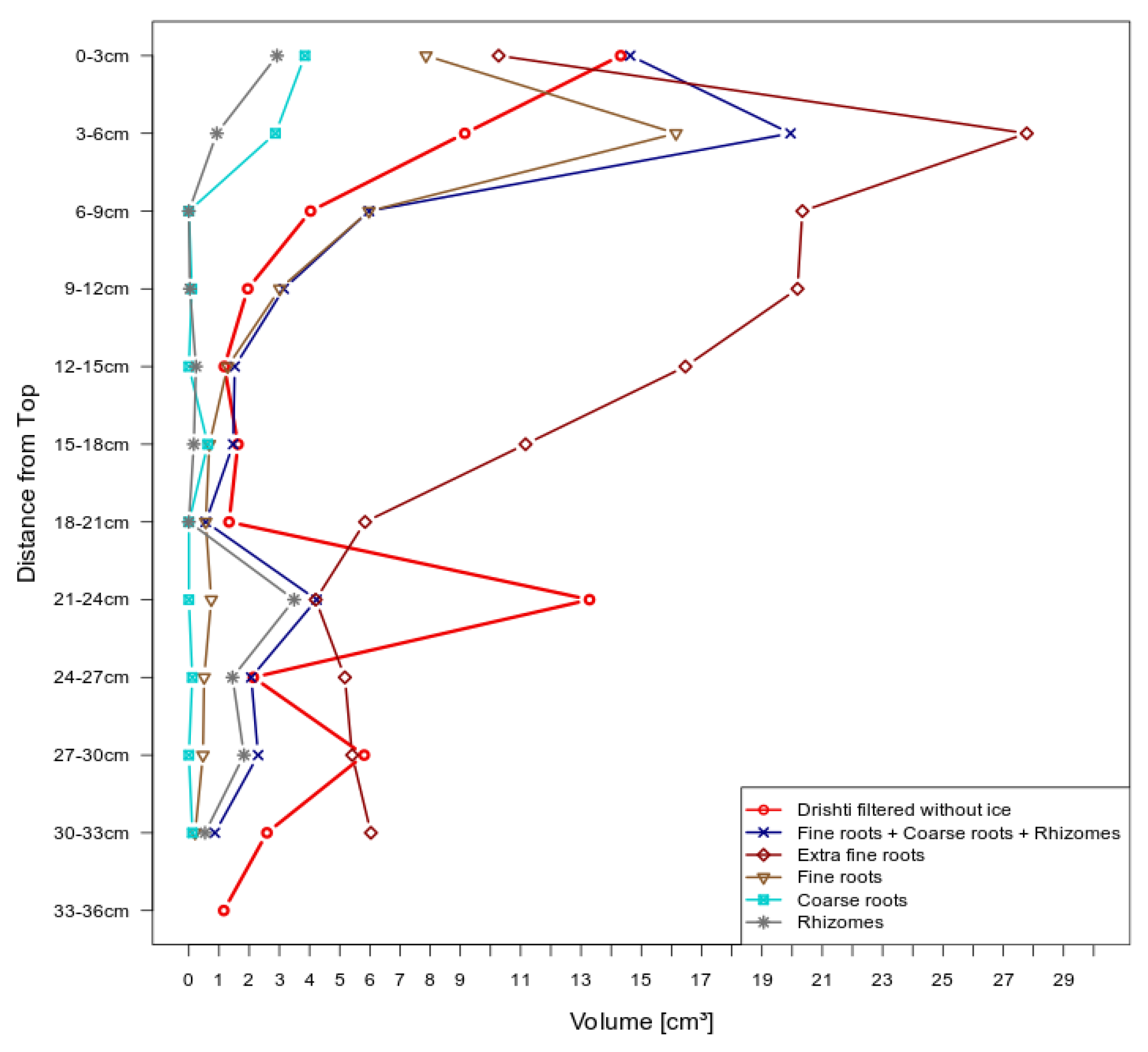
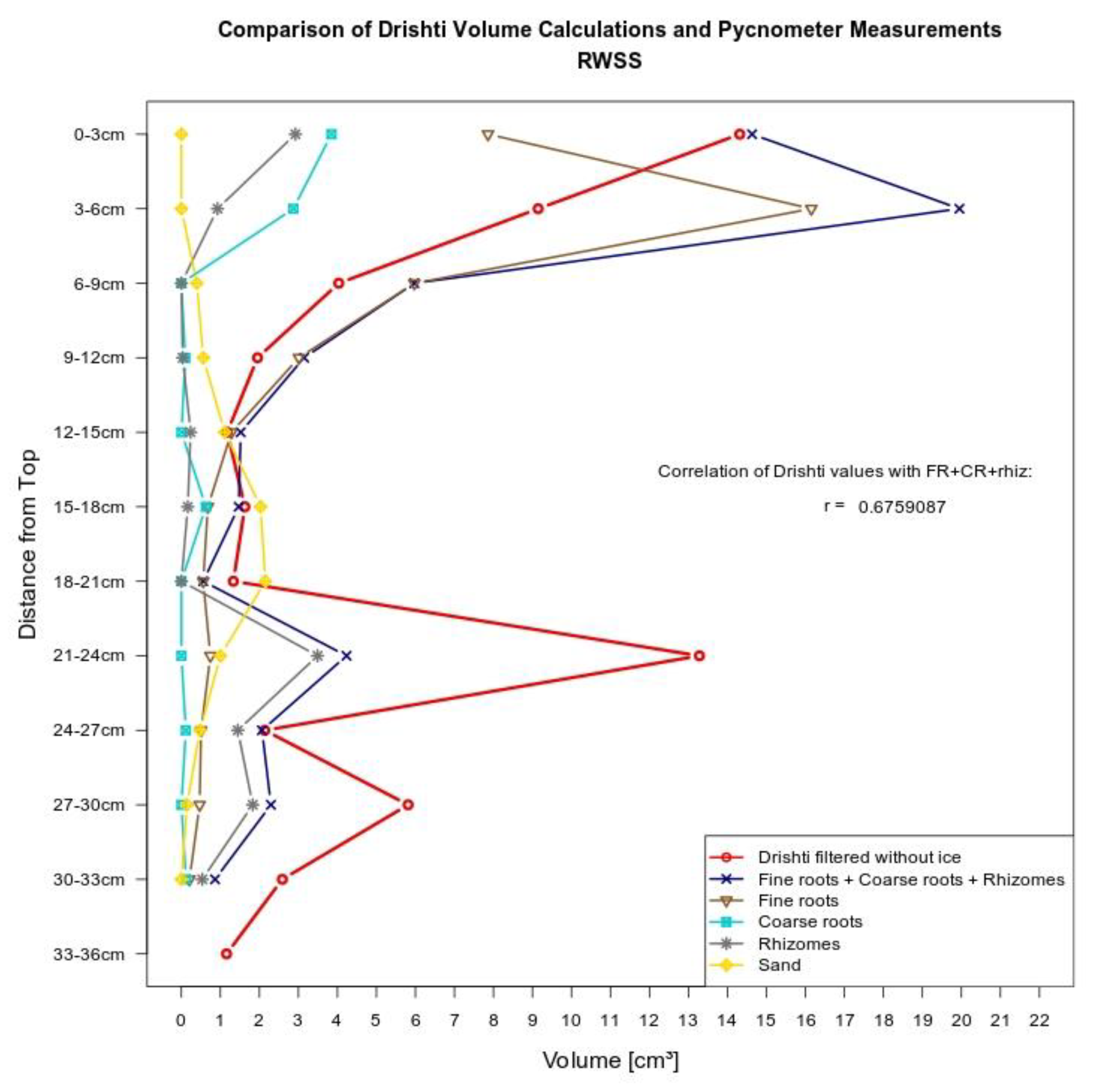
3.2. Volumetric Measurements
3.2.1. Manual Root Washing
3.2.2. Model Measurements
4. Discussion
4.1. Technical Aspects
4.2. Volumetric Measurement Methods
4.3. 3D Structure
4.4. Applications
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
Appendix A
| Sample | Distance 1 in cm | Focal Length in mm | ISO-Value | Exposure Time in s | F-Number | External Studio Flash with Diffuser |
|---|---|---|---|---|---|---|
| RWSS (crosswise) | 25 | 69 | 1600 | 1 | F/13 | - |
| RWSN (lengthwise) | 27 | 18 | 100 | 2 | F/5 | yes |
Appendix B
Appendix C
| Parameter | RWSN | RWSS |
|---|---|---|
| Membrane thickness | 1 | 1 |
| Membrane patch size | 19 | 19 |
| Minimum sigma value | 0.5 | 0.5 |
| Maximum sigma value | 20.0 | 30.0 |
| Applied filters | Gaussian blur, Hessian, Membrane projections, Sobel, Difference of Gaussians, Kuwahara | Gaussian blur, Hessian, Membrane projections, Sobel, Difference of Gaussians, Kuwahara |
Appendix D
| Parameter | RWSN | RWSS |
|---|---|---|
| Method | simple | simple |
| Input | 0 | 0 |
| Smooth | 0,0; 0,0; 0,0 | 0,0; 0,0; 0,0 |
| Threshold | 0.29 | 0.24 |
| Minimum size filter | 95 | 65 |
| Maximum size filter | 10,000,000 | 10,000,000 |
Appendix E
Appendix F
Appendix G
| Core Name | Calculated Measures of 3D Model | Measures of Rectangular Slabs for Washing out Roots |
|---|---|---|
| RWSN | 10.1 cm × 6.0 cm × 44.6 cm | 10.0 cm × 6.0 cm × 3.0 cm |
| RWSS | 12.5 cm × 6.1 cm × 36.6 cm | 12.5 cm × 6.0 cm × 3.0 cm |
Appendix H
Appendix I
References
- Joosten, H.; Sirin, A.; Couwenberg, J.; Laine, J.; Smitz, P. The Role of Peatlands in Climate Regulation. In Peatland Restoration and Ecosystem Services: Science, Policy and Practice; Bonn, A., Allot, T., Evans, M., Joosten, H., Stoneman, R., Eds.; Cambridge University Press: Cambridge, UK, 2016; pp. 63–76. [Google Scholar]
