Optimal Histopathological Magnification Factors for Deep Learning-Based Breast Cancer Prediction
Abstract
:1. Introduction
Problem Statement
2. Literature Review
3. Preliminaries
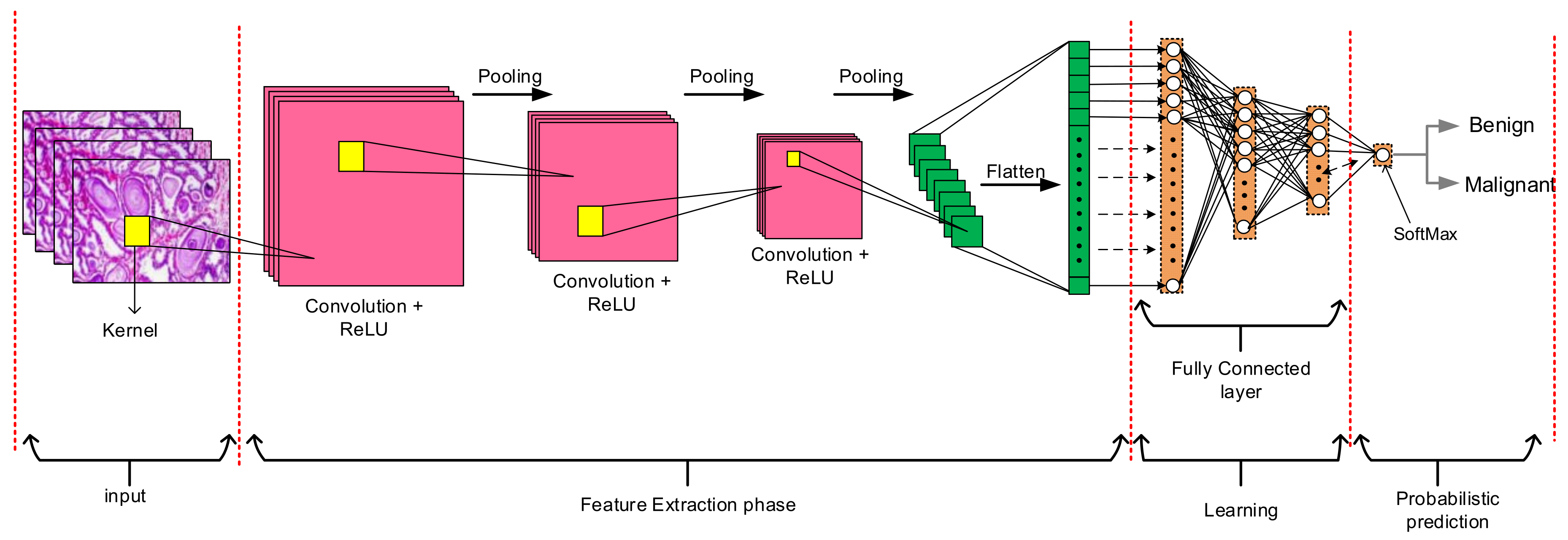
3.1. Convolutional Neural Networks (CNNs)
3.2. Measurement and Performance Evaluation Methods
3.2.1. Area Under ROC Curve
3.2.2. Training and Validation Loss
3.2.3. Precision, Recall, and F1-score Performance Measure
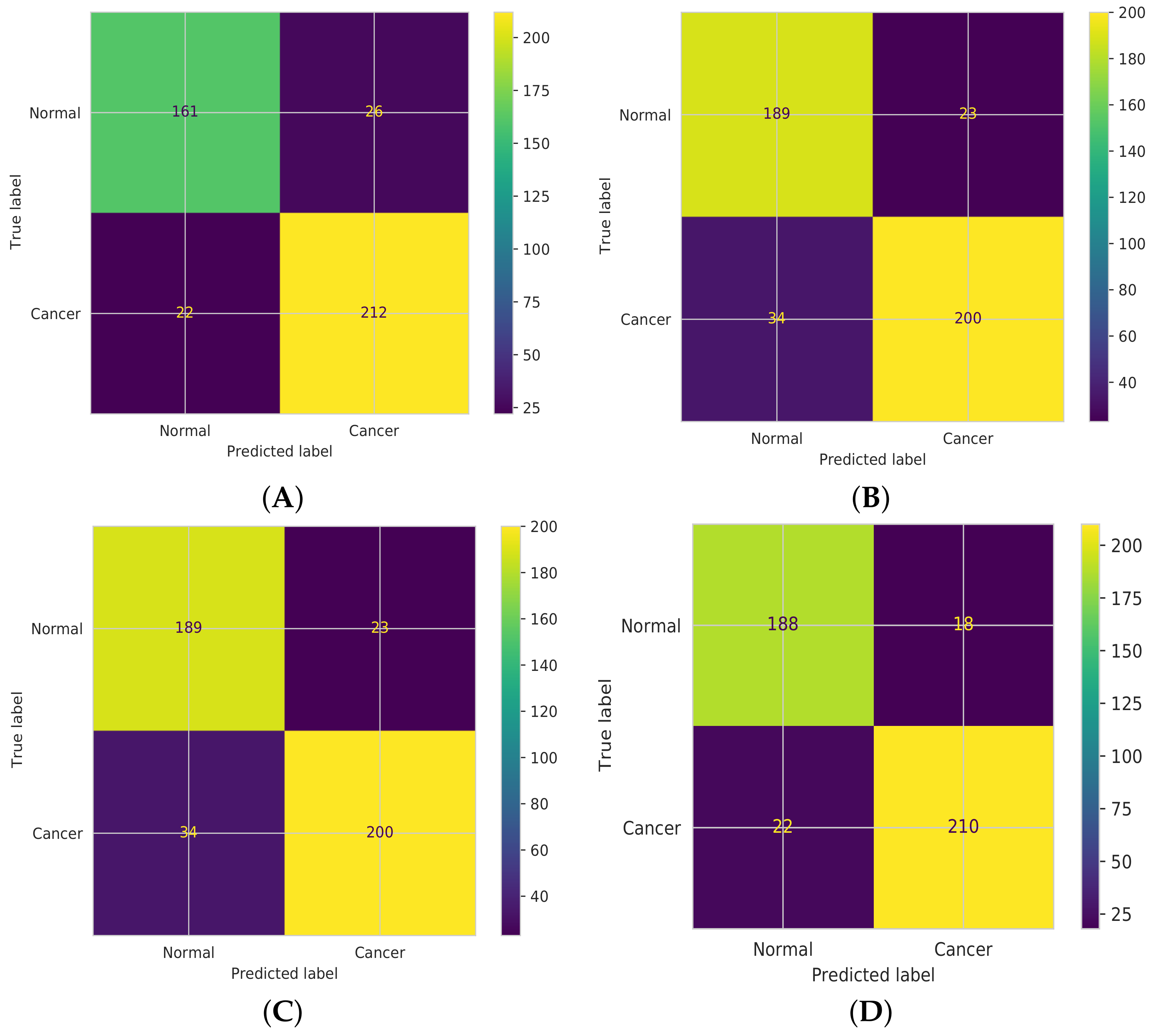
3.2.4. Confusion Matrix
4. Methodology
4.1. Dataset Description
4.2. Dataset Preprocessing
Skew Data
4.3. Image Scaling
4.4. The Dataset Splitting
4.5. Data Transformation
- Horizontal flipping = 0.4
- Vertical flipping = 0.4
- Image rotation = 20
