Abstract
The clinical presentation of COVID-19 is non-specific, and to improve and limit the spread of the SARS-CoV-2 virus, an accurate diagnosis with a robust method is needed. A total of 500 nasopharyngeal swab specimens were tested for SARS-CoV-2. Of these, 184 samples were found to be positive with Allplex™ 2019-nCoV Assay, which is fully automated. All the positive samples were retested with TaqPath™ COVID-19 CE-IVD RT-PCR Kit (after this, referred to as TaqPath™ COVID-19), semi-automated. The comparison of RT-qPCR for SARS-CoV-2 genes target points shows only one target point in common, the N gene. Therefore, the N gene was used to compare both assays. We noticed different Ct values between the tests. Therefore, samples were divided into four groups depending to the Ct value results: (1) Ct < 25, (2) Ct 25–30, (3) Ct 30–35, (4) Ct > 35. TaqPath™ COVID-19 Kit reconfirmed the results obtained from Allplex™ 2019-nCoV Assay. In conclusion, both the Allplex™ 2019-nCoV assay and TaqPath™ COVID-19 tests accurately confirm the diagnosis of SARS-CoV-2 infection. Even if TaqPath™ COVID-19 has a semi-automated workflow, it does not introduce bias in the diagnostic screening of SARS-CoV-2, and it supports the indirect identification of variants of concern to undergo sequencing.
1. Introduction
Two years have passed since the first coronavirus disease 2019 (COVID-19) case in December 2019. According to the exponential progression in the number of infections globally, COVID-19 has become one of the most significant pandemics in modern history. The clinical relationship between COVID-19 and symptoms is non-specific, and to improve and limit the spread of the SARS-CoV-2 virus, an accurate diagnosis through a robust method is mandatory [,]. Molecular epidemiology of SARS-CoV-2 offers new avenues to investigate associations between genetic and environmental factors of the disease []. Real-time reverse-transcription polymerase chain reaction (RT-qPCR) tests are performed on respiratory samples. Therefore, nasopharyngeal, nasal, oropharyngeal, and/or, when hospitalized, bronchoalveolar lavage, are analyzed. RT-qPCR represents the gold standard, and viral presence could appear earlier, even before clinical symptoms [,]. The viral genome consists of several genes encoding non-structural, structural, and accessory proteins. In the viral genome, genes for four structural proteins (spike surface glycoprotein S, envelope E, membrane M, and nucleocapsid N) and several accessory proteins are located []. For diagnostic and screening monitoring, ORF1ab/RdRp, E, N, and S genes are the targets most frequently used by the RT-qPCR method []. However, according to Coronavirus COVID-19 Global Cases by the Center for Systems Science and Engineering (CSSE) at Johns Hopkins University (JHU), virus variants of COVID-19 are originating, even in our region (Figure 1) (accessed on 22 January 2022) []. Therefore, in the present study, we compared two commercial Reverse Transcriptase (RT) polymerase chain reaction (PCR) kits for COVID-19 diagnosis, all having the N gene in common. Therefore, the N gene was used to compared both kits, the Allplex™ 2019-nCoV assay, and TaqPath™ COVID-19 tests. By this latter, a peculiar drop-off of the S gene was evidenced and indicative of a possible variant of concern (VOC) that was sequenced by whole-genome-based next-generation sequencing (NGS) [,].
Figure 1.
Adapted from COVID-19 Map–Johns Hopkins Coronavirus Resource Center (accessed on 22 January 2022).
2. Materials and Methods
We performed a prospective study on specimens referred to the Department of Microbiology and Virology of Pugliese Ciaccio’s Hospital in Catanzaro from 4 to 24 December 2021. All study participants underwent a nasopharyngeal swab and real-time reverse-transcription polymerase chain reaction (RT-PCR) analysis for SARS-CoV-2. This study is part of the clinical trial recorded in clinicaltrial.gov (accessed on 25 October 2021) (NCT04322513) and was conducted in compliance with the Institutional Review Board/Human Subjects Research Committee requirements.
Experimental protocol
At the time of the study, for each patient, samples were collected in Universal Transport Media (UTM), opened in biosafety cabinet class-II, and then 600 μL of the UTM were further processed for viral nucleic acid extraction.
The viral nucleic acid extracted was evaluated through 2 RT-PCR kits specific for COVID-19:
- (i).
- 200 μL of UTM was extracted for Allplex™ 2019-nCoV Assay (Seegene-Seoul, South Korea) with the fully Automated Liquid Handling Workstations NIMBUS which also arrange the PCR plate and the Real-Time PCR System from Bio-Rad CFX96™ Dx (Bio-Rad, Hercules, CA, USA);
- (ii).
