Machine Learning Models for Abnormality Detection in Musculoskeletal Radiographs
Abstract
1. Introduction
2. Experimental Section
2.1. Data
2.2. Study Design
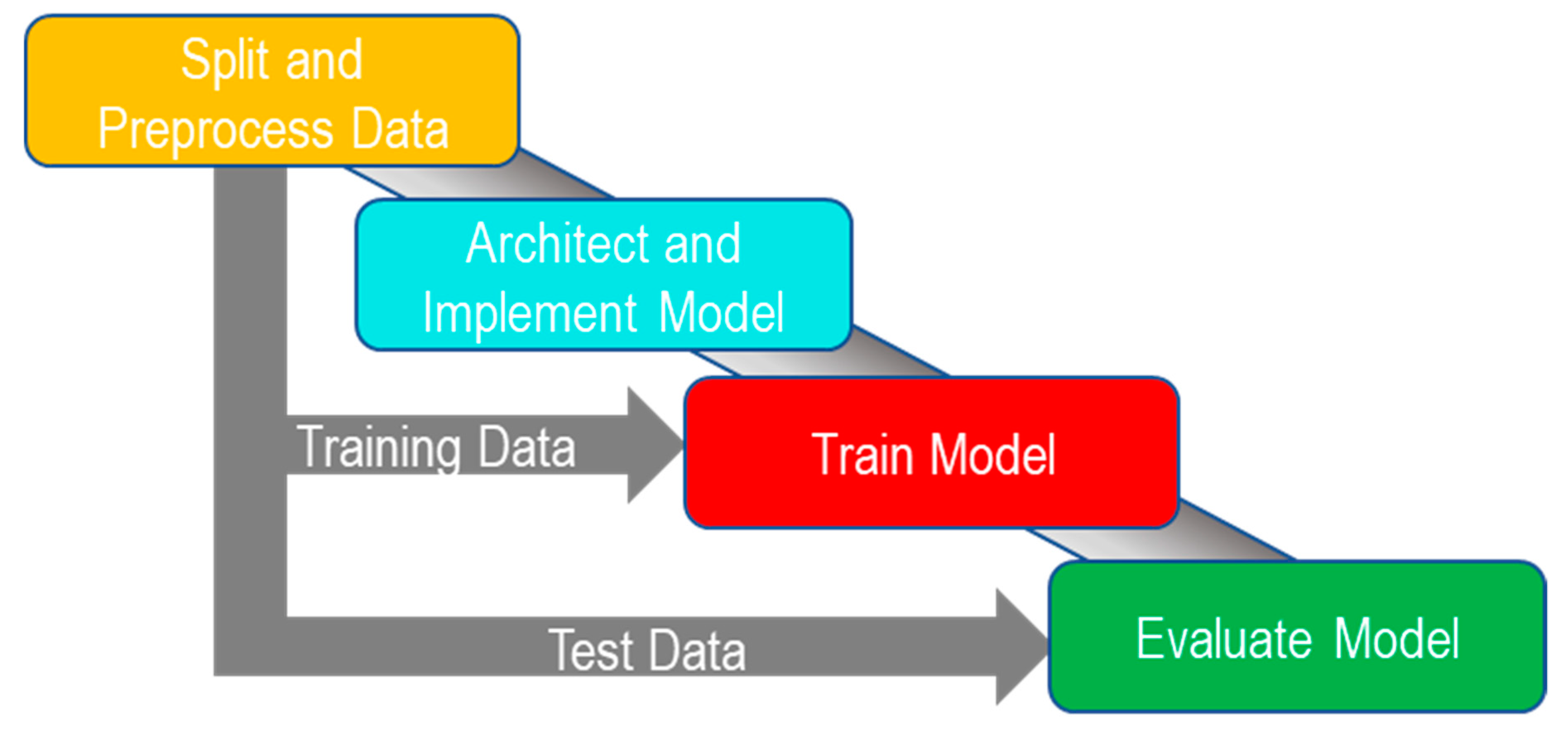
2.2.1. Overview
2.2.2. Splitting and Preprocessing Data
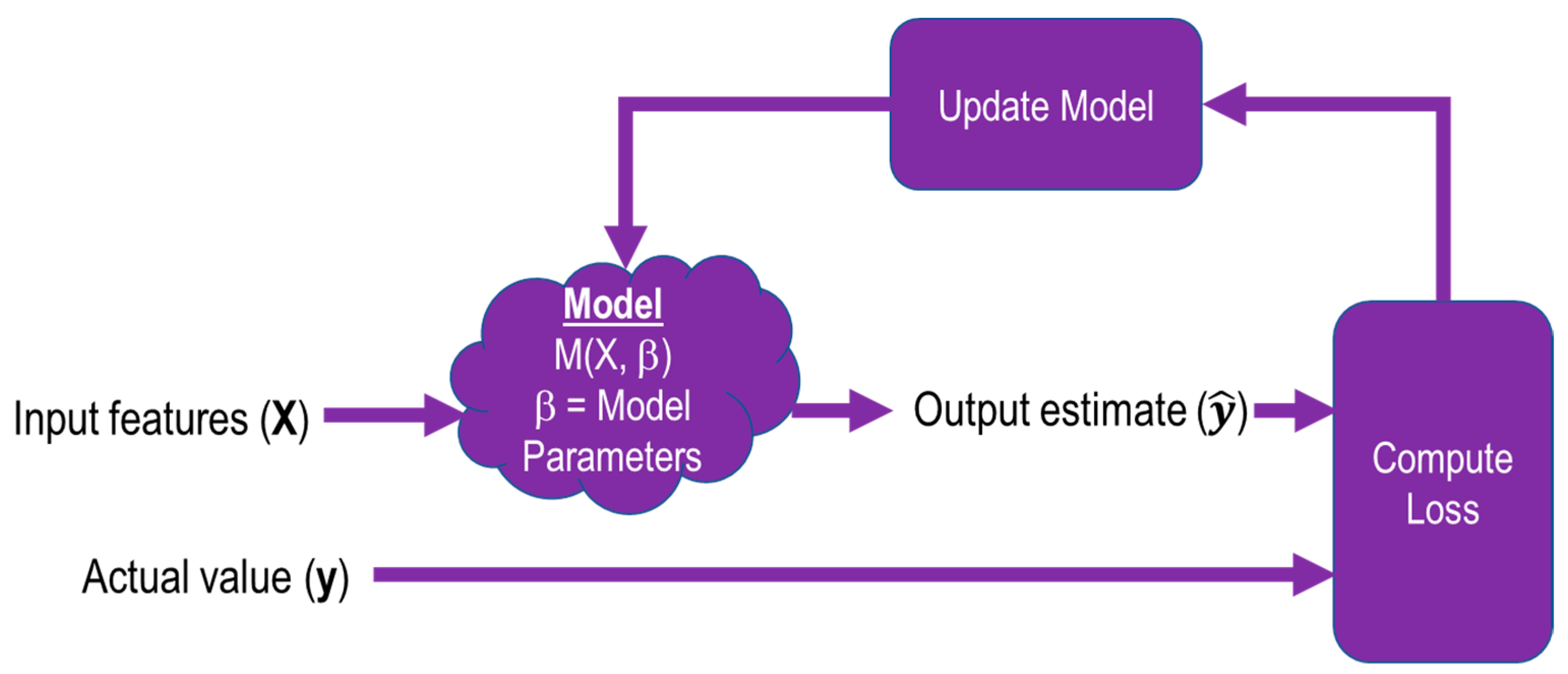
2.2.3. Architecture and Implementation
2.2.4. Training
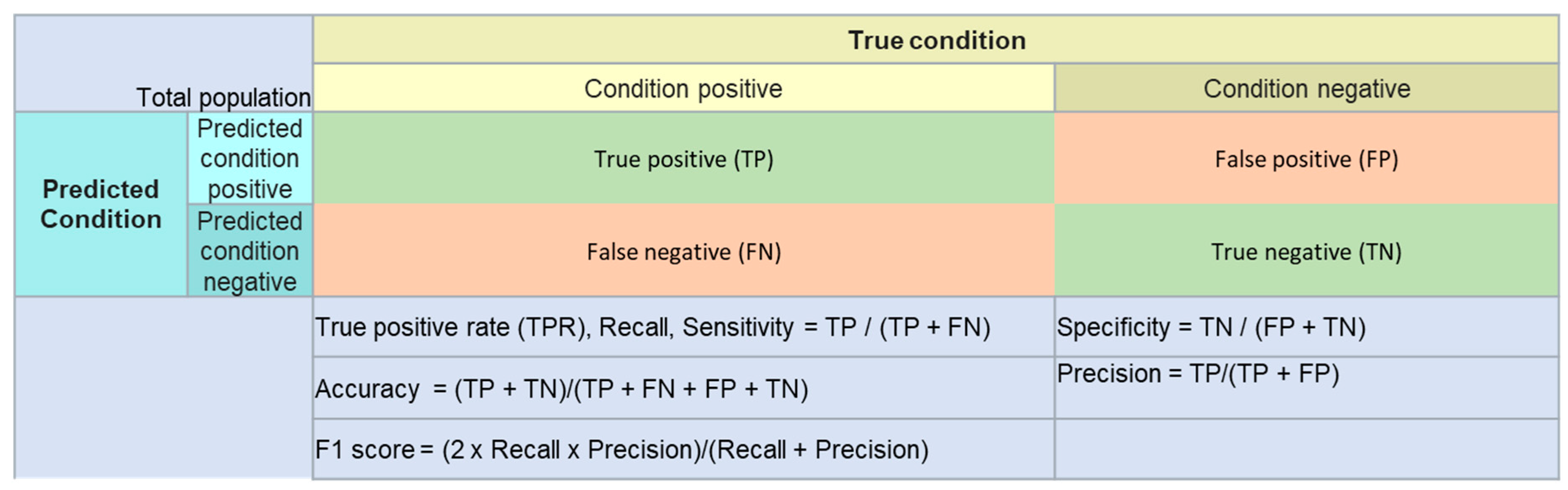
2.3. Model Evaluation
2.4. Software and Hardware
- Keras 2.2.4 and TensorFlow 1.12.0 for model development
- Scikit-Learn 0.20.2 for calculating metrics
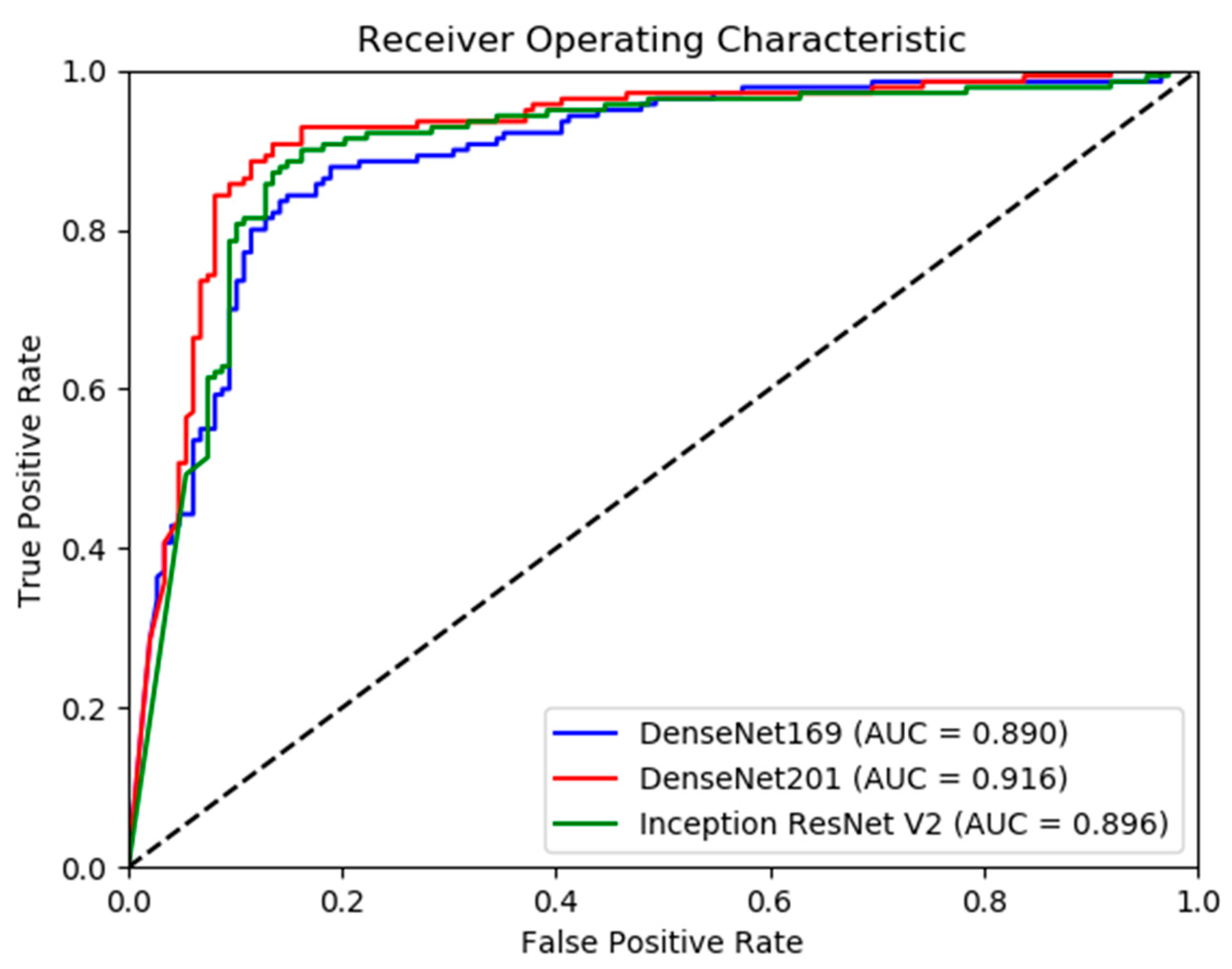
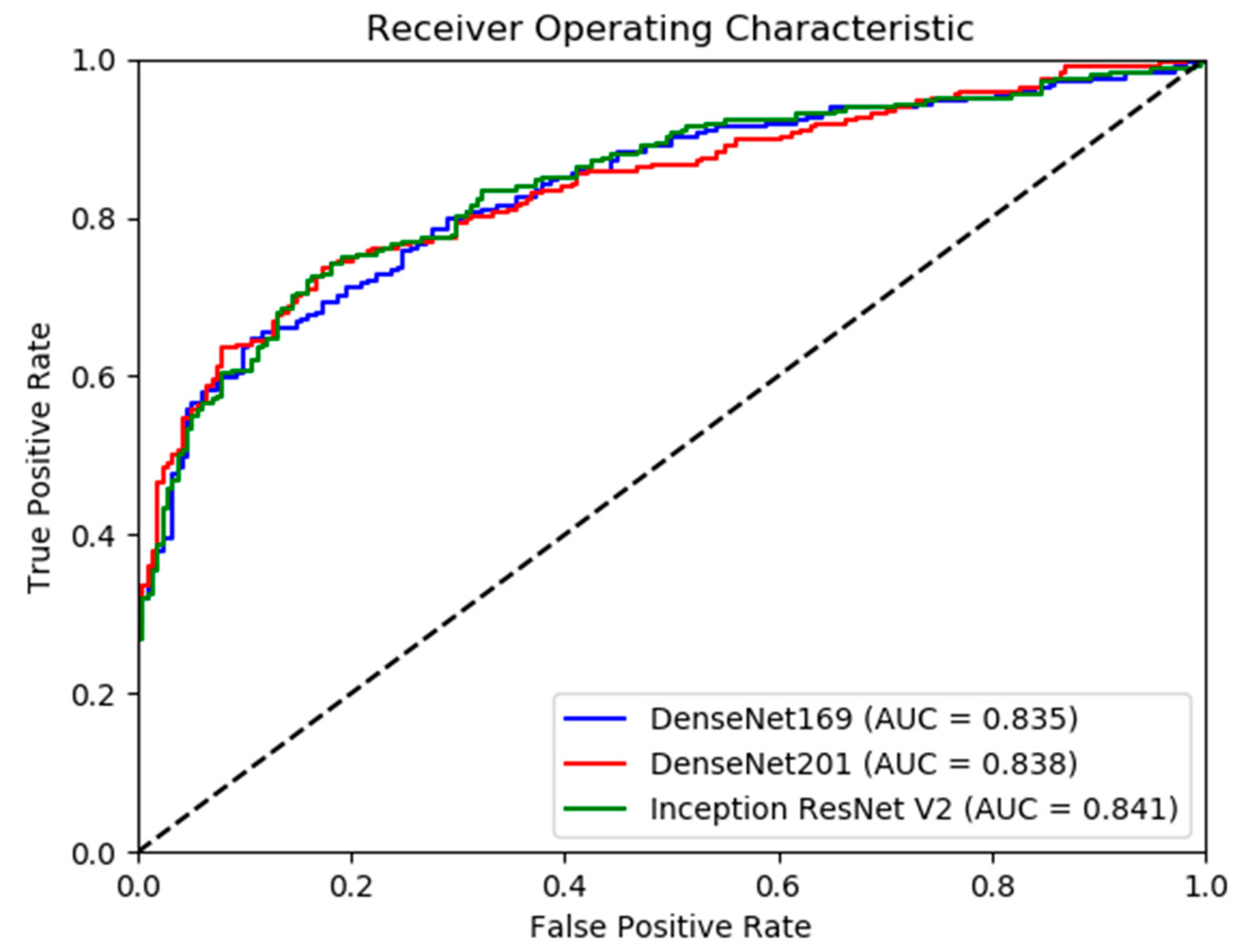
3. Results
Model Performance
4. Discussion
5. Conclusions
Funding
Acknowledgments
Conflicts of Interest
References
- United States Bone and Joint Initiative: The Burden of Musculoskeletal Diseases in the United States (BMUS). Available online: http://www.boneandjointburden.org (accessed on 14 June 2019).
- McDonald, R.J.; Schwartz, K.M.; Eckel, L.J.; Diehn, F.E.; Hunt, C.H.; Bartholmai, B.J.; Erickson, B.J.; Kallmes, D.F. The effects of changes in utilization and technological advancements of cross-sectional imaging on radiologist workload. Acad. Radiol. 2015, 22, 1191–1198. [Google Scholar] [CrossRef] [PubMed]