- For RGB channel normalization, the following values for mean and standard deviation are used: (0.5, 0.5, 0.5) and (0.5, 0.5, 0.5), respectively.
- Image resizing = (700 × 460) pixels
- For image normalization, the following values for mean and standard deviation are used: (0.5, 0.5, 0.5) and (0.5, 0.5, 0.5), respectively.
4.6. Feature Extraction Phase
4.7. Classification Phase
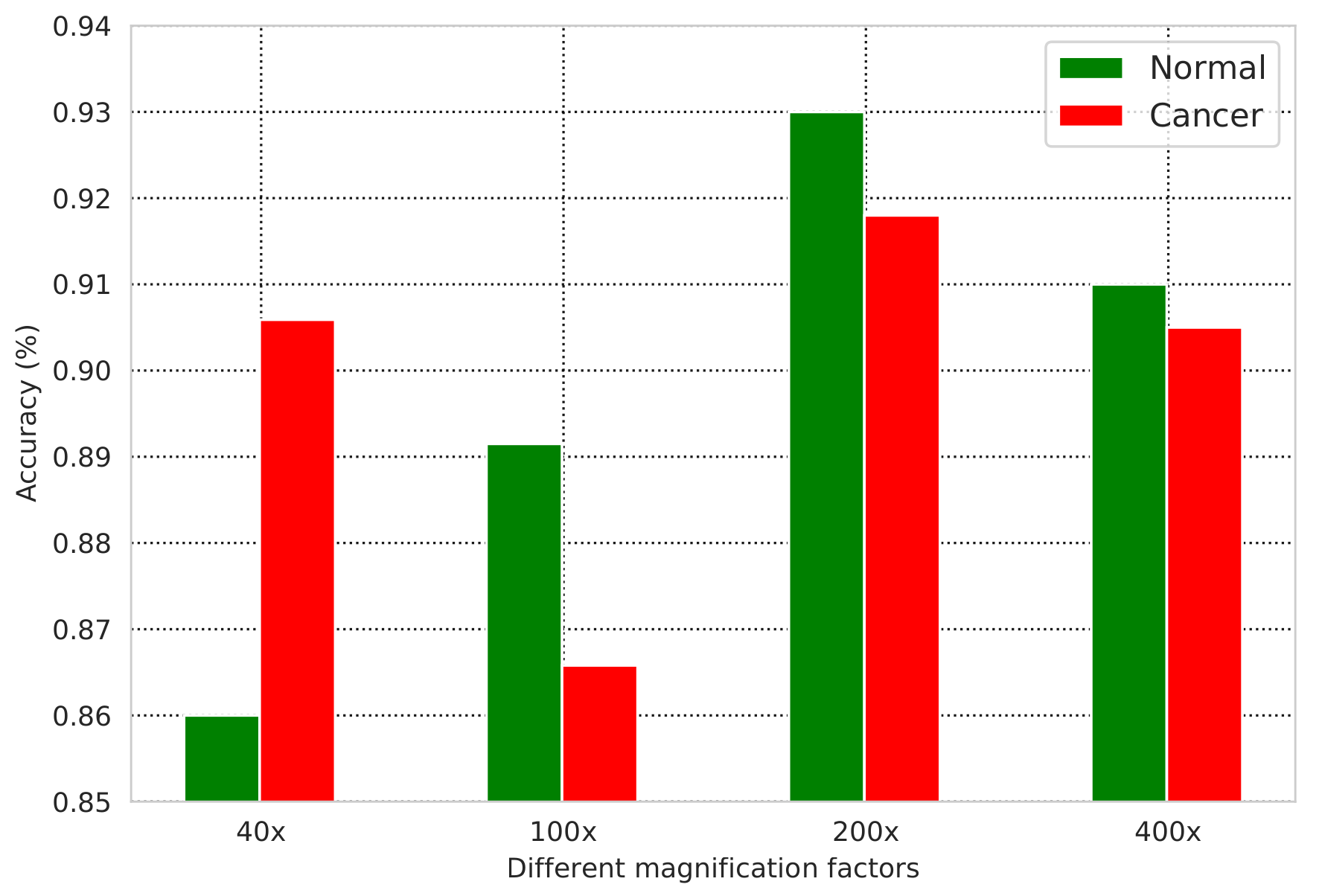
5. Results
5.1. AU-ROC/AUC Curve
5.2. Training Loss and Validation Loss
6. Discussion
7. Conclusions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Valenstein, P.; Sirota, R. Identification Errors in Pathology and Laboratory Medicine. Clin. Lab. Med. 2004, 24, 979–996. [Google Scholar] [CrossRef] [PubMed]
- National Academy of Medicine. Available online: https://nam.edu/ (accessed on 7 November 2021).