- Moen, A.; Joosten, H.; Tanneberger, F. Mire diversity in Europe: Mire regionality. In Mires and Peatlands in Europe—Status, Distribution and Conservation; Joosten, H., Tanneberger, F., Moen, A., Eds.; Schweizerbart Science Publishers: Stuttgart, Germany, 2017; pp. 97–172. [Google Scholar]
- Grosse-Brauckmann, G. Analysis of vegetative plant macrofossils. In Handbook of Holocene Palaeoecology and Palaeohydrology; Berglund, B.E., Ralska-Jasiewiczowa, M., Eds.; Wiley: Berlin/Heidelberg, Germany, 1986; pp. 591–618. [Google Scholar]
- Couwenberg, J.; DeKlerk, P.; Endtmann, E.; Joosten, H.; Michaelis, D. Hydrogenetische Moortypen in der Zeit—Eine Zusammenschau. In Landschaftsökologische Moorkunde, 2nd ed.; Succow, M., Joosten, H., Eds.; Schweizerbart Science Publishers: Stuttgart, Germany, 2001; pp. 399–403. [Google Scholar]
- Pagès, L.; Vercambre, G.; Drouet, J.L.; Lecompte, F.; Collet, C.; Le Bot, J. Root Typ: A generic model to depict and analyse the root system architecture. Plant Soil 2004, 258, 103–119. [Google Scholar] [CrossRef]
- Hodson, M.J.; Bryant, J.A. Roots. In Functional Biology of Plants, 1st ed.; Hodson, M.J., Bryant, J.A., Eds.; John Wiley & Sons, Ltd.: Chichester, UK, 2012; pp. 124–144. [Google Scholar]
- Davey, E.; Wigand, C.; Johnson, R.; Sundberg, K.; Morris, J.; Roman, C.T. Use of computed tomography imaging for quantifying coarse roots, rhizomes, peat, and particle densities in marsh soils. Ecol. Appl. 2011, 21, 2156–2171. [Google Scholar] [CrossRef] [PubMed]
- Taylor, H.M.; Upchurch, D.R.; McMichael, B.L. Applications and limitations of rhizotrons and minirhizotrons for root studies. Plant Soil 1990, 129, 29–35. [Google Scholar] [CrossRef]
