Microbial-Enhanced Heavy Oil Recovery under Laboratory Conditions by Bacillus firmus BG4 and Bacillus halodurans BG5 Isolated from Heavy Oil Fields
Abstract
:1. Introduction
2. Materials and Methods
2.1. Culture Media and Cultivation
2.2. Characterization of Soil and Oil Samples
2.3. Isolation of Spore Forming Bacterial Strains Using Heavy Crude Oil as Carbon Source
2.4. Identification of Bacillus firmus and Bacillus halodurans
2.5. Growth Characteristics during Biotransformation under Aerobic Conditions
2.6. Biotransformation Studies Using GC-MS
2.7. Core Flooding Experiments
2.8. Statistical Analysis
3. Results
3.1. Characterization of Soil and Oil Samples
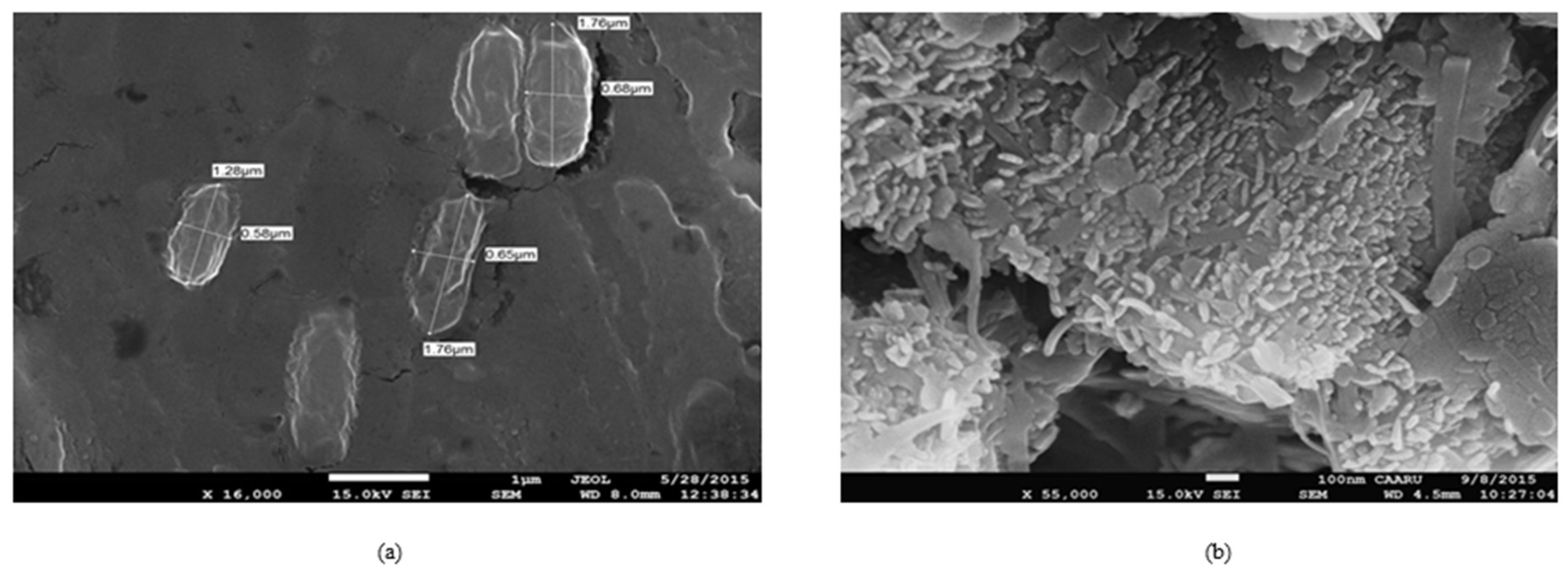
3.2. Isolation and Identification of Oil-Oxidizing Bacteria, Bacillus firmus and Bacillus halodurans
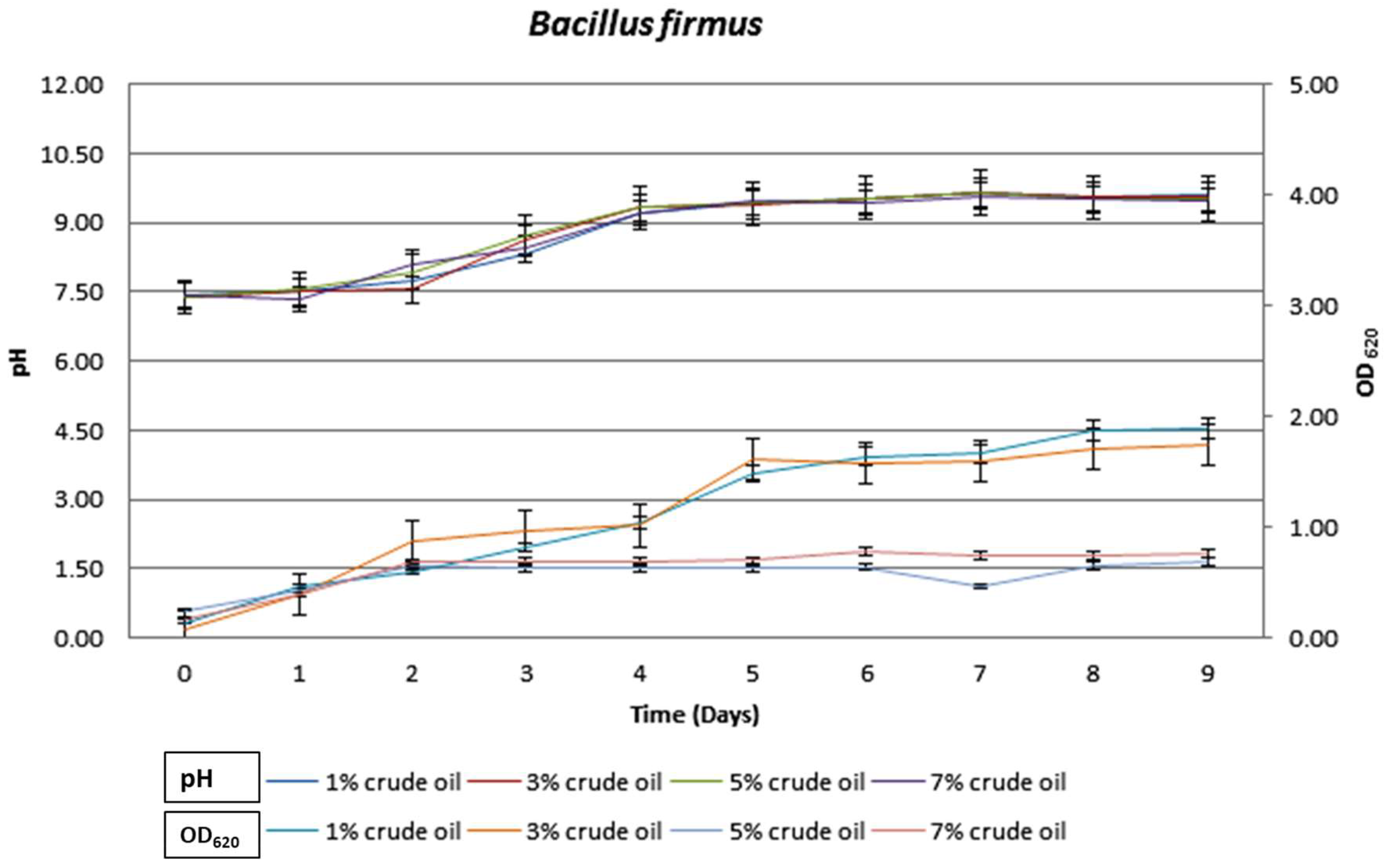
3.3. Growth Characteristics of Bacteria during Crude Oil Degradation under Aerobic Conditions
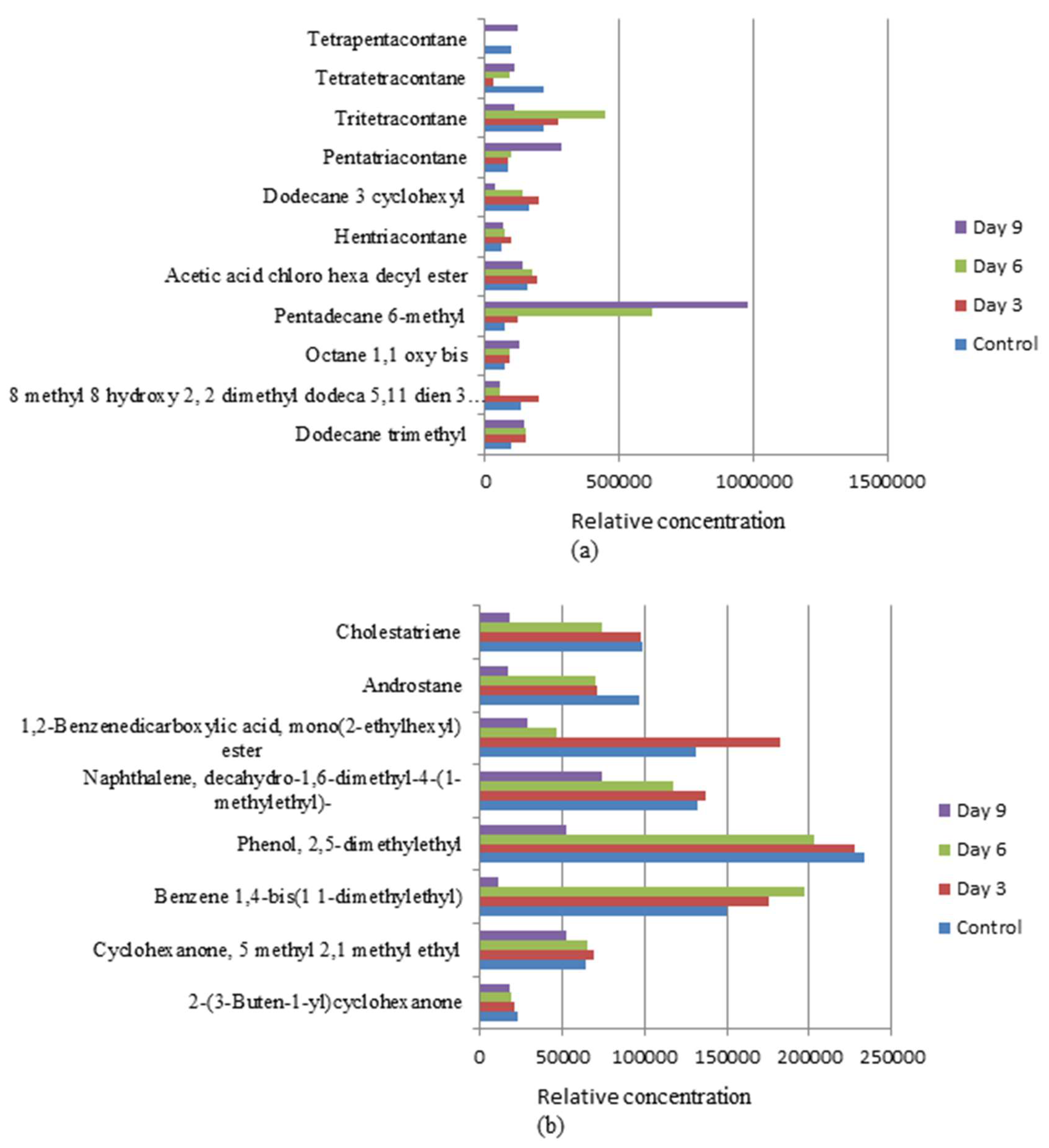
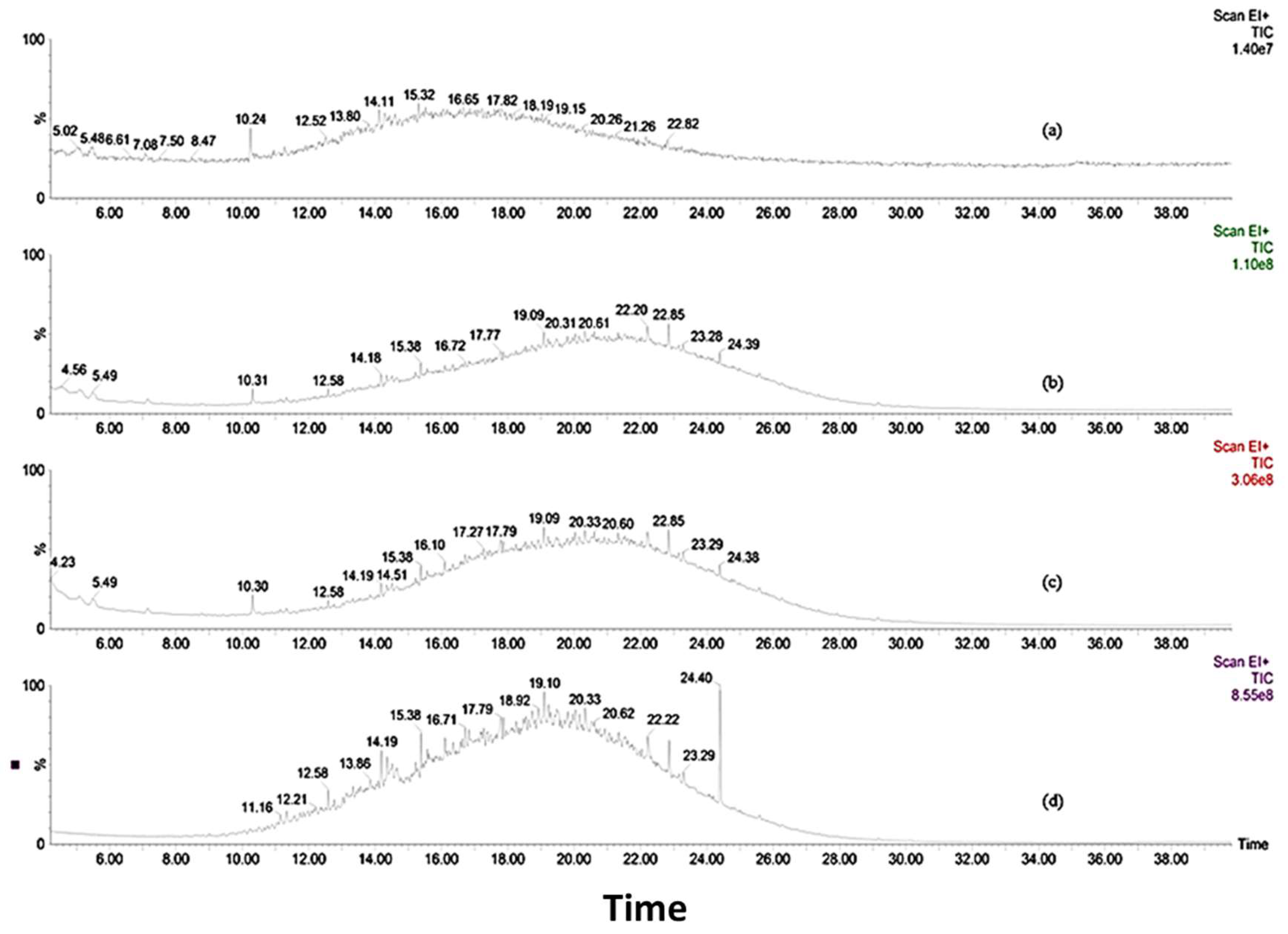
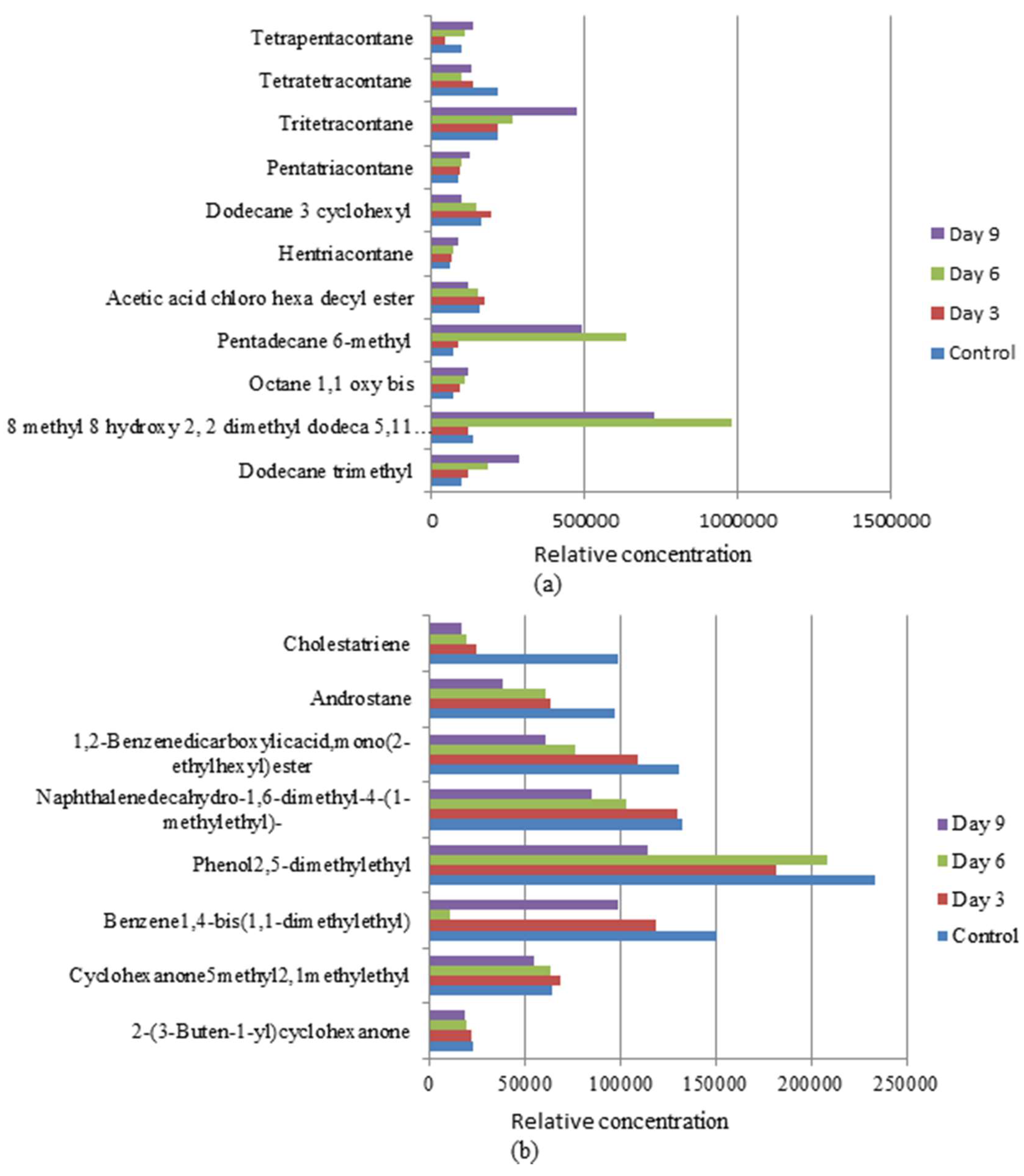
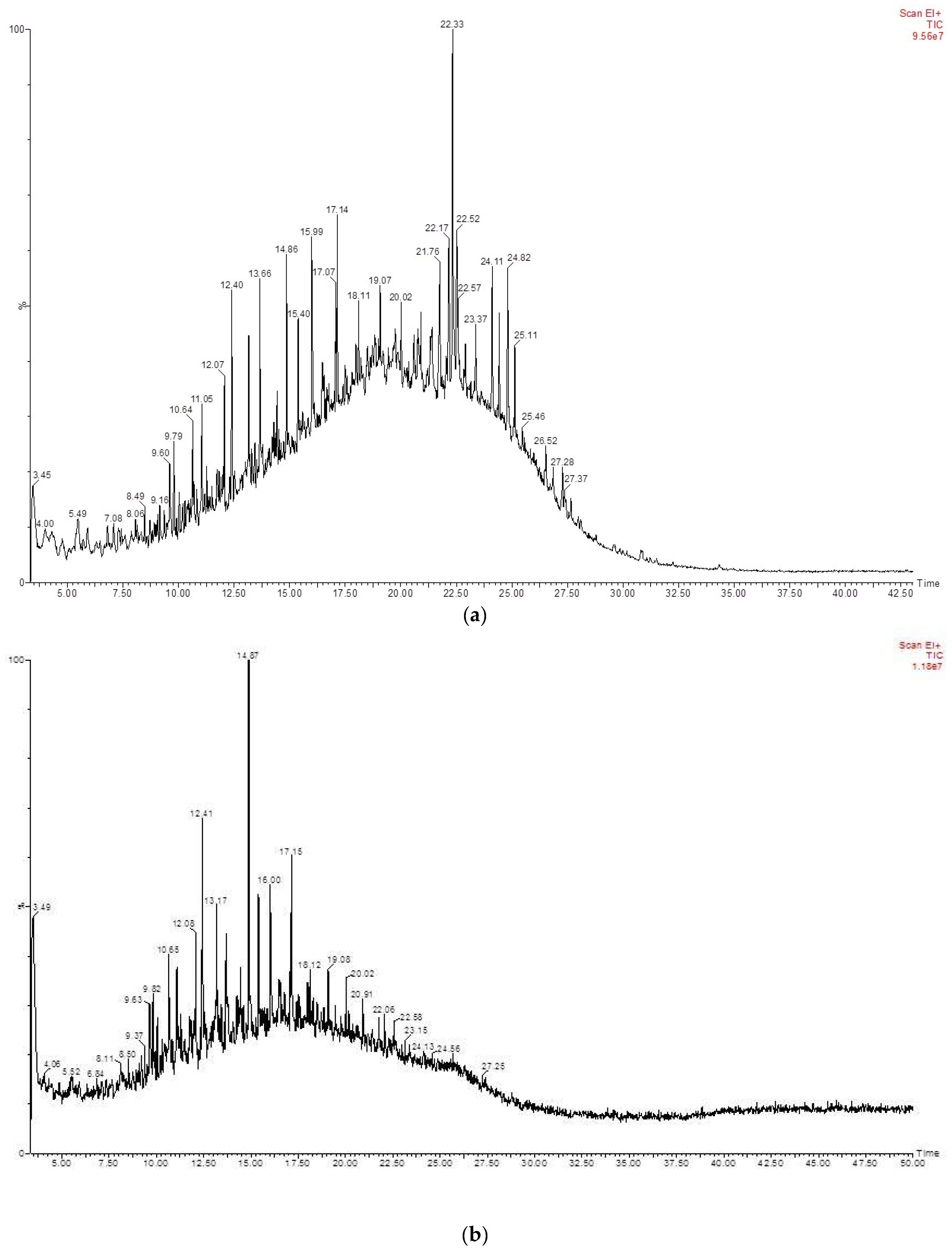
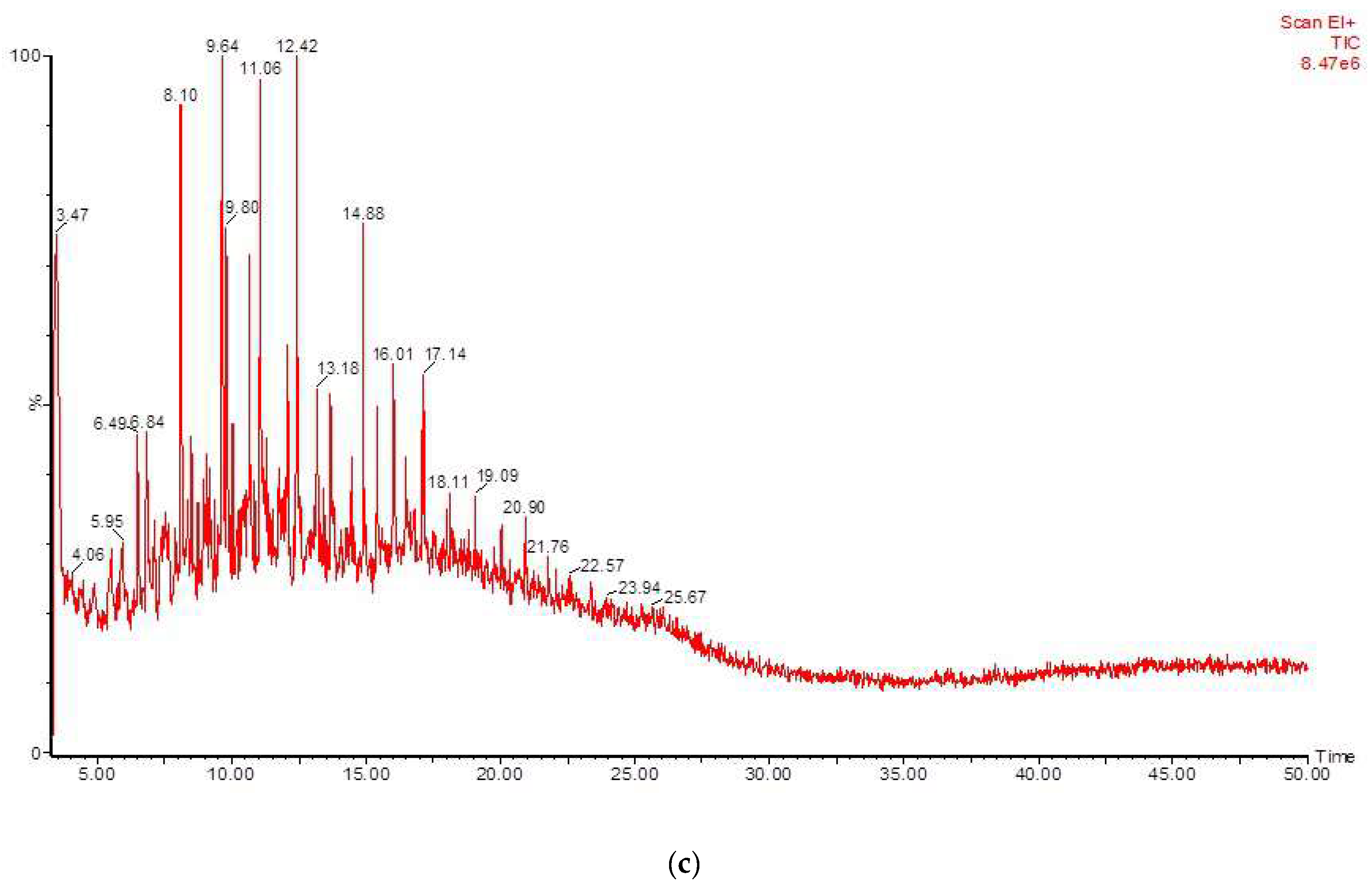
3.4. Biotransformation Studies Using GC-MS
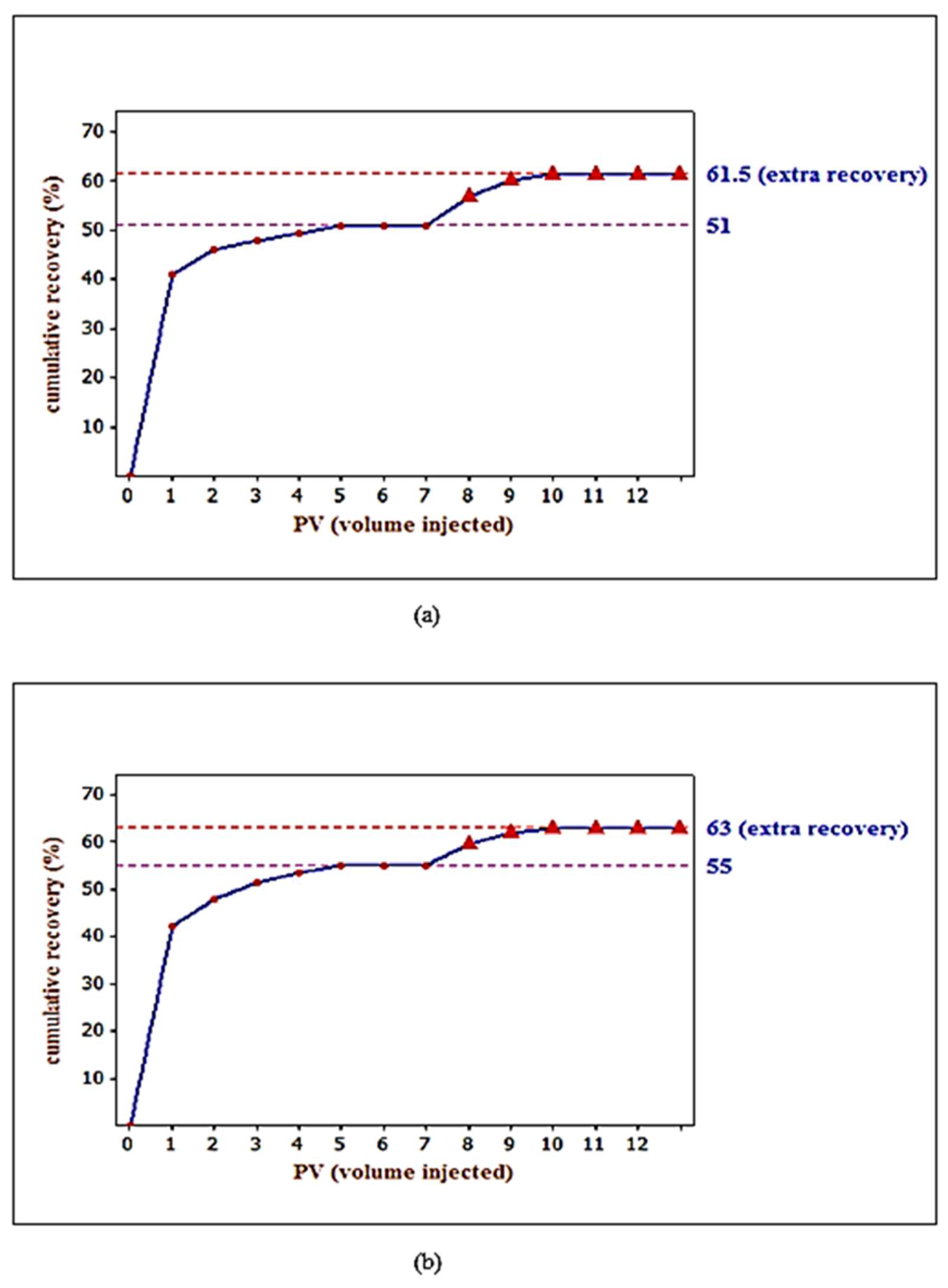
3.5. Core Flooding Experiments
4. Discussion
5. Conclusions
Acknowledgments
Author Contributions
Conflicts of Interest
References
- Babadagli, T. Development of mature oil fields—A review. J. Pet. Sci. Eng. 2007, 57, 221–246. [Google Scholar] [CrossRef]