- Anjum, R.; Rahman; Rashid, R.; Shamin, H. An efficient breast cancer analysis technique by using a combination of HOG and canny edge detection techniques. In Proceedings of the 2021 5th International Conference on Trends in Electronics and Informatics (ICOEI), Tirunelveli, India, 3–5 June 2021; Volume 10, pp. 1290–1295. [Google Scholar]
- Gupta, K.; Chawla, N. Analysis of Histopathological Images for Prediction of Breast Cancer Using Traditional Classifiers with Pre-Trained CNN. Procedia Comput. Sci. 2020, 167, 878–889. [Google Scholar] [CrossRef]
- Yadav, A.; Verma, V.K.; Pal, V.; Jain, V.; Garg, V. Automated detection and classification of breast cancer tmour cells using machine learning and deep learning on histopathological images. In Proceedings of the 2021 6th International Conference for Convergence in Technology (I2CT), Maharashtra, India, 2–4 April 2021; pp. 1–6. [Google Scholar]
- Anwar, F.; Attallah, O.; Ghanem, N.; Ismail, M.A. Automatic breast cancer classification from histopathological images. In Proceedings of the 2019 International Conference on Advances in the Emerging Computing Technologies (AECT), Al Madinah Al Munawwarah, Saudi Arabia, 10 February 2020; pp. 1–6. [Google Scholar]
- Gultekin, T.; Koyuncu, C.F.; Sokmensuer, C.; Gunduz-Demir, C. Two-Tier Tissue Decomposition for Histopathological Image Representation and Classification. IEEE Trans. Med. Imaging 2015, 34, 275–283. [Google Scholar] [CrossRef] [PubMed]
- Vo-Le, C.; Son, N.H.; Van Muoi, P.; Phuong, N.H. Breast cancer detection from histopathological biopsy images using transfer learning. In Proceedings of the 2020 IEEE Eighth International Conference on Communications and Electronics (ICCE), Phu Quoc Island, Vietnam, 13–15 January 2021; pp. 408–412. [Google Scholar]
- Höhn, J.; Henning, E.K.; Jutzi, T.B. Combining CNN-based Histologic Whole Slide Image Analysis and Patient Data to Improve Skin Cancer Classification. Eur. J. Cancer 2021, 149, 94–101. [Google Scholar] [CrossRef] [PubMed]
- Dabeer, S.; Khan, M.M.; Islam, S. Cancer Diagnosis in Histopathological Image: CNN Based Approach. Inform. Med. Unlocked 2019, 16, 100231. [Google Scholar] [CrossRef]
- Wadhwa, G.; Kaur, A. A Deep CNN Technique for Detection of Breast Cancer Using Histopathology Images. In Proceedings of the 2020 Advanced Computing and Communication Technologies for High Performance Applications (ACCTHPA), Cochin, India, 2–4 July 2020; pp. 179–185. [Google Scholar]
- Khuriwal, N.; Mishra, N. Breast Cancer Detection from Histopathological Images Using Deep Learning. In Proceedings of the 2018 3rd International Conference and Workshops on Recent Advances and Innovations in Engineering (ICRAIE), Jaipur, India, 22–25 November 2018; pp. 1–4. [Google Scholar]
- Qi, Q.; Li, Y.; Wang, J.; Zheng, H.; Huang, Y.; Ding, X.; Rohde, G.K. Label-Efficient Breast Cancer Histopathological Image Classification. IEEE J. Biomed. Health Inform. 2018, 23, 2108–2116. [Google Scholar] [CrossRef] [PubMed]
- El-Agouri, H.A.J.A.R.; Azizi, M.; El Attar, H.; El Khannoussi, M.; Ibrahimi, A.; Kabbaj, R.; Kadiri, H.; BekarSabein, S.; EchCharif, S.; Mounjid, C.; et al. Assessment of Deep Learning Algorithms to Predict Histopathological Diagnosis of Breast Cancer: First Moroccan Prospective Study on a Private Dataset. BMC Res. Notes 2022, 15, 66. [Google Scholar] [CrossRef] [PubMed]
- Da, Q.; Deng, S.; Li, J.; Yi, H.; Huang, X.; Yang, X.; Yu, T.; Wang, X.; Liu, J.; Duan, Q.; et al. Quantifying the Cell Morphology and Predicting Biological Behavior of Signet Ring Cell Carcinoma using Deep Learning. Sci. Rep. 2022, 12, 183. [Google Scholar] [CrossRef] [PubMed]
- Chatterjee, C.C.; Krishna, G. A novel method for IDC prediction in breast cancer histopathol- ogy images using deep residual neural networks. In Proceedings of the 2019 2nd International Conference on Intelligent Communication and Computational Techniques (ICCT), Jaipur, India, 28–29 September 2019; pp. 95–100. [Google Scholar]
- Carvalho, R.H.d.O.; Martins, A.S.; Neves, L.A.; Nascimento, M.Z. Analysis of Features for Breast cancer recognition in different magnifications of histopathological images. In Proceedings of the 2020 International Conference on Systems, Signals and Image Processing (IWSSIP), Niteroi, Brazil, 1–3 July 2020; pp. 39–44. [Google Scholar]
- Qidwai, U.; Sadia, M. Color-based fuzzy classifier for automated detection of cancerous cells in histopathological analysis. In Proceedings of the 2014 IEEE Conference on Biomedical Engineering and Sciences (IECBES), Kuala Lumpur, Malaysia, 8–10 December 2014; pp. 103–108. [Google Scholar]
- Chen, C.; Chen, C.; Ma, M.; Ma, X.; Lv, X.; Dong, X.; Yan, Z.; Zhu, M.; Chen, J. Classification of Multi-differentiated Liver Cancer Pathological Image Based on Deep Learning Attention Mechanism. BMC Med. Inform. Decis. Mak. 2022, 22, 176. [Google Scholar] [CrossRef] [PubMed]
- White, B.S.; Park, J.; Sheridan, T.B.; Chuang, J.H. Deep Learning Features Encode Interpretable Morphologies within Histological Images. Sci. Rep. 2022, 12, 9428. [Google Scholar]
- Laboratory of Vision, Robotics and Imaging (VRI). Available online: https://web.inf.ufpr.br/vri/ (accessed on 7 December 2022).