- Busch, J.; Mendelssohn, I.A.; Lorenzen, B.; Brix, H.; Miao, S. A rhizotron to study root growth under flooded conditions tested with two wetland Cyperaceae. Flora Morphol. Distrib. Funct. Ecol. Plants 2006, 201, 429–439. [Google Scholar] [CrossRef]
- Kokkonen, P. Beobachtungen über das Wurzelsystem der Kiefer im Moorböden. Acta For. Fenn. 1923, 25, 1–21. [Google Scholar] [CrossRef]
- Rigg, G.B.; Harrar, E.S. The root systems of trees growing in sphagnum. Am. J. Bot. 1931, 18, 391–397. [Google Scholar] [CrossRef]
- Metsävainio, K. Untersuchungen über das Wurzelsystem der Moorpflanzen. Annales Botanici Societatis Zoologicae-Botanici Fennici Vanamo 1931, 1, 1–422. [Google Scholar]
- Kettridge, N.; Binley, A. X-ray computed tomography of peat soils: Measuring gas content and peat structure. Hydrol. Process. Int. J. 2008, 22, 4827–4837. [Google Scholar] [CrossRef]
- Kettridge, N.; Binley, A. Characterization of peat structure using X-ray computed tomography and its control on the ebullition of biogenic gas bubbles. J. Geophys. Res. Biogeosci. 2001, 116. [Google Scholar] [CrossRef]
- Gharedaghloo, B.; Price, J.S.; Rezanezhad, F.; Quinton, W.L. Evaluating the hydraulic and transport properties of peat soil using pore network modeling and X-ray micro computed tomography. J. Hydrol. 2018, 561, 494–508. [Google Scholar] [CrossRef]
- Clymo, R.S. A high-resolution sampler of surface peat. Funct. Ecol. 1988, 2, 425–431. [Google Scholar] [CrossRef]
- Joosten, H.; DeKlerk, P.D. DAMOCLES: A DAshing MOnolith Cutter for fine sectioning of peats and sediments into LargE Slices. Boreas 2007, 36, 76–81. [Google Scholar] [CrossRef]
- GIMP. GNU Image Manipulation Program. Available online: www.gimp.org (accessed on 13 January 2020).