- Bao, M.; Kong, X.; Jiang, G.; Wang, X.; Li, X. Laboratory study on activating indigenous microorganisms to enhance oil recovery in Shengli Oilfield. J. Pet. Sci. Eng. 2009, 66, 42–46. [Google Scholar] [CrossRef]
- Shibulal, B.; Al-Bahry, S.N.; Al-Wahaibi, Y.M.; Elshafie, A.E.; Al-Bemani, A.S.; Joshi, S.J. Microbial Enhanced Heavy Oil Recovery by the Aid of Inhabitant Spore-Forming Bacteria: An Insight Review. Sci. World J. 2014, 2014. [Google Scholar] [CrossRef] [PubMed]
- Huc, A.-Y. Heavy Crude Oils: From Geology to Upgrading: An Overview; Editions Technip: Paris, France, 2010. [Google Scholar]
- Al-Sayegh, A.; Al-Wahaibi, Y.; Joshi, S.; Al-Bahry, S.; Elshafie, A.; Al-Bemani, A. Bioremediation of Heavy Crude Oil Contamination. Open Biotechnol. J. 2016, 10, 301–311. [Google Scholar] [CrossRef]
- Temizel, C.; Rodriguez, D.; Saldierna, N.; Narinesingh, J. Stochastic Optimization of Steam flooding Heavy Oil Reservoirs. In Proceedings of the Society of Petroleum Engineers (SPE) Trinidad and Tobago Section Energy Resources Conference, Port of Spain, Trinidad and Tobago, 13–15 June 2016. [Google Scholar]
- Joshi, S.J. Isolation and Characterization of Biosurfactant Producing Microorganisms and Their Possible Role in Microbial Enhanced Oil Recovery (MEOR). Ph.D. Thesis, Maharaja Sayajirao University of Baroda, Vadodara, India, 2008. [Google Scholar]