- TaqPath™ COVID-19 CE-IVD RT-PCR Kit (after this referred to as TaqPath™ COVID-19) with semi-automatic KingFisher Duo Prime by Thermofisher. Briefly, by extracting a 200 μL aliquot of specimen in UTM using the MagMAX™ Viral/Pathogen Nucleic Acid isolation kit on the KingFisher Flex Purification system (Thermo Fisher Scientific, Waltham, MA, USA). Before RNA extraction, 10 μL of Proteinase K was added to each well in the KingFisher™ Deep 96-well Plate. In addition, 10 μL of the MS2 Phage Control was added to all specimens together with 10 μL of magnetic beads.
Elution volumes from the two workstations were different, at 100 µL and 50 µL for the Automated Liquid Handling Workstations NIMBUS and KingFisher Duo Prime, respectively. For both kits, a final volume of 25 µL was used in the reaction master mix and dispended as follows: Allplex™ 2019-nCoV Assay 17 µL of its master mix plus 8 µL of RNA-extracted sample; TaqPath™ COVID-19 15 µL of its master mix and 10 µL of RNA extracted sample.
2.1. Allplex™ 2019-nCoV Assay
All the samples included in the study were tested using Allplex™ 2019-nCoV Assay, following the manufacturer’s manual (Ref: RV10284X Lot: RVA321D04). Accordingly, a sample is considered positive if at least N or RdRp targets amplify with cycle threshold (Ct) Ct ≤ 40. If only the E gene target is amplified with Ct ≤ 40, the sample is considered “presumptive positive” []. The range of Ct, only for the N gene, is reported in Table 1.

Table 1.
Gene target points similarity.
2.2. TaqPath™ COVID-19
All the samples included in the study were tested using TaqPath™ COVID-19 CE-IVD RT-PCR Kit, following the manufacturer’s manual (Ref: A48099 Lot: 2101036). Accordingly, a sample is considered positive if any of the targets (either ORF1ab, N or S) amplify with Ct ≤ 37, or the internal control MS2 (Bacteriophage MS2) with amplifying Ct value ≤ 32 []. The range of Ct, for the N gene and MS2, is reported in Table 2.

Table 2.
N gene Ct value tested with Allplex™ 2019-nCoV Assay and TaqPath™ COVID-19.
2.3. Sequencing SARS-CoV-2
The NGS approach by MiSeq System (Illumina, San Diego, CA, USA), provided 2 × 250 bp read data. The SOPHIA DDM Platform analyzed FASTQ reads. Clade analysis was performed by ICOGEN Platform. Then, lineage information was described using the Pangolin nomenclatures [,].
2.4. Statistical Analysis
All data were analyzed using the GraphPad 5.0 statistical program (GraphPad Software Inc., San Diego, CA, USA) by evaluating the standard deviation (SD). Data were analyzed by one-way ANOVA analysis of variance followed by Tukey’s multiple comparison test; HSD (honestly significant difference) was used to estimate difference amongst groups. The online free software from GraphPad Software Inc., CA-USA, was used (https://www.graphpad.com/quickcalcs/kappa1/, accessed on 25 January 2022) for the Cohen’s kappa determination in order to obtain a value of the inter-observer agreement.
3. Results and Discussion
A total of 500 nasopharyngeal swab specimens were tested for SARS-CoV-2. Of these, 184 samples were positive with Allplex™ 2019-nCoV Assay (36.8%), and 316 were negative (63.2%). Positive samples from Allplex™ 2019-nCoV Assay were re-tested with TaqPath™ COVID-19 and 50 negative pooled samples (coming from 10 of each specimen), which were negative. The comparison of RT-qPCR for SARS-CoV-2 gene target points is presented in Table 1 (updated 29 December 2021, online sources) [,]. The common gene was the N gene (Table 1). Therefore, it was used to compare both assays.
The TaqPath™ COVID-19 Kit reconfirmed the results obtained from Allplex™ 2019-nCoV Assay, but we noticed a different distribution of Ct values. Therefore, samples were divided into four groups depending on the cycle threshold (Ct) value results: (1) Ct < 25, (2) Ct 25–30, (3) Ct 30–35, (4) Ct > 35 (Figure 2A,B).
