Deep Learning Methods to Reveal Important X-ray Features in COVID-19 Detection: Investigation of Explainability and Feature Reproducibility
Abstract
:1. Introduction
- The samples are too few for deep model training
- The image information is not accompanied by clinical outcomes.
- There are few multicenter studies to support the conclusions.
- The samples commonly illustrate COVID-19 disease of patients showing disease symptoms. Asymptomatic cases are under-represented.
- The successful state-of-the-art network (MobileNet v2) was extensively evaluated in performing multi-class and two-class classification of X-ray images with the aim of identifying images related to the coronavirus disease. Further, the consistency of the reported metrics was assessed by running a 25-times 10-fold cross-validation
- The explainability algorithm (Grad-CAM) was employed to inspect the consistency of the suggested areas of interest across a three-run experiment.
- We present a staged approach for the detection of COVID-19 from X-ray images that exhibited an accuracy of 89.88 ± 3.66%.
2. COVID-19 Detection Based on X-ray Imaging: Recent Studies
3. Materials and Methods
3.1. Deep Learning with Mobile Networks
- (a)
- The selection of a simpler CNN, which contains less trainable parameters and fits in the particular data well.
- (b)
- Transfer learning [19], utilizing deep and complex CNNs, but freezing their layers, thereby decreasing the trainable parameters and allowing for knowledge transfer, following their training on large image datasets.
- (c)
- Data augmentation methods to increase the training set size, such as geometric transformation (rotation, sheer) and pixel-level transformations (equalizations, grey-level alterations) [20].
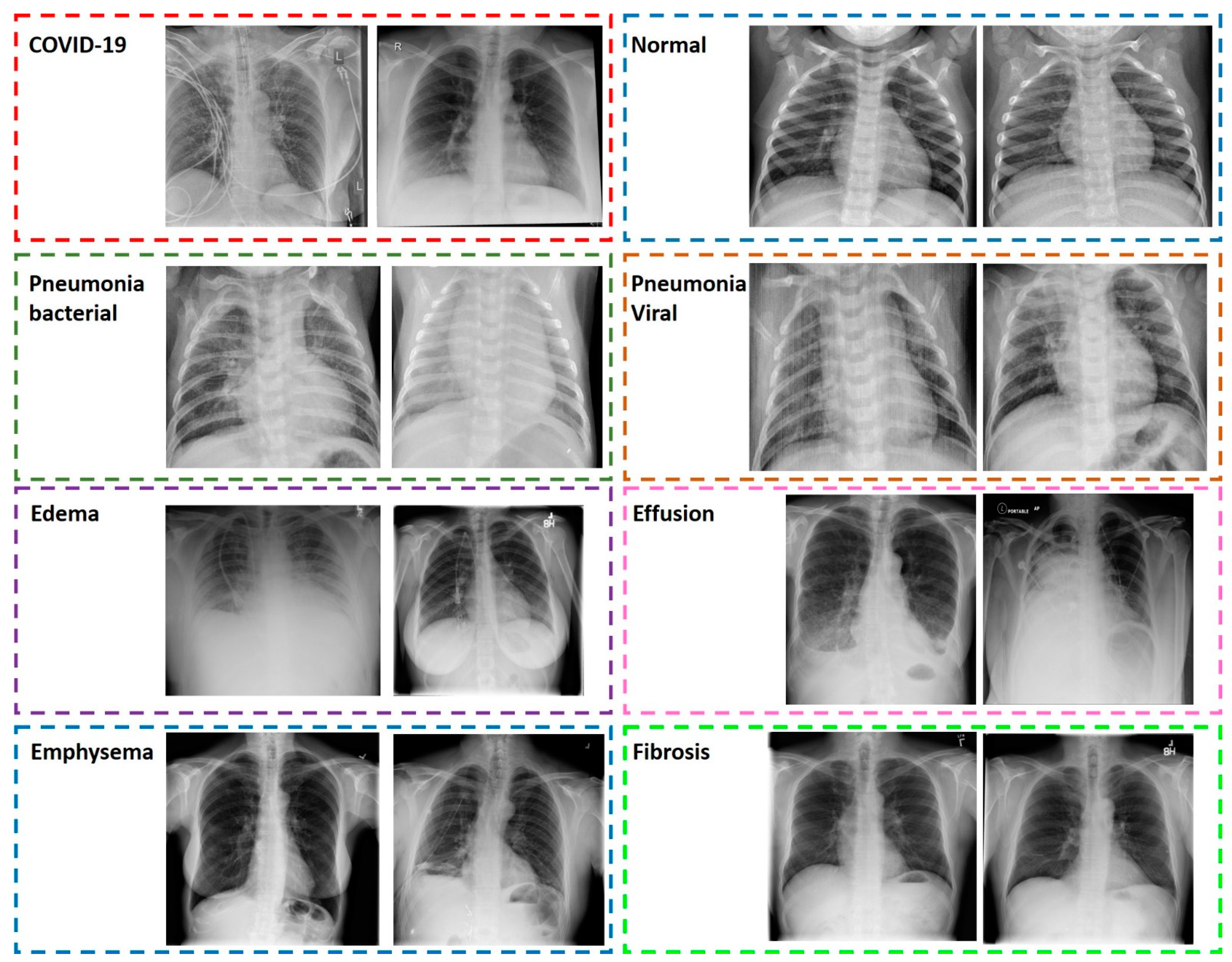
3.2. Image Dataset
3.2.1. COVID-19, Common Bacterial and Viral Pneumonia X-ray Scans
3.2.2. Pulmonary Diseases Detected from X-ray Scans
3.3. Data Augmentation Techniques
- Random rotations;
- Horizontal flips;
- Height and width shifts.