- Sivasankar, P.; Kumar, G.S. Influence of pH on dynamics of microbial enhanced oil recovery processes using biosurfactant producing Pseudomonas putida: Mathematical modelling and numerical simulation. Bioresour. Technol. 2017, 224, 498–508. [Google Scholar] [CrossRef] [PubMed]
- Banat, I.M. Biosurfactants production and possible uses in microbial enhanced oil recovery and oil pollution remediation: A review. Bioresour. Technol. 1995, 51, 1–12. [Google Scholar] [CrossRef]
- Xu, T.; Chen, C.; Liu, C.; Zhang, S.; Wu, Y.; Zhang, P. A novel way to enhance the oil recovery ratio by Streptococcus sp. BT-003. J. Basic Microbiol. 2009, 49, 477–481. [Google Scholar] [CrossRef] [PubMed]
- Brown, L.R. Microbial enhanced oil recovery (MEOR). Curr. Opin. Microbiol. 2010, 13, 316–320. [Google Scholar] [CrossRef] [PubMed]
- Bryant, R.S.; Douglas, J. Survival of MEOR Systems in Porous Media; National Institute for Petroleum and Energy Research: Bartlesville, OK, USA, 1986; pp. 1–32.
- Lazar, I.; Petrisor, I.; Yen, T. Microbial enhanced oil recovery (MEOR). Pet. Sci. Technol. 2007, 25, 1353–1366. [Google Scholar] [CrossRef]
- Nielsen, S.M.; Shapiro, A.A.; Michelsen, M.L.; Stenby, E.H. 1D Simulations for Microbial Enhanced Oil Recovery with Metabolite Partitioning. Transp. Porous Media 2010, 85, 785–802. [Google Scholar] [CrossRef]
- Almeida, P.; Moreira, R.; Almeida, R.; Guimaraes, A.; Carvalho, A.; Quintella, C.; Esperidia, M.; Taft, C. Selection and application of microorganisms to improve oil recovery. Eng. Life Sci. 2004, 4, 319–325. [Google Scholar] [CrossRef]