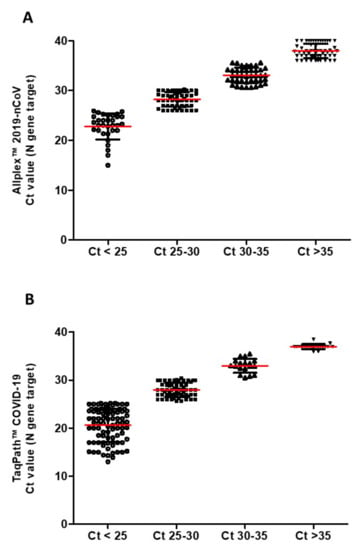
Figure 2.
(A) Allplex™ 2019-nCoV Assay and (B) TaqPath™ COVID-19. In red, the average value is indicated for each group.
Samples that display the N gene with a Ct < 25 (Group 1) are almost three times greater with TaqPath™ COVID-19 compared to Allplex™ 2019-nCoV Assay, and any significant difference is highlighted between them (see Figure 3). Both TaqPath™ COVID-19 and Allplex™ 2019-nCoV Assay have similar samples at Ct 25–30 (Group 2). In contrast, samples with Ct values > 30 (Groups 3 and 4) were reached with Allplex™ 2019-nCoV Assay (Table 2). These results are consistent with the difference in elution volume used in the two workstations: 50 µL in TaqPath™ COVID-19 and 100 µL in Allplex™ 2019-nCoV. This difference could result in more diluted nucleic acids in the second kit (Allplex™ 2019-nCoV). Another reason could be the different complementarity of the primers’ probe, crucial for PCR, where mismatch can give up to 7 Ct of difference [].
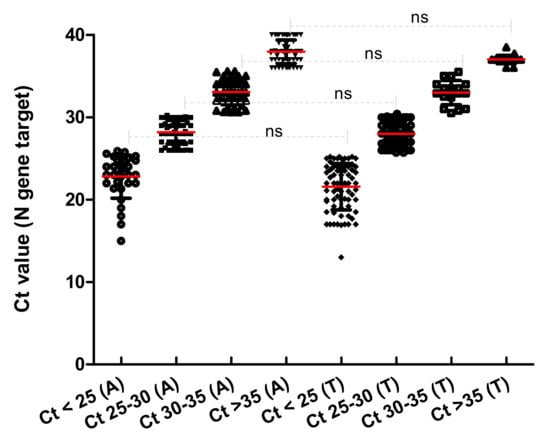
Figure 3.
Comparison of Ct groups data from Allplex™ 2019-nCoV (A) TaqPath™ COVID-19 (T).
Tukey’s multiple comparison test HSD statistical analysis was performed to evaluate the differences amongst the groups. No difference was highlighted between groups and the two analytical tests (see Figure 2).
Both Allplex™ 2019-nCoV Assay and TaqPath™ COVID-19 show similar agreement for the N genes. Kappa Cohen calculation for both Allplex™ 2019-nCoV Assay and TaqPath™ COVID-19 displays the following data. Number of observed agreements: 500 (100.00% of the observations) and number of agreements expected by chance: 267.4 (53.48% of the observations), Kappa = 1.000 and SE of Kappa = 0.000, 95% confidence interval from 1.000 to 1.000. Besides that, the diagnosis made by the infectious disease specialist shows an overall agreement of 100% and a Kappa value of 1 for both tests. Furthermore, in the 184 positive samples re-tested with TaqPath™ COVID-19, drop off of the S gene (indicative of VOC) was observed in four samples. Normally in routine practice, the samples that underwent sequencing are chosen randomly considering only that the Ct value must be lower than 25. By including TaqPath™ COVID-19 in our clinical laboratory setting, we were facilitated in this choice. In fact, the four samples with the drop-off S gene underwent genomic characterization by NGS sequencing, resulting in four Omicron VOC, as reported in Table 3.

Table 3.
NGS sequencing and amino acidic mutations.
Although the N gene in VOC Omicron presents several mutations, the amplification curve for this gene was not different between Allplex™ 2019-nCoV Assay and TaqPath™ COVID-19 (data not shown).
It is worth noting that real-time reverse-transcription polymerase chain reaction (RT-PCR) assays for SARS-CoV-2 RNA detection in clinical specimens are widely used in COVID-19 diagnostic laboratories as the standard-gold method. Although the Allplex™ 2019-nCoV Assay amplifies up to 40 cycles and states that any gene with Ct < 40 is a positive result, there is increasing appreciation that Ct 30–35 is considered borderline [,,]. This issue is almost overcome by TaqPath™ COVID-19, as shown here. On the other hand, it was also reported that, in effect, the screening depends largely on frequency of testing and speed of reporting, and is only marginally improved by high test sensitivity, even in acute infection, therefore even specific biomarker is needed [,,,].