- Hirra, I.; Ahmad, M.; Hussain, A.; Ashraf, M.U.; Saeed, I.A.; Qadri, S.F.; Alghamdi, A.M.; Alfakeeh, A.S. Breast Cancer Classification from Histopathological Images Using Patch-Based Deep Learning Modeling. IEEE Access 2021, 9, 24273–24287. [Google Scholar] [CrossRef]
- Mohalder, R.D.; Sarkar, J.P.; Hossain, K.A.; Paul, L.; Raihan, M. A deep learning based approach to predict lung cancer from histopathological images. In Proceedings of the 2021 International Conference on Electronics, Communications and Information Technology (ICECIT), Khulna, Bangladesh, 14–16 September 2021; pp. 1–4. [Google Scholar]
- Al-Haija, Q.A.; Adebanjo, A. Breast cancer diagnosis in histopathological images using ResNet-50 convolutional neural network. In Proceedings of the 2020 IEEE International IOT, Electronics and Mechatronics Conference (IEMTRONICS), Vancouver, BC, Canada, 9–12 September 2020; pp. 1–7. [Google Scholar]
- Jannesari, M.; Habibzadeh, M.; Aboulkheyr, H.; Khosravi, P.; Elemento, O.; Totonchi, M.; Hajirasouliha, I. Breast cancer histopathological image classification: A deep learning approach. In Proceedings of the 2018 IEEE International Conference on Bioinformatics and Biomedicine (BIBM), Madrid, Spain, 3–6 December 2018; pp. 2405–2412. [Google Scholar]
- Madduri, A.; Adusumalli, S.S.; Katragadda, H.S.; Dontireddy, M.K.R.; Suhasini, P.S. Classification of Breast Cancer Histopathological Images using Convolutional Neural Networks. In Proceedings of the 2021 8th International Conference on Signal Processing and Integrated Networks (SPIN), Noida, India, 26–27 August 2021; pp. 755–759. [Google Scholar]