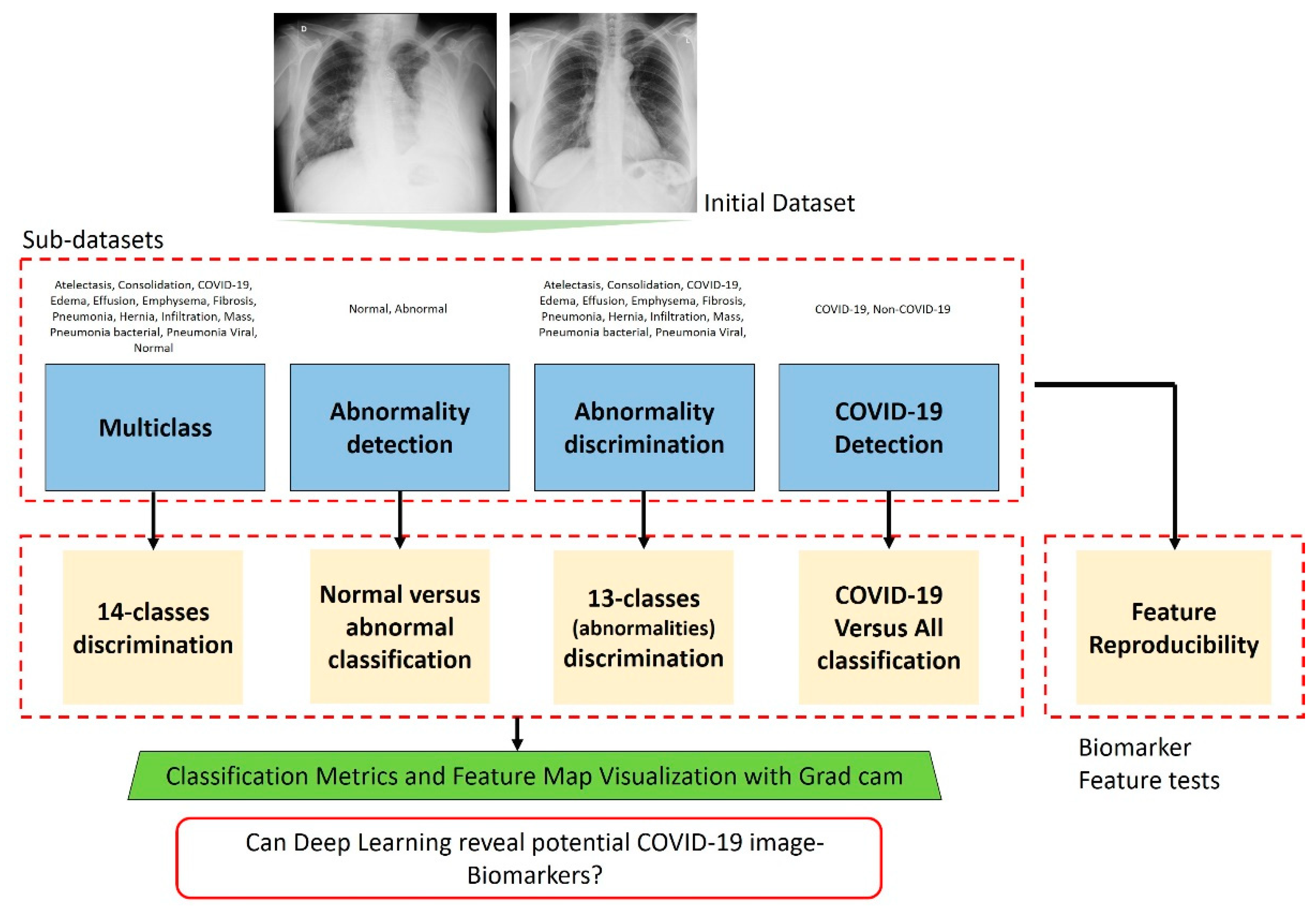
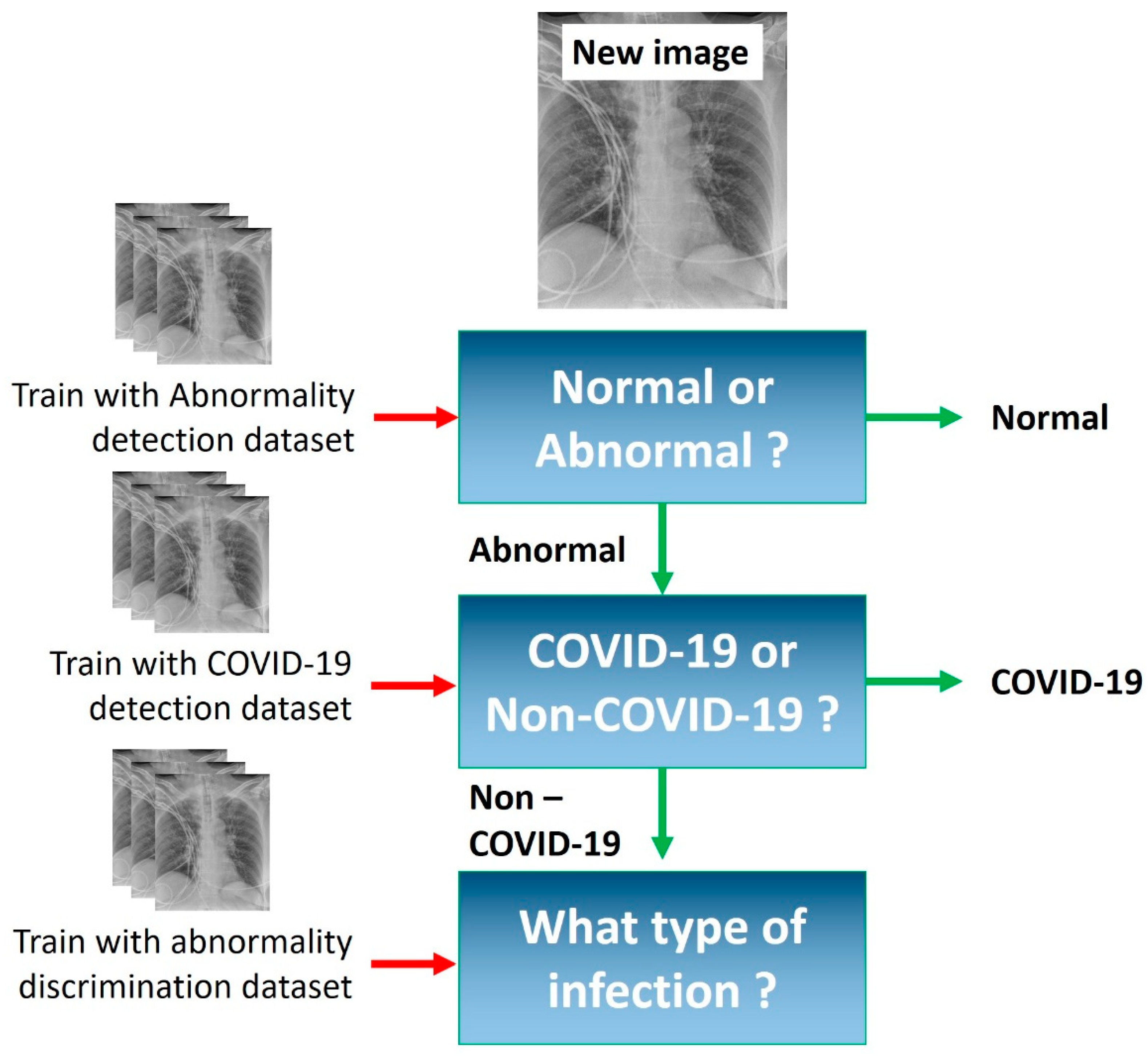
3.4. Experiments
4. Results
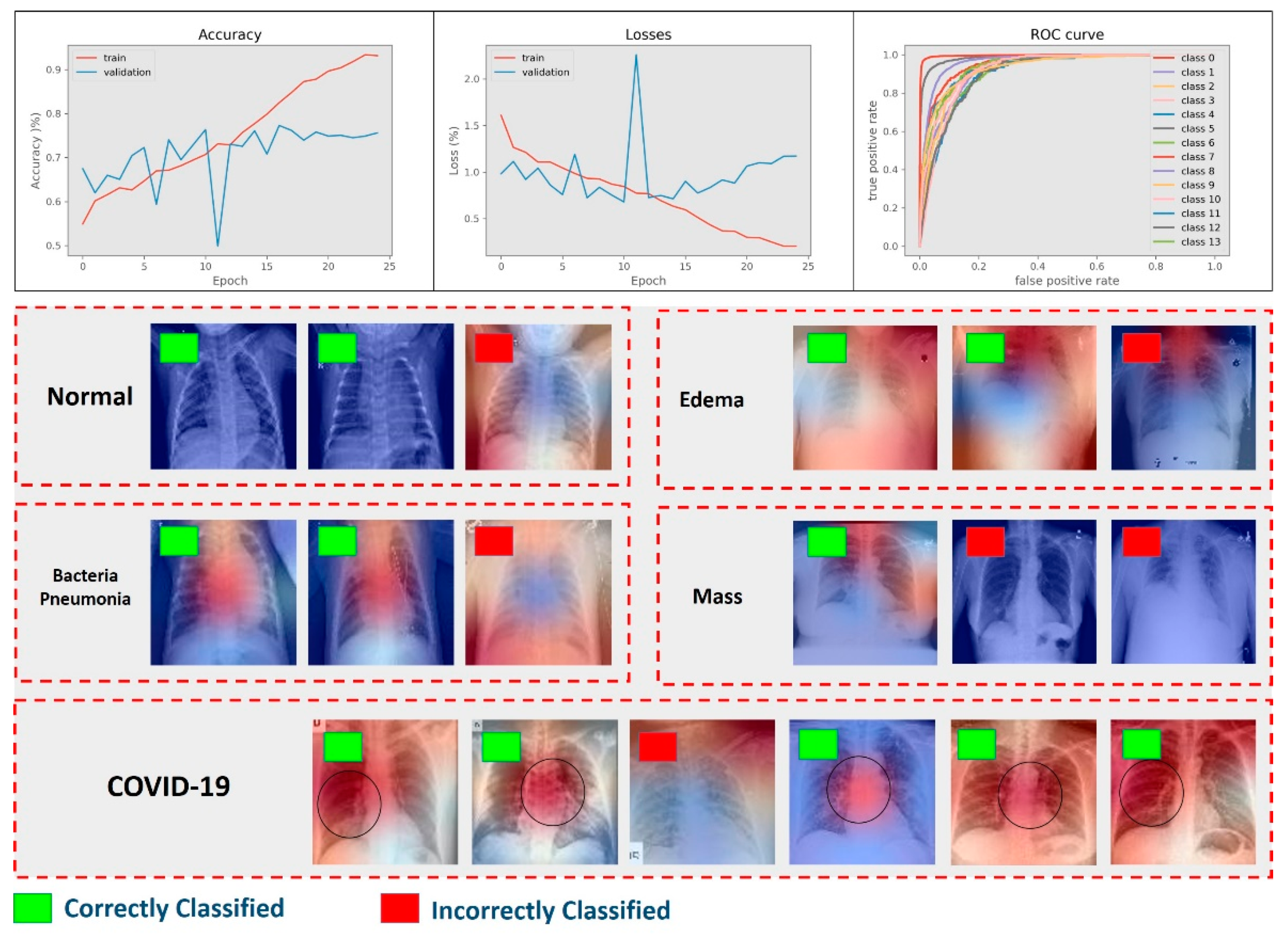
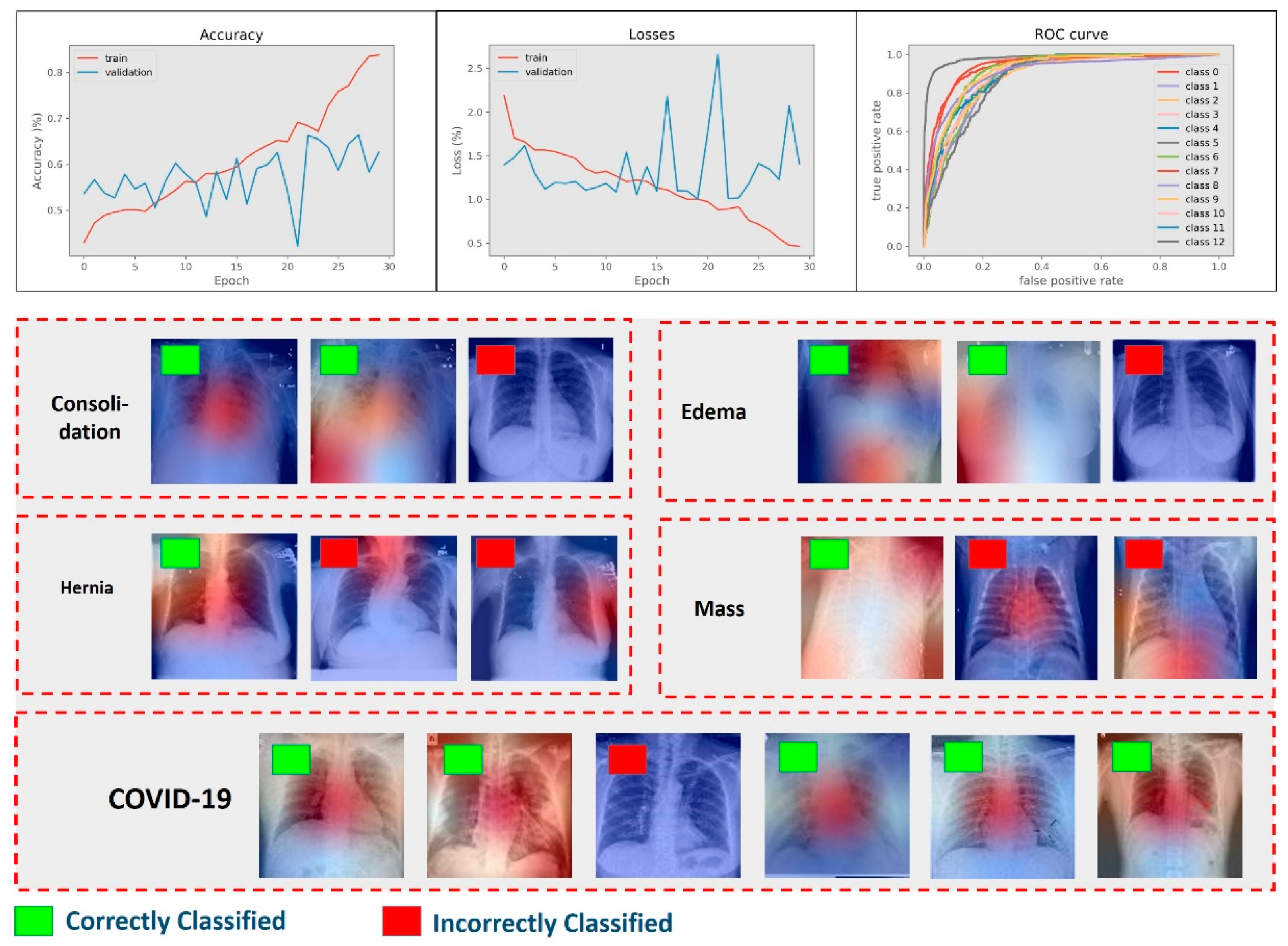
4.1. Results of Multiclass Classification
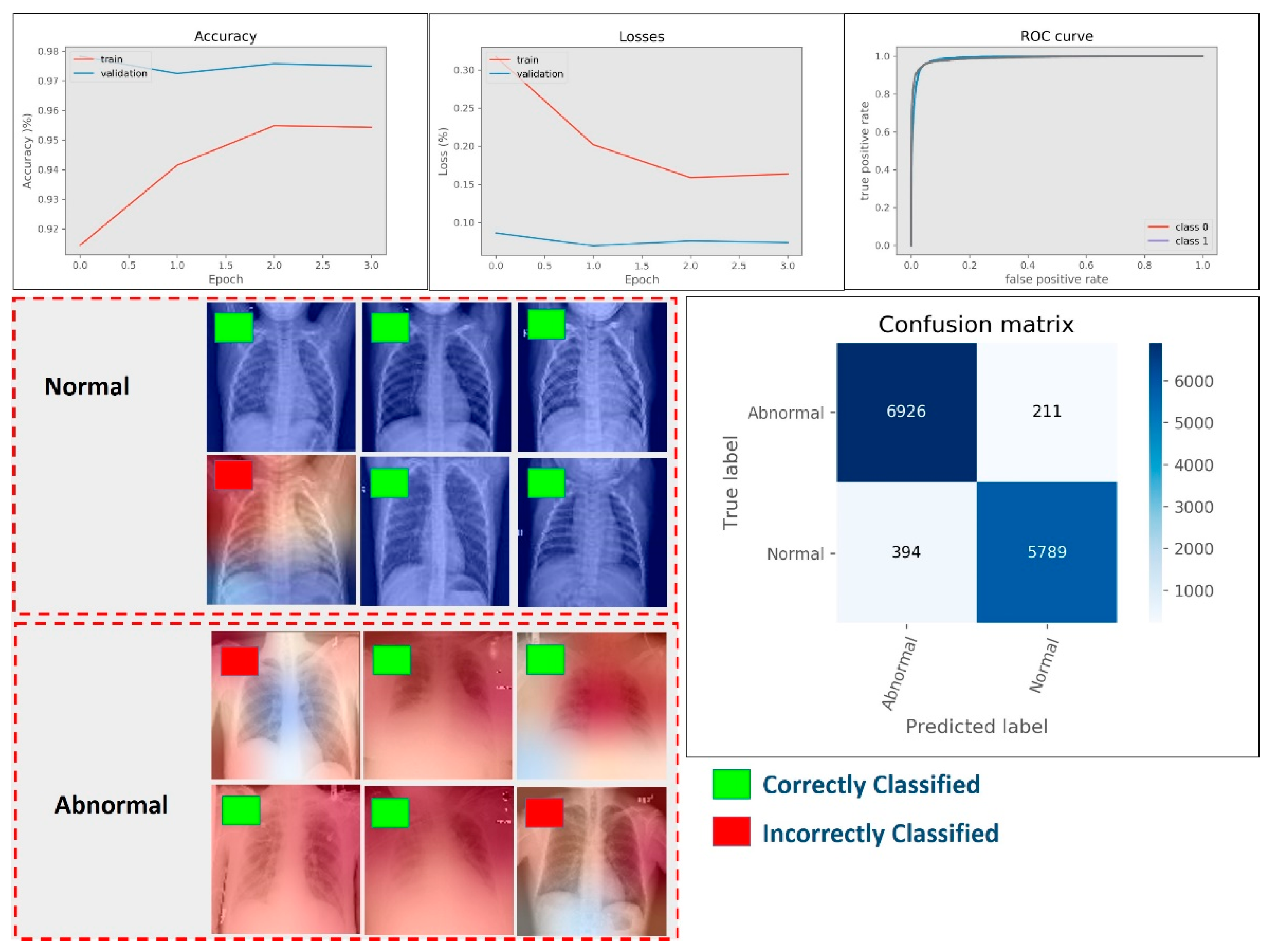
4.2. Results of Abnormality Detection (Two-Class)
4.3. Results of Abnormality Discrimination
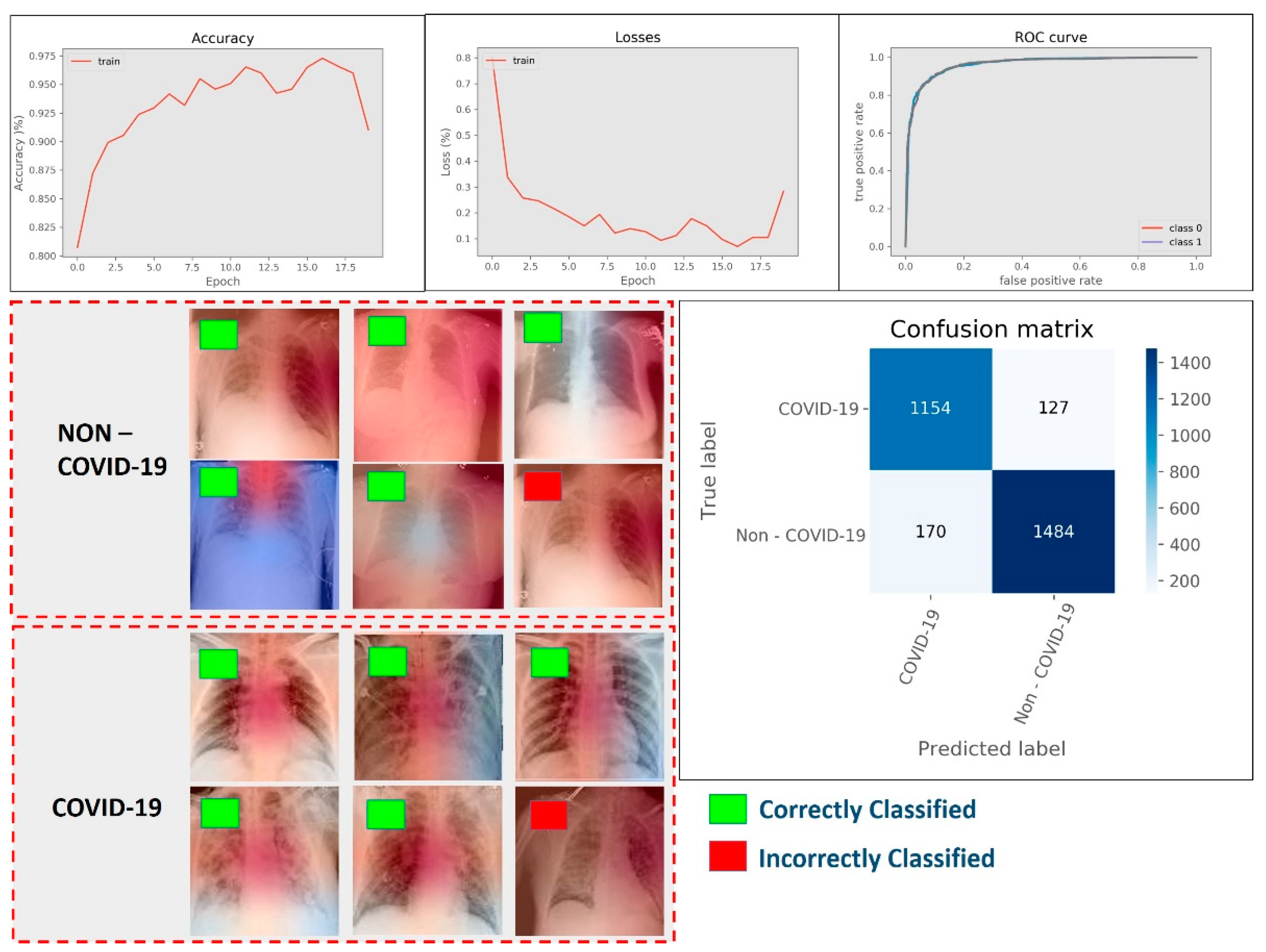
4.4. Results of COVID-19 Detection
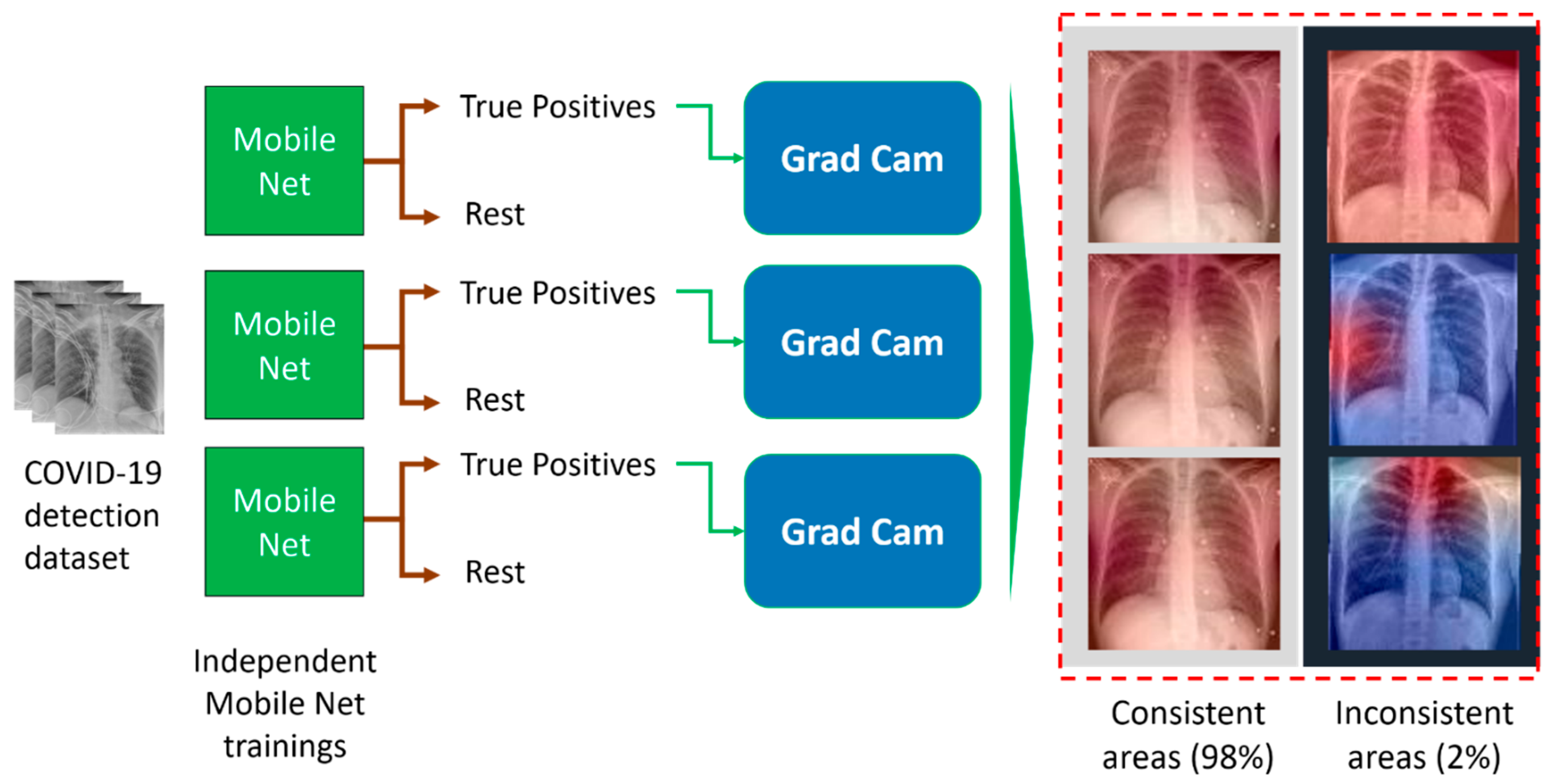
4.5. Results of Feature Reproducibility in COVID-19 Detection
- (a)
- Some of the COVID-19 images may contain annotations that are recognized by the model as features. Although the data were tested, the non-official nature of the dataset source led us to not be completely sure about the origin of the images and the pre-processing that may have taken place.
- (b)
- The learning capacity of MobileNet (v2) is not enough to capture all significant features, leading to the exclusion of some of them.
- (c)
- Data augmentation fails to improve the model’s capability in capturing global and important features completely, thereby allowing for irrelevant feature discovery.
5. Discussion
6. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Sahiner, B.; Pezeshk, A.; Hadjiiski, L.M.; Wang, X.; Drukker, K.; Cha, K.H.; Summers, R.M.; Giger, M.L. Deep learning in medical imaging and radiation therapy. Med. Phys. 2019, 46, e1–e36. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Sedik, A.; Iliyasu, A.M.; El-Rahiem, A.; Abdel Samea, M.E.; Abdel-Raheem, A.; Hammad, M.; Peng, J.; El-Samie, A.; Fathi, E.; El-Latif, A.A.A.; et al. Deploying machine and deep learning models for efficient data-augmented detection of COVID-19 infections. Viruses 2020, 12, 769. [Google Scholar] [CrossRef] [PubMed]
- Kovács, A.; Palásti, P.; Veréb, D.; Bozsik, B.; Palkó, A.; Kincses, Z.T. The Sensitivity and Specificity of Chest CT in the Diagnosis of COVID-19. Eur. Radiol. 2021, 31, 2819–2824. [Google Scholar] [CrossRef] [PubMed]