4. Conclusions
The clinical presentation of COVID-19 is non-specific, and to improve and limit the spread of the SARS-CoV-2 virus, accurate diagnosis with a robust method is needed [], even in light of reinfection []. Herein, we show that both, Allplex™ 2019-nCoV Assay and TaqPath™ COVID-19 tests are accurate enough to confirm the diagnosis of SARS-CoV-2 infection. The two methods tested are similar, with one substantial difference: TaqPath™ COVID-19 is performed in a semi-automated way. Despite this, TaqPath™ COVID-19 does not introduce bias in the diagnostic screening of SARS-CoV-2 and supports clinicians in the choice of VOC for NGS determination.
Author Contributions
Conceptualization, E.C., G.D.M. and P.M.; methodology, I.T.; E.C., R.T., C.P. and M.C.; software, E.C.; writing—original draft preparation, E.C. and M.C.; writing—review and editing, E.C., G.D.M. and L.G.; supervision, P.M. All authors have read and agreed to the published version of the manuscript.
Funding
This research received no external funding.
Institutional Review Board Statement
All the procedures followed the 1964 Helsinki Declaration and later amendments.
Informed Consent Statement
The study was conducted using residual, de-identified specimens, and no clinical or demographic information was collected. According to Italian health public law, this type of study did not require specific, informed consent and ethics committee approval.
Data Availability Statement
Data supporting reported results can be found at the Department of Microbiology and Virology, Pugliese Ciaccio’s Hospital, Catanzaro, Italy.
Conflicts of Interest
The authors declare no conflict of interest.
References
- Clinical Characteristics of COVID-19. Available online: https://www.ecdc.europa.eu/en/covid-19/latest-evidence/clinical (accessed on 25 June 2021).
- Yuen, K.S.; Ye, Z.W.; Fung, S.Y.; Chan, C.P.; Jin, D.Y. SARS-CoV-2 and COVID-19: The most important research questions. Cell Biosci. 2020, 10, 40. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Recommendations for National SARS-CoV-2 Testing Strategies and Diagnostic Capacities. Available online: https://www.who.int/publications/i/item/WHO-2019-nCoV-Surveillance_Case_Definition-2020.2 (accessed on 7 September 2021).
- Kostaki, E.G.; Pavlopoulos, G.A.; Verrou, K.M.; Ampatziadis-Michailidis, G.; Harokopos, V.; Hatzis, P.; Moulos, P.; Siafakas, N.; Pournaras, S.; Hadjichristodoulou, C.; et al. Molecular Epidemiology of SARS-CoV-2 in Greece Reveals Low Rates of Onward Virus Transmission after Lifting of Travel Restrictions Based on Risk Assessment during Summer 2020. mSphere 2021, 6, e0018021. [Google Scholar] [CrossRef] [PubMed]
- Luciani, F.; Cione, E.; Caroleo, M.C.; Colosimo, M.; Zanolini, A.; Barca, A.; Cosimo, S.; Pasqua, P.; Gallelli, L. SARS-CoV-2 Translocate from Nasopharyngeal to Bronchoalveolar Site: A Case Presentation. Reports 2020, 3, 23. [Google Scholar] [CrossRef]
- Kohmer, N.; Eckermann, L.; Böddinghaus, B.; Götsch, U.; Berger, A.; Herrmann, E.; Kortenbusch, M.; Tinnemann, P.; Gottschalk, R.; Hoehl, S.; et al. Self-Collected Samples to Detect SARS-CoV-2: Direct Comparison of Saliva, Tongue Swab, Nasal Swab, Chewed Cotton Pads and Gargle Lavage. J. Clin. Med. 2021, 10, 5751. [Google Scholar] [CrossRef] [PubMed]
- Naqvi, A.A.T.; Fatima, K.; Mohammad, T.; Fatima, U.; Singh, I.K.; Singh, A.; Atif, S.M.; Hariprasad, G.; Hasan, G.M.; Hassan, M.I. Insights into SARS-CoV-2 genome, structure, evolution, pathogenesis and therapies: Structural genomics approach. Biochim. Biophys. Acta Mol. Basis Dis. 2020, 1866, 165878. [Google Scholar] [CrossRef] [PubMed]
- Coronavirus COVID-19 Global Cases by Johns Hopkins. Available online: https://coronavirus.jhu.edu/map.html (accessed on 22 January 2022).