- Cetindag, S.C.; Guran, K.; Bilgin, G. Transfer Learning Methods for Using Textural Features in Histopathological Image Classification. In Proceedings of the 2020 Medical Technologies Congress (TIPTEKNO), Antalya, Turkey, 19–20 November 2020; pp. 1–4. [Google Scholar]
- Das, K.; Conjeti, S.; Chatterjee, J.; Sheet, D. Detection of Breast Cancer from Whole Slide Histopathological Images Using Deep Multiple Instance CNN. IEEE Access 2020, 8, 213502–213511. [Google Scholar] [CrossRef]
- Sun, Y.; Xu, Z.; Strell, C.; Moro, C.F.; Wärnberg, F.; Dong, L.; Zhang, Q. Detection of Breast Tumour Tissue Regions in Histopathological Images using Convolutional Neural Networks. In Proceedings of the 2018 IEEE International Conference on Image Processing, Applications and Systems (IPAS), Sophia Antipolis, France, 12–14 December 2018; pp. 98–103. [Google Scholar]
- Boktor, M.; Ecclestone, B.R.; Pekar, V.; Dinakaran, D.; Mackey, J.R.; Fieguth, P.; Haji Reza, P. Virtual Histological Staining of Label-free Total Absorption Photoacoustic Remote Sensing (TA-PARS). Sci. Rep. 2022, 12, 10296. [Google Scholar] [CrossRef] [PubMed]
- Djouima, H.; Zitouni, A.; Megherbi, A.C.; Sbaa, S. Classification of Breast Cancer Histopathological Images using DensNet201. In Proceedings of the 2022 7th International Conference on Image and Signal Processing and their Applications (ISPA), Mostaganem, Algeria, 8–9 May 2022; pp. 1–6. [Google Scholar]






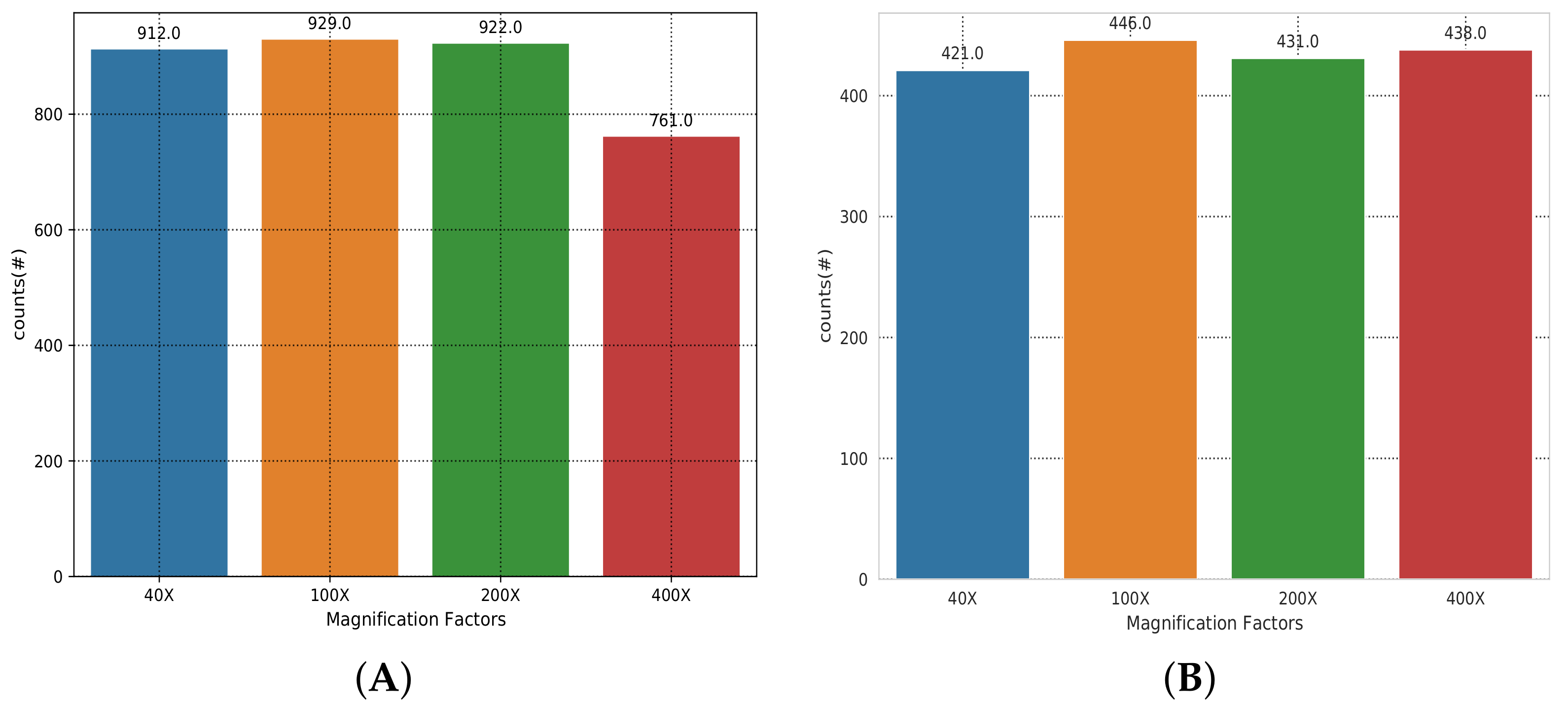

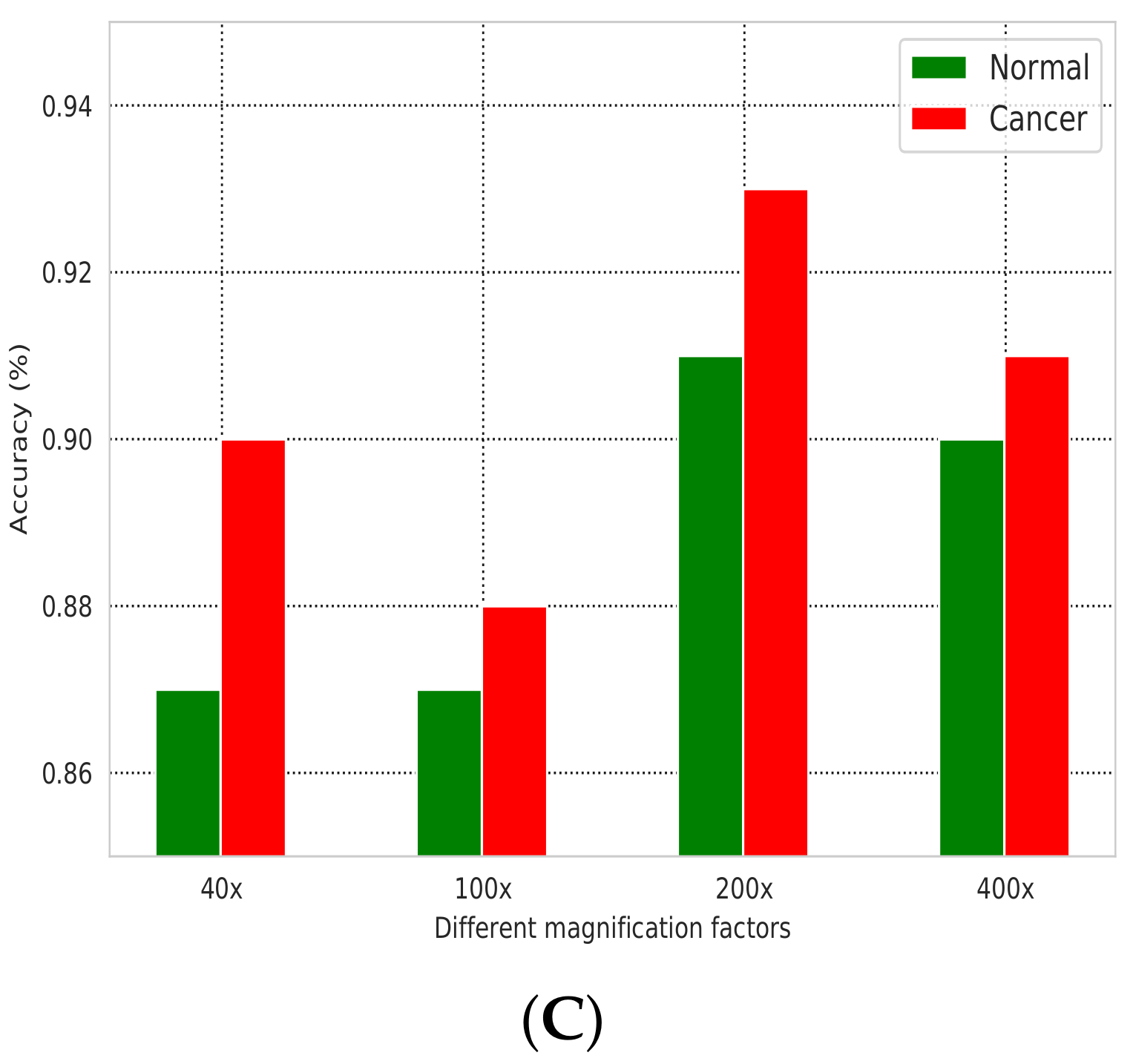

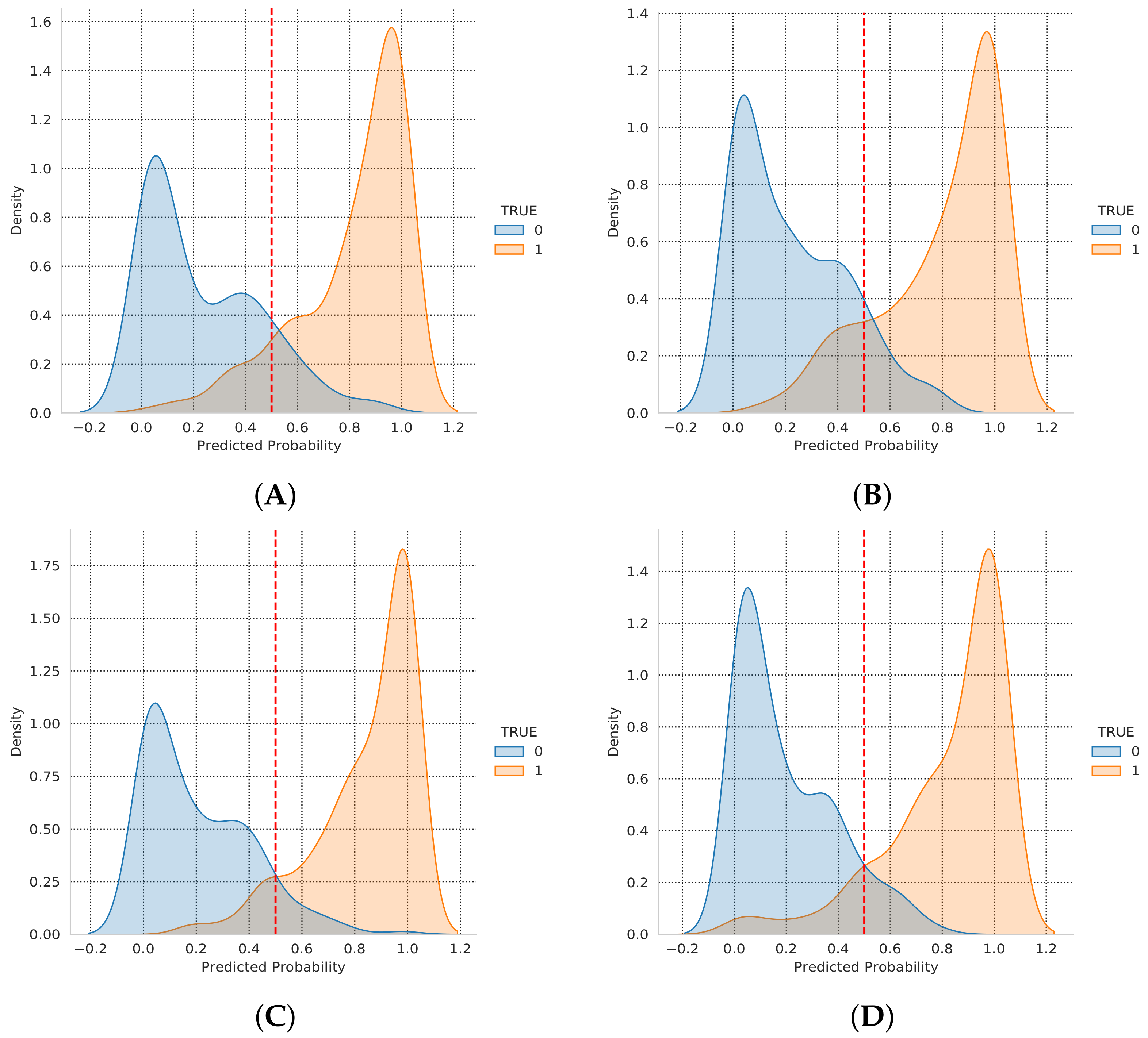
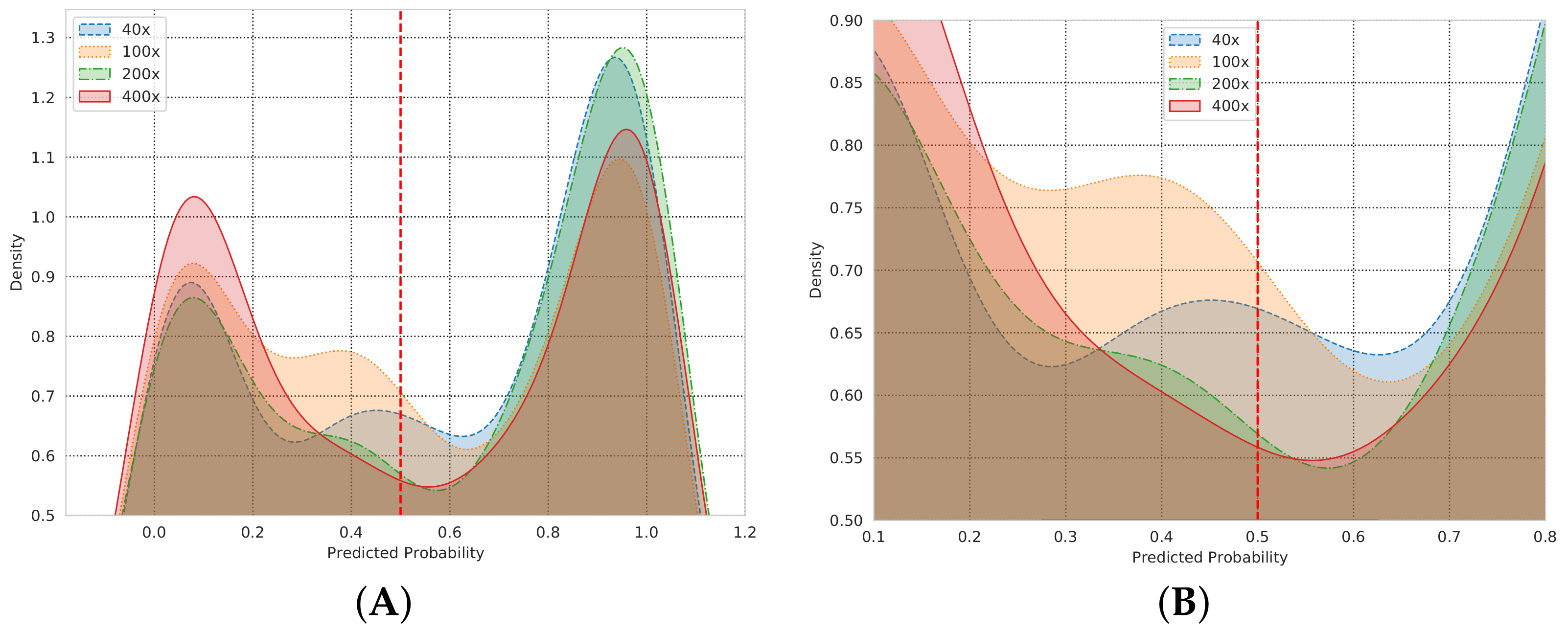