- Jinfeng, L.; Lijun, M.; Bozhong, M.; Rulin, L.; Fangtian, N.; Jiaxi, Z. The field pilot of microbial enhanced oil recovery in a high temperature petroleum reservoir. J. Pet. Sci. Eng. 2005, 48, 265–271. [Google Scholar] [CrossRef]
- Wentzel, A.; Ellingsen, T.E.; Kotlar, H.K.; Zotchev, S.B.; Throne-Holst, M. Bacterial metabolism of long-chain n-alkanes. Appl. Microbiol. Biotechnol. 2007, 76, 1209–1221. [Google Scholar] [CrossRef] [PubMed]
- Grishchenkov, V.G.; Townsend, R.T.; McDonald, T.J.; Autenrieth, R.L.; Bonner, J.S.; Boronin, A.M. Degradation of petroleum hydrocarbons by facultative anaerobic bacteria under aerobic and anaerobic conditions. Process Biochem. 2000, 35, 889–896. [Google Scholar] [CrossRef]
- Li, Q.; Kang, C.; Wang, H.; Liu, C.; Zhang, C. Application of microbial enhanced oil recovery technique to Daqing Oilfield. Biochem. Eng. J. 2002, 11, 197–199. [Google Scholar] [CrossRef]
- Shibulal, B.; Al-Bahry, S.N.; Al-Wahaibi, Y.M.; Elshafie, A.E.; Al-Bemani, A.S.; Joshi, S.J. The potential of indigenous Paenibacillus ehimensis BS1 for recovering heavy crude oil by biotransformation to light fractions. PLoS ONE 2017, 12, e0171432. [Google Scholar] [CrossRef] [PubMed]
- Etoumi, A.; Musrati, I.E.; Gammoudi, B.E.; Behlil, M.E. The reduction of wax precipitation in waxy crude oils by Pseudomonas species. J. Ind. Microbiol. Biotechnol. 2008, 35, 1241–1245. [Google Scholar] [CrossRef] [PubMed]