- Robert, L. Musculoskeletal Hits Top 10 in Primary Care Visits. Available online: https://www.patientcareonline.com/musculoskeletal-disorders/musculoskeletal-hits-top-10-primary-care-visits (accessed on 15 July 2019).
- Scher, D.L.; Ferreira, J.V.; Cote, M.; Abdelgawad, A.; Wolf, J.M. The need for musculoskeletal education in primary care residencies. Orthopedics 2014, 37, 511–513. [Google Scholar] [CrossRef] [PubMed]
- Freedman, K.B.; Bernstein, J. Educational deficiencies in musculoskeletal medicine. J. Bone Jt. Surg. Am. 2002, 84, 604–608. [Google Scholar] [CrossRef] [PubMed]
- Freedman, K.B.; Bernstein, J. The adequacy of medical school education in musculoskeletal medicine. J. Bone Jt. Surg. Am. 1998, 80, 1421–1427. [Google Scholar] [CrossRef]
- Grace, K.; Salvatier, J.; Dafoe, A.; Zhang, B.; Evans, O. When will AI exceed human performance? Evidence from AI experts. J. Artif. Intell. Res. 2018, 62, 729–754. [Google Scholar] [CrossRef]
- LeCun, Y.; Bengio, Y.; Hinton, G. Deep learning. Nature 2015, 521, 436–444. [Google Scholar] [CrossRef]
- Rajpurkar, P.; Irvin, J.; Ball, R.L.; Zhu, K.; Yang, B.; Mehta, H.; Duan, T.; Ding, D.; Bagul, A.; Langlotz, C.P.; et al. Deep learning for chest radiograph diagnosis: A retrospective comparison of the CheXNeXt algorithm to practicing radiologists. PLoS Med. 2018, 15, e1002686. [Google Scholar] [CrossRef]