- Rambaut, A.; Holmes, E.C.; O’Toole, Á.; Hill, V.; McCrone, J.T.; Ruis, C.; du Plessis, L.; Pybus, O.G. A dynamic nomenclature proposal for SARS-CoV-2 lineages to assist genomic epidemiology. Nat. Microbiol. 2020, 5, 1403–1407. [Google Scholar] [CrossRef] [PubMed]
- Peronace, C.; Tallerico, R.; Colosimo, M.; De Fazio, M.; Pasceri, F.; Talotta, I.; Panduri, G.; Pintomalli, L.; Oteri, R.; Calantoni, V.; et al. BA.1 Omicron Variant of SARS-CoV-2: First Case Reported in Calabria Region, Italy. COVID 2022, 2, 211–215. [Google Scholar] [CrossRef]
- Seegene. Allplex 2019-nCoV Assay (Version 2.2; 15 April 2021), Instruction for Use. Available online: https://www.fda.gov/media/137178/download (accessed on 4 December 2021).
- TaqPath™ COVID-19 CE-IVD RT-PCR Kit MAN0019215, Revision F.0. Available online: https://assets.thermofisher.com/TFS-Assets/LSG/manuals/MAN0019215_TaqPathCOVID-19_CE-IVD_RT-PCR%20Kit_IFU.pdf (accessed on 4 December 2021).
- Classification of Omicron (B.1.1.529): SARS-CoV-2 Variant of Concern. Available online: https://www.who.int/news/item/26-11-2021-classification-of-omicron-(b.1.1.529)-sars-cov-2-variant-of-concern (accessed on 1 January 2022).
- COVID-19 Diagnostics Resource Centre. Available online: https://www.finddx.org/covid-19/ (accessed on 29 December 2021).
- Stadhouders, R.; Pas, S.D.; Anber, J.; Voermans, J.; Mes, T.H.; Schutten, M. The effect of primer-template mismatches on the detection and quantification of nucleic acids using the 5′ nuclease assay. J. Mol. Diagn. 2010, 12, 109–117. [Google Scholar] [CrossRef] [PubMed]
- Rabaan, A.A.; Tirupathi, R.; Sule, A.A.; Aldali, J.; Mutair, A.A.; Alhumaid, S.; Muzaheed Gupta, N.; Koritala, T.; Adhikari, R.; Bilal, M.; et al. Viral Dynamics and Real-Time RT-PCR Ct Values Correlation with Disease Severity in COVID-19. Diagnostics 2021, 11, 1091. [Google Scholar] [CrossRef] [PubMed]
- Larremore, D.B.; Wilder, B.; Lester, E.; Shehata, S.; Burke, J.M.; Hay, J.A.; Tambe, M.; Mina, M.J.; Parker, R. Test sensitivity is secondary to frequency and turnaround time for COVID-19 screening. Sci. Adv. 2021, 7, eabd5393. [Google Scholar] [CrossRef] [PubMed]
- Mina, M.J.; Parker, R.; Larremore, D.B. Rethinking Covid-19 Test Sensitivity—A Strategy for Containment. N. Engl. J. Med. 2020, 383, e120. [Google Scholar] [CrossRef] [PubMed]
- Cione, E.; Siniscalchi, A.; Gangemi, P.; Cosco, L.; Colosimo, M.; Longhini, F.; Luciani, F.; De Sarro, G.; G&SPWorking Group Berrino, L.; D’Agostino, B.; et al. Neuron-specific enolase serum levels in COVID-19 are related to the severity of lung injury. PLoS ONE 2021, 16, e0251819. [Google Scholar] [CrossRef] [PubMed]
- Bentivegna, E.; Sentimentale, A.; Luciani, M.; Speranza, M.L.; Guerritore, L.; Martelletti, P. New IgM seroconversion and positive RT-PCR test after exposure to the virus in recovered COVID-19 patient. J. Med. Virol. 2021, 93, 97–98. [Google Scholar] [CrossRef] [PubMed]
- Smith, R.L.; Gibson, L.L.; Martinez, P.P.; Ke, R.; Mirza, A.; Conte, M.; Gallagher, N.; Conte, A.; Wang, L.; Fredrickson, R.; et al. Longitudinal Assessment of Diagnostic Test Performance Over the Course of Acute SARS-CoV-2 Infection. J. Infect. Dis. 2021, 224, 976–982. [Google Scholar] [CrossRef] [PubMed]
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).