- Binazadeh, M.; Karimi, I.A.; Li, Z. Fast biodegradation of long chain n-alkanes and crude oil at high concentration with Rhodococcus sp. Moj-3449. Enzym. Microb. Technol. 2009, 45, 195–202. [Google Scholar] [CrossRef]
- Zhang, Y.; Xu, Z.; Ji, P.; Hou, W.; Dietrich, F. Microbial EOR laboratory studies and application results in Daqing oilfield. In Proceedings of the Society of Petroleum Engineers (SPE) Asia Pacific Oil and Gas Conference and Exhibition, Jakarta, Indonesia, 20–22 April 1999. [Google Scholar]
- Felix, J.A.; Cooney, J.J. Response of spores and vegetative cells of Bacillus spp. in a hydrocarbon–water system. J. Appl. Bacteriol. 1971, 34, 411–416. [Google Scholar] [CrossRef] [PubMed]
- Calvo, C.; Toledo, F.L.; González-López, J. Surfactant activity of a naphthalene degrading Bacillus pumilus strain isolated from oil sludge. J. Biotechnol. 2004, 109, 255–262. [Google Scholar] [CrossRef] [PubMed]
- Al-Bahry, S.N.; Al-Wahaibi, Y.M.; Al-Hinai, B.; Joshi, S.J.; Elshafie, A.E.; Al-Bemani, A.S.; Al-Sabahi, J. Potential in heavy oil biodegradation via enrichment of spore forming bacterial consortia. J. Pet. Explor. Prod. Technol. 2016, 6, 787–799. [Google Scholar] [CrossRef]
- Zhuang, W.O.; Tay, J.H.; Maszenan, A.M.; Krumholz, L.R.; Tay, S.L. Importance of Gram-positive naphthalene-degrading bacteria in oil-contaminated tropical marine sediments. Lett. Appl. Microbiol. 2003, 36, 251–257. [Google Scholar] [CrossRef] [PubMed]