- Taylor, A.G.; Mielke, C.; Mongan, J. Automated detection of moderate and large pneumothorax on frontal chest X-rays using deep convolutional neural networks: A retrospective study. PLoS Med. 2018, 15, e1002697. [Google Scholar] [CrossRef]
- Zech, J.R.; Badgeley, M.A.; Liu, M.; Costa, A.B.; Titano, J.J.; Oermann, E.K. Variable generalization performance of a deep learning model to detect pneumonia in chest radiographs: A cross-sectional study. PLoS Med. 2018, 15, e1002683. [Google Scholar] [CrossRef]
- Rajpurkar, P.; Irvin, J.; Bagul, A.; Ding, D.; Duan, T.; Mehta, H.; Yang, B.; Zhu, K.; Laird, D.; Ball, R.L.; et al. MURA: Large Dataset for Abnormality Detection in Musculoskeletal Radiographs. arXiv 2017, arXiv:1712.06957. [Google Scholar]
- Deng, J.; Dong, W.; Socher, R.; Li, L.-J.; Li, K.; Fei-Fei, L. Imagenet: A large-scale hierarchical image database. In Proceedings of the 2009 IEEE Conference on Computer Vision and Pattern Recognition, Miami, FL, USA, 20–25 June 2009; pp. 248–255. [Google Scholar]
- Mormont, R.; Geurts, P.; Marée, R. Comparison of deep transfer learning strategies for digital pathology. In Proceedings of the IEEE Conference on Computer Vision and Pattern Recognition Workshops, Salt Lake City, UT, USA, 18–22 June 2018; pp. 2262–2271. [Google Scholar]
- Esteva, A.; Kuprel, B.; Novoa, R.A.; Ko, J.; Swetter, S.M.; Blau, H.M.; Thrun, S. Dermatologist-level classification of skin cancer with deep neural networks. Nature 2017, 542, 115. [Google Scholar] [CrossRef] [PubMed]