- Brunese, L.; Mercaldo, F.; Reginelli, A.; Santone, A. Explainable deep learning for pulmonary disease and coronavirus COVID-19 detection from X-rays. Comput. Methods Progr. Biomed. 2020, 196, 105608. [Google Scholar] [CrossRef]
- Apostolopoulos, I.D.; Mpesiana, T.A. COVID-19: Automatic Detection from X-Ray Images Utilizing Transfer Learning with Convolutional Neural Networks. Phys. Eng. Sci. Med. 2020, 43, 635–640. [Google Scholar] [CrossRef] [Green Version]
- Das, N.N.; Kumar, N.; Kaur, M.; Kumar, V.; Singh, D. Automated deep transfer learning-based approach for detection of COVID-19 infection in chest X-rays. IRBM 2020, 43, 114–119. [Google Scholar]
- Gozes, O.; Frid-Adar, M.; Greenspan, H.; Browning, P.D.; Zhang, H.; Ji, W.; Bernheim, A.; Siegel, E. Rapid ai development cycle for the coronavirus (COVID-19) pandemic: Initial results for automated detection & patient monitoring using deep learning ct image analysis. arXiv 2020, arXiv:2003.05037. [Google Scholar]
- Afshar, P.; Heidarian, S.; Naderkhani, F.; Oikonomou, A.; Plataniotis, K.N.; Mohammadi, A. Covid-caps: A capsule network-based framework for identification of COVID-19 cases from X-ray images. arXiv 2020, arXiv:2004.02696. [Google Scholar] [CrossRef]
- Apostolopoulos, I.D.; Aznaouridis, S.I.; Tzani, M.A. Extracting Possibly Representative COVID-19 Biomarkers from X-ray Images with Deep Learning Approach and Image Data Related to Pulmonary Diseases. J. Med. Biol. Eng. 2020, 40, 462–469. [Google Scholar] [CrossRef]
- Ozturk, T.; Talo, M.; Yildirim, E.A.; Baloglu, U.B.; Yildirim, O.; Acharya, U.R. Automated detection of COVID-19 cases using deep neural networks with X-ray images. Comput. Biol. Med. 2020, 121, 103792. [Google Scholar] [CrossRef]