- Ijah, U.; Ukpe, L. Biodegradation of crude oil by Bacillus strains 28A and 61B isolated from oil spilled soil. Waste Manag. 1992, 12, 55–60. [Google Scholar] [CrossRef]
- Da Cunha, C.D.; Rosado, A.S.; Sebastián, G.V.; Seldin, L.; von der Weid, I. Oil biodegradation by Bacillus strains isolated from the rock of an oil reservoir located in a deep-water production basin in Brazil. Appl. Microbiol. Biotechnol. 2006, 73, 949–959. [Google Scholar] [CrossRef] [PubMed]
- Cooper, D.G.; Macdonald, C.R.; Duff, S.J.B.; Kosaric, N. Enhanced production of surfactin from Bacillus subtilis by continuous product removal and metal cation additions. Appl. Environ. Microbiol. 1981, 42, 408–412. [Google Scholar] [PubMed]
- Alinnor, I.J.; Nwachukwu, M.A. Determination of total petroleum hydrocarbon in soil and groundwater samples in some communities in Rivers State, Nigeria. J. Environ. Chem. Ecotoxicol. 2013, 5, 292–297. [Google Scholar]
- EPA. gov: Method 3541 (SW-846): Automated Soxhlet Extraction. 1994. Available online: https://www.epa.gov/sites/production/files/2015-06/documents/epa-3541.pdf (accessed on 6 November 2016).
- Kato, T.; Haruki, M.; Imanaka, T. Isolation and characterization of psychotrophic bacteria from oil-reservoir water and oil sands. Appl. Microbiol. Biotechnol. 2001, 55, 794–800. [Google Scholar] [CrossRef] [PubMed]
