Discovering the Underlying Analytic Structure Within Standard Model Constants Using Artificial Intelligence
Abstract
1. Introduction
2. The Method
- The first step is a traditional symbolic regression [6], which derives analytical expressions from SM parameter data, generating a large set of candidate relations. This step is the most computationally demanding and produces large datasets containing analytical relationships among SM constants. Such datasets inevitably include spurious analytic expressions that arise purely from numerical coincidences rather than genuine physical connections.
- The second step operates on the datasets generated in the first stage. At this stage, dimensional analysis and general expectations from the SM are applied to identify the most plausible analytical structures within a high-dimensional functional space. We expect that some constraints can be applied to filter out numerical noise and retain only relations that satisfy certain principles. The obtained relations may provide hints of symmetries or dynamical models underlying the SM. However, since dimensional analysis has not yet been implemented in the GP algorithm, this step cannot currently be performed in a straightforward manner.
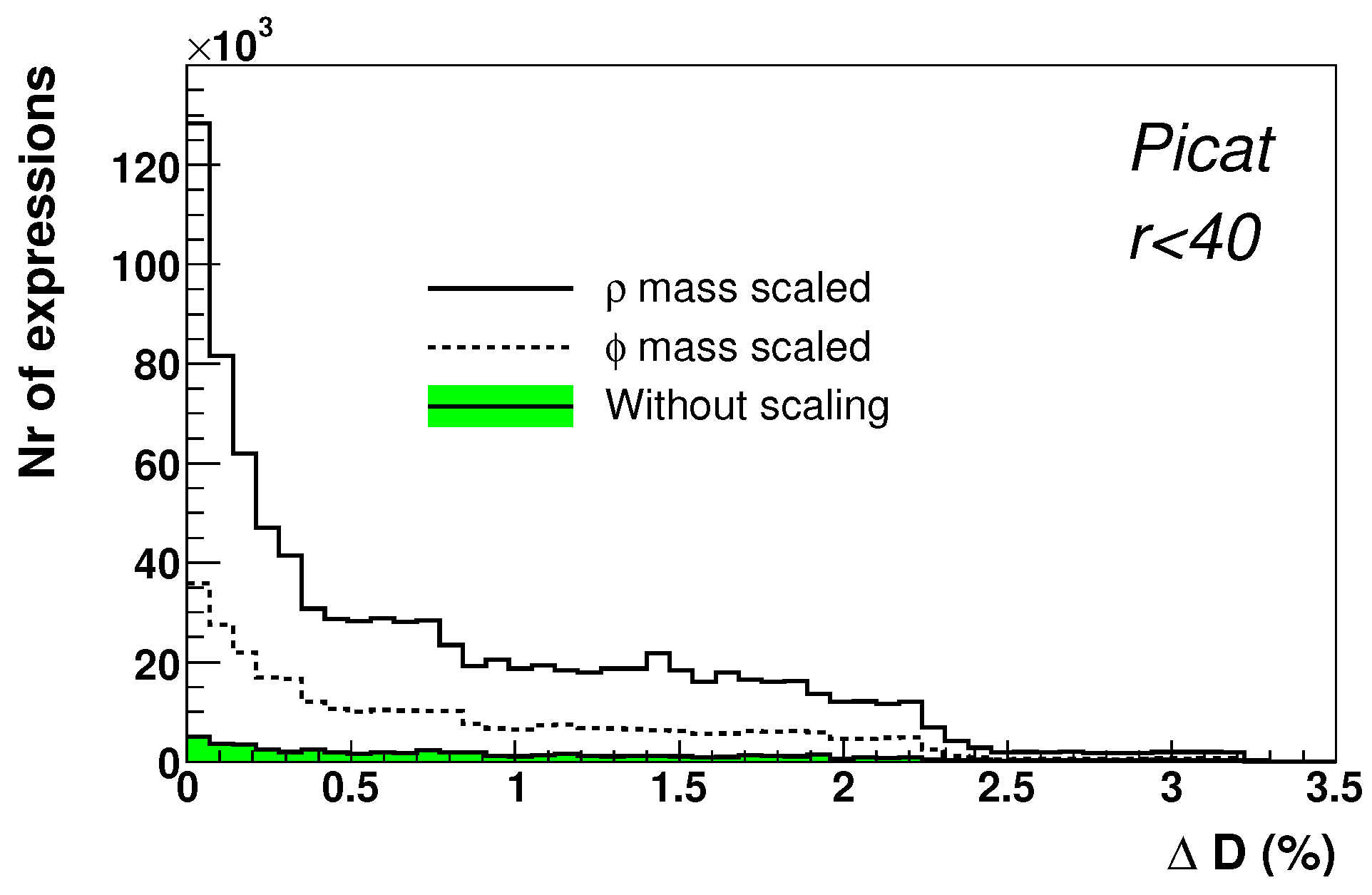
3. Technical Details
- The data set for a single run/experiment consists of a single input-output pair: , where a is an input value, and b is a value of the expected output.