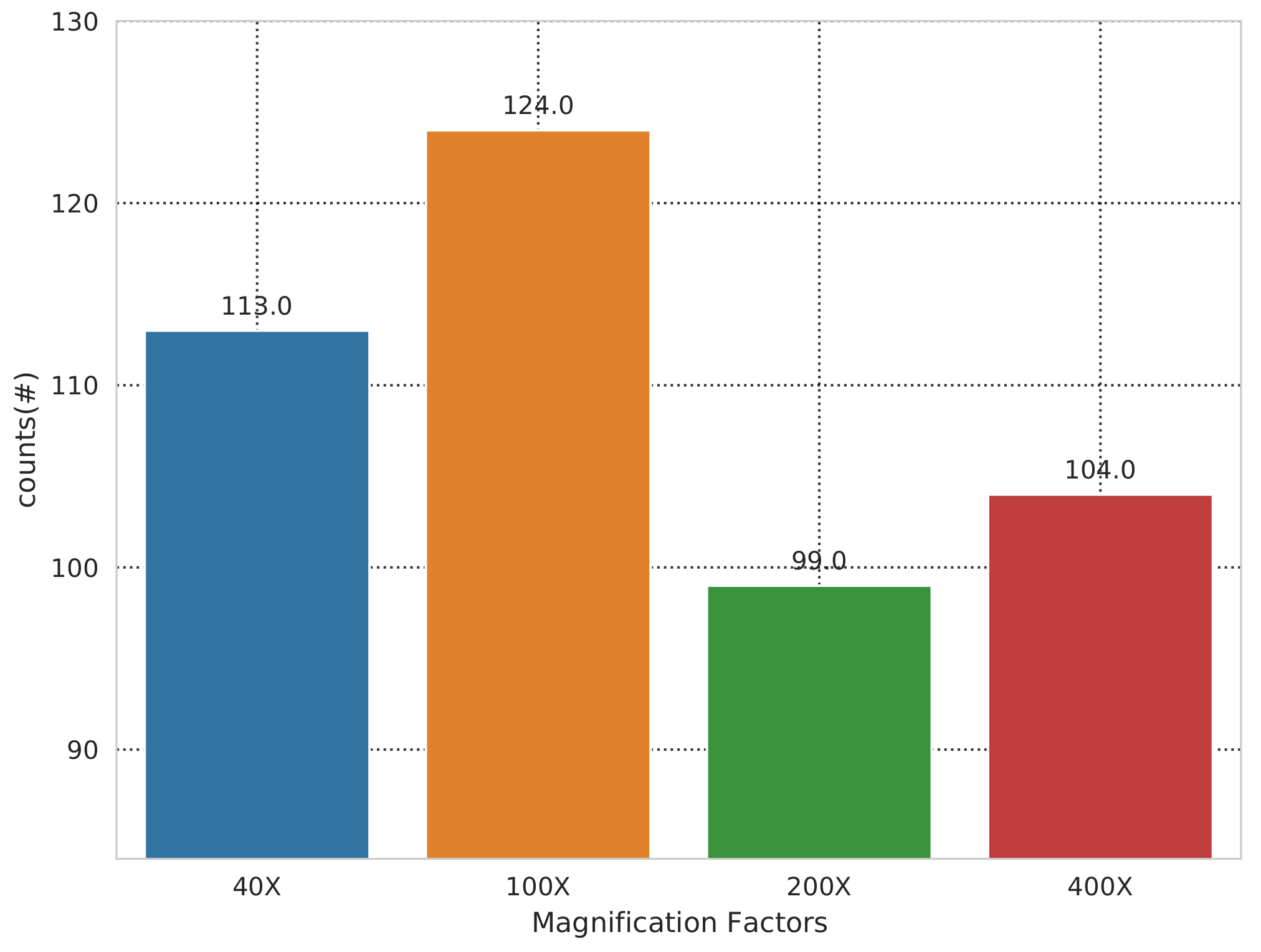
| Magnification Factor Used | Benign | Malignant | Total |
|---|---|---|---|
| 40x | 652 | 1370 | 1995 |
| 100x | 644 | 1437 | 2081 |
| 200x | 623 | 1390 | 2013 |
| 400x | 588 | 1232 | 1820 |
| Total of images | 2480 | 5429 | 7909 |
| Precision | Recall | F1-Score | |
|---|---|---|---|
| Normal | 0.88 | 0.9 | 0.89 |
| Cancer | 0.91 | 0.9 | 0.9 |
| # | Histopahological Image Name | T Label | Pred. Label | Pred. Probability |
|---|---|---|---|---|
| 1 | SOB_B_F-14-23060AB-40-011.png | 0 | 0 | 0.064793758 |
| 2 | SOB_B_F-14-9133-200-038.png | 0 | 0 | 0.111493707 |
| 3 | SOB_M_DC-14-13412-100-004.png | 1 | 1 | 0.802534401 |
| 4 | SOB_M_PC-14-15704-200-024.png | 1 | 1 | 0.945152342 |
| 5 | SOB_B_F-14-9133-200-017.png | 0 | 0 | 0.069869727 |
| 6 | SOB_M_DC-14-11520-100-008.png | 1 | 1 | 0.998456001 |
| 7 | SOB_B_TA-14-19854C-200-016.png | 0 | 0 | 0.020913977 |
| 8 | SOB_B_F-14-25197-400-034.png | 0 | 1 | 0.631648719 |
| 9 | SOB_M_DC-14-11031-40-010.png | 1 | 1 | 0.534332812 |
| 10 | SOB_M_PC-14-19440-400-028.png | 1 | 1 | 0.998934567 |
| Auth. | Dataset | Modality | Methodology | Preprocessing | Objective |
|---|---|---|---|---|---|
| [3] | Kaggle invasive ductal carcinoma | HI | SVM and LR | Feature reduction using PCA and HOG and Canny edge detection | BC classification, i.e., B or M |
| [4] | BreakHis [21] | HI | SVM and LR | CNN (for feature extraction) + SVM (for classification) or CNN (for feature extraction) + LR (for classification) | BC classification, i.e., B or M |
| [5] | Kaggle dataset | HI | SVM and CNN for feature extraction and K-means for tumor extraction. | Segmentation algorithm GA and K-means | locate and extract tumor cells |