- Selvaraju, R.R.; Cogswell, M.; Das, A.; Vedantam, R.; Parikh, D.; Batra, D. Grad-CAM: Visual Explanations from Deep Networks via Gradient-Based Localization. Int. J. Comput. Vis. 2020, 128, 336–359. [Google Scholar] [CrossRef] [Green Version]
- Hou, J.; Gao, T. Explainable DCNN Based Chest X-ray Image Analysis and Classification for COVID-19 Pneumonia Detection. Sci. Rep. 2021, 11, 16071. [Google Scholar] [CrossRef] [PubMed]
- Ahsan, M.M.; Nazim, R.; Siddique, Z.; Huebner, P. Detection of COVID-19 Patients from CT Scan and Chest X-ray Data Using Modified MobileNetV2 and LIME. Healthcare 2021, 9, 1099. [Google Scholar] [CrossRef] [PubMed]
- Palatnik de Sousa, I.; Maria Bernardes Rebuzzi Vellasco, M.; Costa da Silva, E. Local Interpretable Model-Agnostic Explanations for Classification of Lymph Node Metastases. Sensors 2019, 19, 2969. [Google Scholar] [CrossRef] [Green Version]
- Wang, L.; Lin, Z.Q.; Wong, A. COVID-Net: A Tailored Deep Convolutional Neural Network Design for Detection of COVID-19 Cases from Chest X-ray Images. Sci. Rep. 2020, 10, 19549. [Google Scholar] [CrossRef] [PubMed]
- Lin, Z.Q.; Shafiee, M.J.; Bochkarev, S.; Jules, M.S.; Wang, X.Y.; Wong, A. Do Explanations Reflect Decisions? A Machine-Centric Strategy to Quantify the Performance of Explainability Algorithms. arXiv 2019, arXiv:1910.07387. [Google Scholar]
- van Ginneken, B. Fifty years of computer analysis in chest imaging: Rule-based, machine learning, deep learning. Radiol. Phys. Technol. 2017, 10, 23–32. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- LeCun, Y.; Bengio, Y.; Hinton, G. Deep Learning. Nature 2015, 521, 436–444. [Google Scholar] [CrossRef]
- Huh, M.; Agrawal, P.; Efros, A.A. What makes ImageNet good for transfer learning? arXiv 2016, arXiv:1608.08614. [Google Scholar]
- Chlap, P.; Min, H.; Vandenberg, N.; Dowling, J.; Holloway, L.; Haworth, A. A Review of Medical Image Data Augmentation Techniques for Deep Learning Applications. J. Med. Imaging Radiat. Oncol. 2021, 65, 545–563. [Google Scholar] [CrossRef]