- Hilyard, E.J.; Jones-Meehan, J.M.; Spargo, B.J.; Hill, R.T. Enrichment, Isolation, and Phylogenetic Identification of Polycyclic Aromatic Hydrocarbon-Degrading Bacteria from Elizabeth River Sediments. Appl. Environ. Microbiol. 2008, 74, 1176–1182. [Google Scholar] [CrossRef] [PubMed]
- Kleinheinz, G.T.; Bagley, S.T. A filter-plate method for the recovery and cultivation of microorganisms utilizing volatile organic compounds. J. Microbiol. Methods 1997, 29, 139–144. [Google Scholar] [CrossRef]
- Koubek, J.; Uhlik, O.; Jecna, K.; Junkova, P.; Vrkoslavova, J.; Lipov, J.; Kurzawova, V.; Macek, T.; Mackova, M. Whole-cell MALDI-TOF: Rapid screening method in environmental microbiology. Int. Biodeterior. Biodegrad. 2012, 69, 82–86. [Google Scholar] [CrossRef]
- Wunschel, S.C.; Jarman, K.H.; Petersen, C.E.; Valentine, N.B.; Wahl, K.L.; Schauki, D.; Jackman, J.; Nelson, C.P.; White, E. Bacterial analysis by MALDI-TOF mass spectrometry: An inter-laboratory comparison. J. Am. Soc. Mass Spectrom. 2005, 16, 456–462. [Google Scholar] [CrossRef] [PubMed]
- Minai-Tehrani, D.; Herfatmanesh, A. Biodegradation of Aliphatic and Aromatic Fractions of Heavy Crude Oil–Contaminated Soil: A Pilot Study. Bioremediat. J. 2007, 11, 71–76. [Google Scholar] [CrossRef]
- United States Environmental Protection Agency (USEPA): Method 8270D. Semivolatile Organic Compounds by Gas Chromatography/Mass Spectrophotometry; Revision 5; (SW-846 Update V); USEPA: Washington, DC, USA, July 2014.