- Schindelin, J.; Arganda-Carreras, I.; Frise, E.; Kaynig, V.; Longair, M.; Pietzsch, T.; Preibisch, S.; Rueden, C.; Saalfeld, S.; Schmid, B.; et al. Fiji: An open-source platform for biological-image analysis. Nat. Methods 2012, 9, 676–682. [Google Scholar] [CrossRef] [PubMed]
- Sommer, C.; Straehle, C.; Koethe, U.; Hamprecht, F.A. Ilastik: Interactive learning and segmentation toolkit. In Proceedings of the 2011 IEEE International Symposium on Biomedical Imaging: From Nano to Macro, Chicago, IL, USA, 30 March–2 April 2011; IEEE: New York, NY, USA, 2011; pp. 230–233. [Google Scholar] [CrossRef]
- Limaye, A. Drishti: A volume exploration and presentation tool. In Developments in X-Ray Tomography VIII; SPIE: Washington, DC, USA, 2012; p. 85060X. [Google Scholar] [CrossRef]
- Cignoni, P.; Callieri, M.; Corsini, M.; Dellepiane, M.; Ganovelli, F.; Ranzuglia, G. Meshlab: An open-source mesh processing tool. In Proceedings of the Eurographics Italian Chapter Conference, Salerno, Italy, 2–4 July 2008; pp. 129–136. [Google Scholar]
- Bengtsson, E.; Wahlby, C.; Lindblad, J. Robust cell image segmentation methods. Pattern Recognit. Image Anal. C/c Raspozn. Obraz. Anal. Izobr. 2004, 14, 157–167. [Google Scholar]
- Arganda-Carreras, I.; Kaynig, V.; Rueden, C.; Eliceiri, K.W.; Schindelin, J.; Cardona, A.; Sebastian Seung, H. Trainable Weka Segmentation: A machine learning tool for microscopy pixel classification. Bioinformatics 2017, 33, 2424–2426. [Google Scholar] [CrossRef] [PubMed]
- Hall, M.; Frank, E.; Holmes, G.; Pfahringer, B.; Reutemann, P.; Witten, I.H. The WEKA data mining software: An update. ACM SIGKDD Explor. Newsl. 2009, 11, 10–18. [Google Scholar] [CrossRef]
- Hodge, A.; Berta, G.; Doussan, C.; Merchan, F.; Crespi, M. Plant root growth, architecture and function. Plant Soil 2009, 321, 153–187. [Google Scholar] [CrossRef]







| Sample Core | Length of Cutting Distance [cm] | Number of Cuts/Images | Number of Pixels in 1 mm | Voxel Depth [mm] |
|---|---|---|---|---|
| RWSN | 10.1 | 101 | 11 | 1.0 |
| RWSS | 44.9 | 944 1 | 19 | 0.48 |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Gribbe, S.; Blume-Werry, G.; Couwenberg, J. Digital, Three-Dimensional Visualization of Root Systems in Peat. Soil Syst. 2020, 4, 13. https://doi.org/10.3390/soilsystems4010013
Gribbe S, Blume-Werry G, Couwenberg J. Digital, Three-Dimensional Visualization of Root Systems in Peat. Soil Systems. 2020; 4(1):13. https://doi.org/10.3390/soilsystems4010013
Chicago/Turabian StyleGribbe, Stella, Gesche Blume-Werry, and John Couwenberg. 2020. "Digital, Three-Dimensional Visualization of Root Systems in Peat" Soil Systems 4, no. 1: 13. https://doi.org/10.3390/soilsystems4010013
APA StyleGribbe, S., Blume-Werry, G., & Couwenberg, J. (2020). Digital, Three-Dimensional Visualization of Root Systems in Peat. Soil Systems, 4(1), 13. https://doi.org/10.3390/soilsystems4010013