- Howard, A.G.; Zhu, M.; Chen, B.; Kalenichenko, D.; Wang, W.; Weyand, T.; Andreetto, M.; Adam, H. MobileNets: Efficient Convolutional Neural Networks for Mobile Vision Applications. arXiv 2017, arXiv:1704.04861. [Google Scholar]
- Szegedy, C.; Ioffe, S.; Vanhoucke, V.; Alemi, A.A. Inception-v4, inception-resnet and the impact of residual connections on learning. In Proceedings of the Thirty-First AAAI Conference on Artificial Intelligence, San Francisco, CA, USA, 4–9 February 2017. [Google Scholar]
- Chollet, F. Xception: Deep learning with depthwise separable convolutions. In Proceedings of the IEEE Conference on Computer Vision and Pattern Recognition, Honolulu, HI, USA, 21–26 July 2017; pp. 1251–1258. [Google Scholar]
- Deng, J.; Dong, W.; Socher, R.; Li, L.-J.; Li, K.; Fei-Fei, L. Imagenet: A large-scale hierarchical image database. In Proceedings of the 2009 IEEE Conference on Computer Vision and Pattern Recognition, Miami, FL, USA; 2009; pp. 248–255. [Google Scholar]
- Lin, M.; Chen, Q.; Yan, S. Network in Network. arXiv 2013, arXiv:1312.4400. [Google Scholar]
- Sait, U. Curated Dataset for COVID-19 Posterior-Anterior Chest Radiography Images (X-rays); Mendeley Data: London, UK, 2020. [Google Scholar]
- Wang, X.; Peng, Y.; Lu, L.; Lu, Z.; Bagheri, M.; Summers, R.M. Chestx-ray8: Hospital-scale chest x-ray database and benchmarks on weakly-supervised classification and localization of common thorax diseases. In Proceedings of the IEEE Conference on Computer Vision and Pattern Recognition, Honolulu, HI, USA, 21–26 July 2017; pp. 2097–2106. [Google Scholar]
- Wang, J.; Perez, L. The effectiveness of data augmentation in image classification using deep learning. arXiv 2017, arXiv:1712.04621. [Google Scholar]
- Shorten, C.; Khoshgoftaar, T.M. A survey on image data augmentation for deep learning. J. Big Data 2019, 6, 60. [Google Scholar] [CrossRef]