- The algorithm then tries to learn some relation between the constant a and b, generating an analytic expression. See below for more on the specific genetic programming method.
- The found expression does not contain variable names as described above for the general symbolic regression approach. In our version of constant identification, everything in the generated expression is either a function or constants (a physical constant or an integer).
- The equation for the physical constants that is shown below in the paper contains the name of the physical constants, but that is handled by post-processing after the symbolic regression part is completed.
- The number of expressions in each population is 1000. In the first run, the expressions are generated randomly from:
- –
- functions: +, −, *, /, , , , , , , , , , ;
- –
- the integers from 1 to 10. The use of only small integers instead of larger integers or random floats is for simplicity and for the interpretability of an expression;
- –
- and the two or three physical constants involved, the output and input value(s).
- The analytic formulas are evaluated for each expression in a population, and the results that are closest to the output value are kept for the next generation. The formulas that are too far from the output value are replaced with previous expressions that are combined from other expressions using the standard genetic programming operators crossover and mutation (with some specific probability). New random formulas may also be added (from the same set as shown above).
- When an expression is sufficiently close to the value of output c—within the allowed error of the output—then this is considered a solution. All solutions found within the allotted number of runs (1000) are reported. If no solution is found within the allotted number of runs, the program continues to run until a solution is found (and then reported), or until the program times out. The timeout for each experiment, based on a given output–input configuration, was set to 60 s.
- An important feature of the program is that it enforces the inclusion of all input value(s) in each expression. Any expression that does not contain the required input value(s) is removed from the population.
4. Inputs for the Genetic Algorithm
4.1. Inputs with Units
4.2. Inputs Without Physical Units
4.3. The GP Search Algorithm
4.4. Validation of the GP Algorithm
5. Results of the GP Algorithm
5.1. Output for Inputs with Physical Units
5.2. Output for Dimensionless Inputs
5.3. Discussion of the Results
6. Next Steps
7. Conclusions
Author Contributions
Funding
Data Availability Statement
Acknowledgments
Conflicts of Interest
Appendix A. Additional Tables
| Nr | Expression for Rank 10 | (%) |
|---|---|---|
| 1 | 0.54 | |
| 2 | 0.24 | |
| 3 | 0.72 | |
| 4 | 0.72 | |
| 5 | 0.06 | |
| 6 | 0.03 | |
| 7 | 0.43 | |
| 8 | 0.24 | |
| 9 | 0.87 | |
| 10 | 0.18 | |
| 11 | 0.009 | |
| 12 | 0.61 | |
| 13 | 0.88 | |
| 14 | 0.99 | |
| 15 | 0.30 | |
| 16 | 0.93 | |
| 17 | 0.61 | |
| 18 | 0.80 | |
| 19 | 0.53 | |
| 20 | 0.71 | |
| 21 | 0.15 | |