- Litjens, G.; Kooi, T.; Bejnordi, B.E.; Setio, A.A.A.; Ciompi, F.; Ghafoorian, M.; Van Der Laak, J.A.; Van Ginneken, B.; Sánchez, C.I. A survey on deep learning in medical image analysis. Med. Image Anal. 2017, 42, 60–88. [Google Scholar] [CrossRef] [PubMed]
- Kensert, A.; Harrison, P.J.; Spjuth, O. Transfer Learning with Deep Convolutional Neural Networks for Classifying Cellular Morphological Changes. SLAS DISCOVERY Adv. Life Sci. R D 2019, 24, 466–475. [Google Scholar] [CrossRef] [PubMed]
- Gulshan, V.; Peng, L.; Coram, M.; Stumpe, M.C.; Wu, D.; Narayanaswamy, A.; Venugopalan, S.; Widner, K.; Madams, T.; Cuadros, J.; et al. Development and Validation of a Deep Learning Algorithm for Detection of Diabetic Retinopathy in Retinal Fundus PhotographsAccuracy of a Deep Learning Algorithm for Detection of Diabetic RetinopathyAccuracy of a Deep Learning Algorithm for Detection of Diabetic Retinopathy. JAMA 2016, 316, 2402–2410. [Google Scholar] [CrossRef] [PubMed]
- Huang, G.; Liu, Z.; van der Maaten, L.; Weinberger, K.Q. Densely Connected Convolutional Networks. arXiv 2016, arXiv:1608.06993. [Google Scholar]
- Szegedy, C.; Ioffe, S.; Vanhoucke, V.; Alemi, A. Inception-v4, Inception-ResNet and the Impact of Residual Connections on Learning. arXiv 2016, arXiv:1602.07261. [Google Scholar]