- Tartaglione, E.; Barbano, C.A.; Berzovini, C.; Calandri, M.; Grangetto, M. Unveiling COVID-19 from CHEST X-ray with Deep Learning: A Hurdles Race with Small Data. Int. J. Environ. Res. Public Health 2020, 17, 6933. [Google Scholar] [CrossRef]
- Adebayo, J.; Gilmer, J.; Muelly, M.; Goodfellow, I.; Hardt, M.; Kim, B. Sanity Checks for Saliency Maps. Adv. Neural Inf. Process. Syst. 2018, 31, 9505–9515. [Google Scholar]
- Binder, A.; Montavon, G.; Lapuschkin, S.; Müller, K.-R.; Samek, W. Layer-Wise Relevance Propagation for Neural Networks with Local Renormalization Layers. In Proceedings of the International Conference on Artificial Neural Networks, Barcelona, Spain, 6–9 September 2016; Springer: Cham, Switzerland, 2016; pp. 63–71. [Google Scholar]

| Dataset Name | Classes | Description | Total Number of Images |
|---|---|---|---|
| Multiclass | 14 | Huge dataset including normal, COVID-19, and 12 categories of abnormal X-ray scans. | 11,984 |
| Abnormality detection | 2 | Huge dataset consisting of normal and abnormal X-ray scans. In the abnormal class, X-rays corresponding to COVID-19 were also included. | 13,320 |
| Abnormality discrimination | 13 | Dataset containing 13 classes corresponding to 13 abnormalities, including COVID-19. | 8714 |
| COVID-19 detection | 2 | Dataset containing COVID-19 X-ray scans and a second class of both normal and abnormal X-ray scans (selected samples). | 2935 |
| Experiment Name | Aim | Classes Utilized |
|---|---|---|
| Multiclass | Evaluate the effectiveness of MobileNet (v2) in multiclass discrimination | 13 respiratory infections and the class normal. |
| Abnormality detection | Evaluate the effectiveness of MobileNet (v2) in abnormality detection | All respiratory infection classes, including COVID-19, were joined together into a big class. Normal X-ray scans constituted the second class. |