| [6] | BreakHis [21] | HI | SVM, CNN, RF, QDA | HOG, WPT, ResNet, and PCA for feature extraction | BC classification, i.e., B or M |
| [7] | locally prepared | HI | SVM | Tissue decomposition to locate multityped objects | image decomposition toward colon cancer classification |
| [8] | Vietnamese Dataset VBCan | HI | SVM, LR, NB, DT, RF | For feature extraction they use VGG16, ResNet50, GoogleNet | BC classification, i.e., B or M |
| [9] | 430 patients collected from two laboratories | HI + age, sex, lesion site | CNNs | Fusion of patient information with an image | BC classification, i.e., B or M |
| [10] | BreakHis [21] | HI | CNNs | Using original CNN architecture | BC classification, i.e., B or M |
| [11] | BreakHis [21] | HI | CNNs | Feature extraction using DenseNet-201 | BC classification, i.e., B or M |
| [12] | MIAS mammograms | HI | CNN | Feature extraction using the Entropy Function | BC classification, i.e., B or M |
| [13] | BreakHis [21] | HI | Active learning | image labeling using Entropy-based query strategy | BC labeling |
| [16] | Kaggle BC HI | HI | Deep RNNs | Convert RGB image to 7 channels | BC classification, i.e., B or M |
| [17] | BreakHis [21] | HI | RF, SMO | Intensity Histogram Co-occurrence matrix | BC classification, i.e., B or M |
| [18] | —– | HI | Fussy classification | Geometric transformation from RGB to HSV | Cancer Cells classification, i.e., B or M |
| [22] | Hospital of the University of Pennsylvania | HI | DBNs + LR | Applying RoI extraction | BC classification, i.e., B or M |