| 22 | 0.93 | |
| 23 | 0.93 | |
| 24 | 0.06 | |
| 25 | 0.93 | |
| 26 | 0.41 | |
| 27 | 0.93 | |
| 28 | 0.38 | |
| 29 | 0.70 | |
| 30 | 0.93 | |
| 31 | 0.23 | |
| 32 | 0.93 | |
| 33 | 0.69 | |
| 34 | 0.93 | |
| 35 | 0.93 | |
| 36 | 0.93 | |
| 37 | 0.93 | |
| 38 | 0.93 | |
| 39 | 0.93 | |
| 40 | 0.93 | |
| 41 | 0.93 | |
| 42 | 0.40 | |
| 43 | 0.93 | |
| 44 | 0.93 | |
| 45 | 0.93 | |
| 46 | 0.76 | |
| 47 | 0.16 | |
| 48 | 0.22 | |
| 49 | 0.71 | |
| 50 | 0.36 | |
| 51 | 0.59 | |
| 52 | 0.78 | |
| 53 | 0.02 | |
| 54 | 0.03 | |
| 55 | 0.01 | |
| 56 | 0.03 | |
| 57 | 0.08 | |
| 58 | 0.06 | |
| 59 | 0.11 | |
| 60 | 0.02 | |
| 61 | 0.005 | |
| 62 | 0.20 | |
| 63 | 0.26 | |
| 64 | 0.33 | |
| 65 | 0.08 | |
| 66 | 0.30 | |
| 67 | 0.20 | |
| 68 | 0.36 | |
| 69 | 0.86 | |
| 70 | 0.94 | |
| 71 | 0.68 | |
| 72 | 0.70 | |
| 73 | 0.06 | |
| 74 | 0.07 | |
| 75 | 0.76 | |
| 76 | 0.32 | |
| 77 | 0.79 | |
| 78 | 0.57 | |
| 79 | 0.03 | |
| 80 | 0.13 | |
| 81 | 0.004 | |
| 82 | 0.12 | |
| 83 | 0.02 | |
| 84 | 0.24 |
References
- Navas, S.; Amsler, C.; Gutsche, T.; Hanhart, C.; Hernández-Rey, J.J.; Lourenço, C.; Masoni, A.; Mikhasenko, M.; Mitchell, R.E.; Patrignani, C.; et al. Review of particle physics. Phys. Rev. D 2024, 110, 030001. [Google Scholar] [CrossRef]
- Chekanov, S.V. Estimation of the chances to find new phenomena at the LHC in a model-agnostic combinatorial analysis. Universe 2024, 10, 414. [Google Scholar] [CrossRef]
- Weinberg, S. Dreams of a Final Theory; Pantheon Books: New York, NY, USA, 1994. [Google Scholar]
- Nielsen, H.B.; Surlykke, C.; Rugh, S.E. Seeking inspiration from the standard model in order to go beyond it. arXiv 1994, arXiv:hep-th/9407012. [Google Scholar] [CrossRef]
- Froggatt, C.D.; Nielsen, H.B. Trying to understand the standard model parameters. Surv. High Energy Phys. 2003, 18, 55–75. [Google Scholar] [CrossRef]
- Koza, J.R. Genetic Programming; MIT Press: Cambridge, MA, USA, 1992. [Google Scholar]
- Zhou, N.F.; Kjellerstrand, H.; Fruhman, J. Picat Programming Language. 2013–2025. Available online: https://picat-lang.org (accessed on 25 November 2025).
- Kjellerstrand, H. symbolic_regression.pi. 2025. Available online: https://hakank.org/picat/symbolic_regression.pi (accessed on 25 November 2025).
- Esteban, I.; Gonzalez-Garcia, M.C.; Maltoni, M.; Martinez-Soler, I.; Pinheiro, J.P.; Schwetz, T. NuFit-6.0: Updated global analysis of three-flavor neutrino oscillations. J. High Energy Phys. 2024, 12, 216. [Google Scholar] [CrossRef]
- Duff, M. How fundamental are fundamental constants? Contemp. Phys. 2014, 56, 35–47. [Google Scholar] [CrossRef]
- Roskies, R.; Peres, A. A new pastime–calculating alpha to one part in a million. Phys. Today 1971, 24, 9. [Google Scholar] [CrossRef]
- Torrente-Lujan, E. The Higgs and top mass coincidence problem. EPJ Web Conf. 2015, 95, 05015. [Google Scholar] [CrossRef]
- Chekanov, S.V.; Kjellerstrand, H. GC4PhysicalConstants—Genetic Computing for Physical Constants. GitHub Repository. 2025. Available online: https://github.com/chekanov/GC4PhysicalConstants (accessed on 25 November 2025).