| Abnormality discrimination | Evaluate the effectiveness of MobileNet (v2) in distinguishing between various diseases, including COVID-19 | 13 classes of X-rays corresponding to 13 respiratory infections |
| COVID-19 detection | Evaluate the effectiveness of MobileNet (v2) in distinguishing between COVID-19 and non-COVID-19 X-ray scans | Selected samples from the 12 respiratory diseases constituted the first class, whereas the second class referred to COVID-19. |
| Reproducibility | Evaluate the reproducibility of features when MobileNet is trained distinguishing between COVID-19 and non-COVID-19 X-ray scans | Selected samples from the 12 respiratory diseases constituted the first class, whereas the second class referred to COVID-19. |
| Dataset | Accuracy (%) | AUC Score (%) |
|---|---|---|
| Multiclass | 73.11 ± 2.21 | 94.07 ± 1.45 |
| Abnormality detection | 95.45 ± 1.54 | 98.92 ± 0.83 |
| Abnormality discrimination | 62.26 ± 4.21 | 90.93 ± 1.57 |
| COVID-19 detection | 89.88 ± 3.66 | 96.26 ± 2.14 |
| Run | Mean Accuracy (%) |
|---|---|
| 1 | 89.88 |
| 2 | 91.23 |
| 3 | 88.54 |
| 4 | 92.14 |
| 5 | 89.24 |
| 6 | 89.36 |
| 7 | 88.53 |
| 8 | 88.86 |
| 9 | 90.76 |
| 10 | 88.86 |
| 11 | 91.23 |
| 12 | 90.37 |
| 13 | 92.43 |
| 14 | 89.02 |
| 15 | 89.67 |
| 16 | 88.54 |
| 17 | 90.79 |
| 18 | 89.36 |
| 19 | 87.13 |
| 20 | 91.23 |
| 21 | 86.98 |
| 22 | 88.86 |
| 24 | 92.41 |
| 24 | 90.66 |
| 25 | 91.23 |
| Overall | 89.89 |
| Std | ±1.49 |
| Factor | Result for Accuracy |
|---|---|
| Mean | 89.89 |
| Variance | 2.24 |
| Observations | 25 |
| T-statistic | 0.0413 |
| p-value | 0.4836 |
| Study | Method | Test Data Size | Classes | Accuracy |
|---|---|---|---|---|
| Hou and Gao [12] | Deep CNN | 400 | 4 (normal, bacterial pneumonia, viral pneumonia, COVID-19) | 96% (COVID-19 vs. ALL) |
| Ahsan et al. [13] | VGG, MobilNet (v2) | 518 | 2 (COVID-19, non-COVID-19) | 95% |