| [24] | BreakHis [21] | HI | CNN | Feature extraction using ResNet50 | BC classification, i.e., B or M |
| [25] | Tissue Micro Array database and BreakHis [21] | HI | CNN | Feature extraction using ResNet152 | BC classification, i.e., B or M |
| [26] | 100 images from unknown source | HI | Origin CNN or CNN + LBP (feature extraction) | First RGB to gray conversion followed by LBP | BC classification, i.e., B or M |
| [28] | BreakHis [21] | HI | DMI based on CNN | images reduced to bag of labels | BC classification, i.e., B or M |
| [29] | —- | HI | AlexNet + CNN, VGG-11 + CNN, and ResNet-18 + CNN | RoI annotation then extract image patches | predict and localize tumour tissue region |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the author. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Ashtaiwi, A. Optimal Histopathological Magnification Factors for Deep Learning-Based Breast Cancer Prediction. Appl. Syst. Innov. 2022, 5, 87. https://doi.org/10.3390/asi5050087
Ashtaiwi A. Optimal Histopathological Magnification Factors for Deep Learning-Based Breast Cancer Prediction. Applied System Innovation. 2022; 5(5):87. https://doi.org/10.3390/asi5050087
Chicago/Turabian StyleAshtaiwi, Abduladhim. 2022. "Optimal Histopathological Magnification Factors for Deep Learning-Based Breast Cancer Prediction" Applied System Innovation 5, no. 5: 87. https://doi.org/10.3390/asi5050087
APA StyleAshtaiwi, A. (2022). Optimal Histopathological Magnification Factors for Deep Learning-Based Breast Cancer Prediction. Applied System Innovation, 5(5), 87. https://doi.org/10.3390/asi5050087