- He, K.; Zhang, X.; Ren, S.; Sun, J. Deep Residual Learning for Image Recognition. arXiv 2015, arXiv:1512.03385. [Google Scholar]
- Szegedy, C.; Vanhoucke, V.; Ioffe, S.; Shlens, J.; Wojna, Z. Rethinking the Inception Architecture for Computer Vision. arXiv 2015, arXiv:1512.00567. [Google Scholar]
- Bianco, S.; Cadene, R.; Celona, L.; Napoletano, P. Benchmark Analysis of Representative Deep Neural Network Architectures. arXiv 2018, arXiv:1810.00736. [Google Scholar] [CrossRef]
- Kingma, D.P.; Ba, J. Adam: A Method for Stochastic Optimization. arXiv 2014, arXiv:1412.6980. [Google Scholar]
- McHugh, M.L. Interrater reliability: the kappa statistic. Biochem. Med. (Zagreb) 2012, 22, 276–282. [Google Scholar] [CrossRef] [PubMed]
- Bolon-Canedo, V.; Ataer-Cansizoglu, E.; Erdogmus, D.; Kalpathy-Cramer, J.; Fontenla-Romero, O.; Alonso-Betanzos, A.; Chiang, M.F. Dealing with inter-expert variability in retinopathy of prematurity: A machine learning approach. Comput. Methods Programs Biomed. 2015, 122, 1–15. [Google Scholar] [CrossRef] [PubMed]
- Sun, C.; Shrivastava, A.; Singh, S.; Gupta, A. Revisiting Unreasonable Effectiveness of Data in Deep Learning Era. arXiv 2014, arXiv:1412.6980. [Google Scholar]