| Brunese et al. [4] | VGG-16 | 1100 | 2 (COVID-19, other disease) | 97% |
| Apostolopoulos et al. [5] | MobileNet (v2) | 1428 | 3 (normal, pneumonia, COVID-19) | 93% |
| Apostolopoulos et al. [5] | MobileNet (v2) | 1428 | 2 (COVID-19, non-COVID-19) | 93% |
| Apostolopoulos et al. [9] | MobileNet (v2) | 3905 | 2 (COVID-19, non-COVID-19) | 99% |
| Apostolopoulos et al. [9] | MobileNet (v2) | 3905 | 7 (COVID-19, normal, 6 abnormal classes) | 87% |
| Wang et al. [15] | tailored CNN (COVID-Net) | 300 | 3 (normal, pneumonia, COVID-19) | 93% |
| This study | MobileNet (v2) | 13,320 | 2 (COVID-19, other abnormal X-ray) | 90% |
| This study | MobileNet (v2) | 11,984 | 7 (normal, COVID-19, 5 abnormal classes) | 73% |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Apostolopoulos, I.D.; Apostolopoulos, D.J.; Papathanasiou, N.D. Deep Learning Methods to Reveal Important X-ray Features in COVID-19 Detection: Investigation of Explainability and Feature Reproducibility. Reports 2022, 5, 20. https://doi.org/10.3390/reports5020020
Apostolopoulos ID, Apostolopoulos DJ, Papathanasiou ND. Deep Learning Methods to Reveal Important X-ray Features in COVID-19 Detection: Investigation of Explainability and Feature Reproducibility. Reports. 2022; 5(2):20. https://doi.org/10.3390/reports5020020
Chicago/Turabian StyleApostolopoulos, Ioannis D., Dimitris J. Apostolopoulos, and Nikolaos D. Papathanasiou. 2022. "Deep Learning Methods to Reveal Important X-ray Features in COVID-19 Detection: Investigation of Explainability and Feature Reproducibility" Reports 5, no. 2: 20. https://doi.org/10.3390/reports5020020
APA StyleApostolopoulos, I. D., Apostolopoulos, D. J., & Papathanasiou, N. D. (2022). Deep Learning Methods to Reveal Important X-ray Features in COVID-19 Detection: Investigation of Explainability and Feature Reproducibility. Reports, 5(2), 20. https://doi.org/10.3390/reports5020020