- Weinberg, S. A Model of Leptons. Phys. Rev. Lett. 1967, 19, 1264–1266. [Google Scholar] [CrossRef]
- Chekanov, S.V.; Kjellerstrand, H. Evidence of Relationships Among Fundamental Constants of the Standard Model. arXiv 2025, arXiv:2509.07713. [Google Scholar] [CrossRef]
| Constant | Name | Value | (%) | |
|---|---|---|---|---|
| PI | 3.14159 | 0.0003 | ||
| Fine-struct. (inv) | 137.036 | 0.001 | 0.0007 | |
| at | 0.1180 | 0.0009 | 0.7627 | |
| CKM constants | no units | (%) | ||
| 12-mix angle | 0.22501 | 0.00068 | 0.3022 | |
| 23-mix angle | 0.04183 | 0.00079 | 1.8886 | |
| 13-mix angle | 0.003732 | 2.4116 | ||
| CP-viol. phase | 1.147 | 0.026 | 2.2668 | |
| Particle masses | MeV | (%) | ||
| electron mass | 0.510998 | 0.0002 | ||
| muon mass | 105.658 | 0.001 | 0.0009 | |
| mass | 1776.93 | 0.09 | 0.0051 | |
| u-quark mass | 2.16 | 0.07 | 3.2407 | |
| d-quark mass | 4.70 | 0.07 | 1.4894 | |
| s-quark mass | 93.5 | 0.8 | 0.8556 | |
| c-quark mass | 1273.0 | 4.6 | 0.3614 | |
| b-quark mass | 4183 | 7 | 0.1673 | |
| t-quark mass | 172,560 | 310 | 0.1796 | |
| Z-boson mass | 91,188.0 | 2.0 | 0.0022 | |
| W-boson mass | 80,369.2 | 13.3 | 0.0165 | |
| H-boson mass | 125,200 | 110 | 0.0879 |
| Constant | Name | Value | (%) | |
|---|---|---|---|---|
| electron mass | 0.0006591 | 2 | 0.0303 | |
| muon mass | 0.1363 | 4 | 0.0293 | |
| mass | 2.292 | 0.0007 | 0.0305 | |
| u-quark mass | 0.002786 | 9 | 3.2304 | |
| d-quark mass | 0.006062 | 9 | 1.4847 | |
| s-quark mass | 0.1206 | 0.0010 | 0.8292 | |
| c-quark mass | 1.642 | 0.006 | 0.3593 | |
| b-quark mass | 5.3956 | 0.0092 | 0.1705 | |
| t-quark mass | 222.583 | 0.405 | 0.1820 | |
| Z-boson mass | 117.622 | 0.035 | 0.0298 | |
| W-boson mass | 103.667 | 0.035 | 0.0338 | |
| H-boson mass | 161.494 | 0.149 | 0.0923 |
| Value | Mathematical Token |
|---|---|
| 1 | physics constant (float) and |
| 2 | plain integer () |
| 3 | addition and subtraction |
| 4 | multiplication and division |
| 5 | , , and |
| 6 | and (log base of e of x), , , , |
| Nr | Expression for Rank 6 | (%) |
|---|---|---|
| 1 | 0.71 | |
| 2 | 0.93 | |
| 3 | 0.47 | |
| 4 | 0.70 | |
| 5 | 0.30 |
| Nr | Expression for Rank 7 | (%) |
|---|---|---|
| 1 | 0.08 | |
| 2 | 0.20 | |
| 3 | 0.09 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Chekanov, S.V.; Kjellerstrand, H. Discovering the Underlying Analytic Structure Within Standard Model Constants Using Artificial Intelligence. Particles 2025, 8, 95. https://doi.org/10.3390/particles8040095
Chekanov SV, Kjellerstrand H. Discovering the Underlying Analytic Structure Within Standard Model Constants Using Artificial Intelligence. Particles. 2025; 8(4):95. https://doi.org/10.3390/particles8040095
Chicago/Turabian StyleChekanov, Sergei V., and Håkan Kjellerstrand. 2025. "Discovering the Underlying Analytic Structure Within Standard Model Constants Using Artificial Intelligence" Particles 8, no. 4: 95. https://doi.org/10.3390/particles8040095
APA StyleChekanov, S. V., & Kjellerstrand, H. (2025). Discovering the Underlying Analytic Structure Within Standard Model Constants Using Artificial Intelligence. Particles, 8(4), 95. https://doi.org/10.3390/particles8040095