- Hestness, J.; Narang, S.; Ardalani, N.; Diamos, G.; Jun, H.; Kianinejad, H.; Patwary, M.M.A.; Yang, Y.; Zhou, Y. Deep Learning Scaling is Predictable, Empirically. arXiv 2017, arXiv:1712.00409. [Google Scholar]
- DiCarlo, J.J.; Zoccolan, D.; Rust, N.C. How does the brain solve visual object recognition? Neuron 2012, 73, 415–434. [Google Scholar] [CrossRef]
- Joulin, A.; van der Maaten, L.; Jabri, A.; Vasilache, N. Learning visual features from large weakly supervised data. In Proceedings of the European Conference on Computer Vision, Amsterdam, The Netherlands, 8–16 October 2016; pp. 67–84. [Google Scholar]
| Image Type | Training | Final Model Validation (Test) | Total | |||
|---|---|---|---|---|---|---|
| Normal | Abnormal | Normal | Abnormal | Normal | Abnormal | |
| Finger | 3138 | 1968 | 214 | 247 | 3352 | 2215 |
| Humerus | 673 | 599 | 148 | 140 | 821 | 739 |
| Model | Accuracy (95% CI) | Sensitivity (Recall) | Specificity | Precision | F1 Score |
|---|---|---|---|---|---|
| DenseNet-169 | 84.03% (79.80–88.26) | 0.81 | 0.86 | 0.85 | 0.83 |
| DenseNet-201 | 88.19% (84.47–91.92) | 0.93 | 0.84 | 0.84 | 0.88 |
| InceptionResNetV2 | 86.46% (82.51–90.41) | 0.90 | 0.83 | 0.83 | 0.87 |
| Model | Kappa | 95% Confidence Interval |
|---|---|---|
| Rajpurkar et. al’s Model | 0.600 | 0.558–0.642 |
| DenseNet-169 | 0.680 | 0.595–0.765 |
| DenseNet-201 | 0.764 | 0.690–0.839 |
| InceptionResNetV2 | 0.730 | 0.651–0.808 |
| Model | Accuracy (95% CI) | Sensitivity (Recall) | Specificity | Precision | F1 Score |
|---|---|---|---|---|---|
| DenseNet-169 | 75.70% (71.79–79.62) | 0.63 | 0.90 | 0.88 | 0.74 |
| DenseNet-201 | 76.57% (72.71–80.44) | 0.69 | 0.85 | 0.84 | 0.76 |
| InceptionResNetV2 | 77.66% (73.85–81.46) | 0.72 | 0.84 | 0.84 | 0.78 |
| Model | Kappa | 95% Confidence Interval |
|---|---|---|
| Rajpurkar et. al’s Model | 0.389 | 0.332–0.446 |
| DenseNet-169 | 0.522 | 0.445–0.599 |
| DenseNet-201 | 0.535 | 0.459–0.612 |
| InceptionResNetV2 | 0.555 | 0.480–0.631 |
© 2019 by the author. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Chada, G. Machine Learning Models for Abnormality Detection in Musculoskeletal Radiographs. Reports 2019, 2, 26. https://doi.org/10.3390/reports2040026
Chada G. Machine Learning Models for Abnormality Detection in Musculoskeletal Radiographs. Reports. 2019; 2(4):26. https://doi.org/10.3390/reports2040026
Chicago/Turabian StyleChada, Govind. 2019. "Machine Learning Models for Abnormality Detection in Musculoskeletal Radiographs" Reports 2, no. 4: 26. https://doi.org/10.3390/reports2040026
APA StyleChada, G. (2019). Machine Learning Models for Abnormality Detection in Musculoskeletal Radiographs. Reports, 2(4), 26. https://doi.org/10.3390/reports2040026