- Raiders, R.A.; Knapp, R.M.; McInerney, M.J. Microbial selective plugging and enhanced oil recovery. J. Ind. Microbial. 1989, 4, 215–229. [Google Scholar] [CrossRef]
- Davey, M.E.; Gevertz, D.; Wood, W.A.; Clark, J.B.; Jenneman, G.E. Microbial selective plugging of sandstone through stimulation of indigenous bacteria in a hypersaline oil reservoir. Geomicrobiol. J. 1998, 15, 335–352. [Google Scholar] [CrossRef]
- Al-Hattali, R.; Al-Sulaimani, H.; Al-Wahaibi, Y.; Al-Bahry, S.; Elshafie, A.; Al-Bemani, A.; Joshi, S.J. Fractured carbonate reservoirs sweep efficiency improvement using microbial biomass. J. Pet. Sci. Eng. 2013, 112, 178–184. [Google Scholar] [CrossRef]
- Beckman, J.W. The Action of Bacteria on Mineral Oil. Ind. Eng. Chem. News Ed. 1926, 4, 10. [Google Scholar]
- Dibble, J.T.; Bartha, R. Effect of environmental parameters on the biodegradation of oil sludge. Appl. Environ. Microbiol. 1979, 37, 729–739. [Google Scholar] [PubMed]
- Hambrick, G.A.; DeLaune, R.D.; Patrick, W.H. Effect of Estuarine Sediment pH and Oxidation-Reduction Potential on Microbial Hydrocarbon Degradation. Appl. Environ. Microbiol. 1980, 40, 365–369. [Google Scholar] [PubMed]
- Rahman, K.S.; Thahira-Rahman, J.; Lakshmanaperumalsamy, P.; Banat, I.M. Towards efficient crude oil degradation by a mixed bacterial consortium. Bioresour. Technol. 2002, 85, 257–261. [Google Scholar] [CrossRef] [Green Version]
- Sepahi, A.A.; Golpasha, I.D.; Emami, M.; Nakhoda, A. Isolation and Characterization of Crude Oil Degrading Bacillus Spp. J. Environ. Health Sci. Eng. 2008, 5, 149–154. [Google Scholar]
- Ojo, O. Petroleum-hydrocarbon utilization by native bacterial population from a wastewater canal Southwest Nigeria. Afr. J. Biotechnol. 2006, 5, 333. [Google Scholar]
- Stackebrandt, E.; Goebel, B. Taxonomic note: A place for DNA-DNA reassociation and 16S rRNA sequence analysis in the present species definition in bacteriology. Int. J. Syst. Evol. Microbiol. 1994, 44, 846–849. [Google Scholar] [CrossRef]
- Head, I.M.; Jones, D.M.; Larter, S.R. Biological activity in the deep subsurface and the origin of heavy oil. Nature 2003, 426, 344–352. [Google Scholar] [CrossRef] [PubMed]
- Liu, H.; Xu, J.; Liang, R.; Liu, J. Characterization of the Medium- and Long-Chain n-Alkanes Degrading Pseudomonas aeruginosa Strain SJTD-1 and Its Alkane Hydroxylase Genes. PLoS ONE 2014, 9, e105506. [Google Scholar] [CrossRef] [PubMed]
- Kostka, J.E.; Prakash, O.; Overholt, W.A.; Green, S.J.; Freyer, G.; Canion, A.; Delgardio, J.; Norton, N.; Hazen, T.C.; Huettel, M. Hydrocarbon-Degrading Bacteria and the Bacterial Community Response in Gulf of Mexico Beach Sands Impacted by the Deepwater Horizon Oil Spill. Appl. Environ. Microbiol. 2011, 77, 7962–7974. [Google Scholar] [CrossRef] [PubMed]
- Brooijmans, R.J.W.; Pastink, M.I.; Siezen, M.J. Hydrocarbon-degrading bacteria: The oil-spill clean-up crew. Microb. Biotechnol. 2009, 2, 587–594. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Sorkhoh, N.; Ibrahim, A.; Ghannoum, M.A.; Radwan, S. High-temperature hydrocarbon degradation by Bacillus stearothermophilus from oil-polluted Kuwaiti desert. Appl. Microbiol. Biotechnol. 1993, 39, 123–126. [Google Scholar] [CrossRef]
- Van Beilen, J.B.; Marin, M.M.; Smits, T.H.; Röthlisberger, M.; Franchini, A.G.; Witholt, B.; Rojo, F. Characterization of two alkane hydroxylase genes from the marine hydrocarbonoclastic bacterium Alcanivorax borkumensis. Environ. Microbial. 2004, 6, 264–273. [Google Scholar] [CrossRef]
- Mittal, A.; Singh, P. Isolation of hydrocarbon degrading bacteria from soils contaminated with crude oil spills. Indian J. Exp. Boil. 2009, 47, 760. [Google Scholar]
- Palanisamy, N.; Ramya, J.; Kumar, S.; Vasanthi, N.S.; Chandran, P.; Khan, S. Diesel biodegradation capacities of indigenous bacterial species isolated from diesel contaminated soil. J. Environ. Health Sci. Eng. 2014, 12, 142. [Google Scholar] [CrossRef] [PubMed]
- Chhatre, S.; Purohit, H.; Shanker, R.; Khanna, P. Bacterial consortia for crude oil spill remediation. Water Sci. Technol. 1996, 34, 187–193. [Google Scholar] [CrossRef]
- Vasudevan, N.; Rajaram, P. Bioremediation of oil sludge-contaminated soil. Environ. Int. 2001, 26, 409–411. [Google Scholar] [CrossRef]
- Gudiña, E.J.; Pereira, J.F.B.; Rodrigues, L.R.; Coutinho, J.A.P.; Teixeira, J.A. Isolation and study of microorganisms from oil samples for application in Microbial Enhanced Oil Recovery. Int. Biodeterior. Biodegrad. 2012, 68, 56–64. [Google Scholar] [CrossRef] [Green Version]
- She, Y.-H.; Zhang, F.; Xia, J.-J.; Kong, S.-Q.; Wang, Z.-L.; Shu, F.-C.; Hu, J.-M. Investigation of biosurfactant-producing indigenous microorganisms that enhance residue oil recovery in an oil reservoir after polymer flooding. Appl. Biochem. Biotechnol. 2011, 163, 223–234. [Google Scholar] [CrossRef] [PubMed]
- Hao, R.; Lu, A.; Wang, G. Crude-oil-degrading thermophilic bacterium isolated from an oil field. Can. J. Microbiol. 2004, 50, 175–182. [Google Scholar] [CrossRef] [PubMed]
- Wu, L.; Yao, J.; Jain, A.K.; Radhika, C.; Xudong, D.; Hans, R.H. An efficient thermotolerant and halophilic biosurfactant-producing bacterium isolated from Dagang oil field for MEOR application. Int. J. Curr. Microbiol. Appl. Sci. 2014, 3, 586–599. [Google Scholar]
- Al-Sayegh, A.; Al-Wahaibi, Y.; Al-Bahry, S.; Elshafie, A.; Al-Bemani, A.; Joshi, S. Microbial enhanced heavy crude oil recovery through biodegradation using bacterial isolates from an Omani oil field. Microb. Cell Fact. 2015, 14, 141. [Google Scholar] [CrossRef] [PubMed]
- Al-Sayegh, A.; Al-Wahaibi, Y.; Al-Bahry, S.; Elshafie, A.; Al-Bemani, A.; Joshi, S. Enhanced oil recovery using biotransformation technique on heavy crude oil. Int. J. GEOMATE 2017, 13, 75–79. [Google Scholar] [CrossRef]
- Youssef, N.; Elshahed, M.S.; McInerney, M.J. Microbial processes in oil fields: Culprits, problems, and opportunities. Adv. Appl. Microbial. 2009, 66, 141–251. [Google Scholar]



| Mineral | Soil Samples | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| SA | SB | SC | SD | SE | SF | SG | SH | SI | SJ | |
| albite | 1 | 1 | 0 | 1 | 0 | 1 | 1 | 1 | 1 | 1 |
| anorthite | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 0 |
| calcite | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 |
| dolomite | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| gypsum | 1 | 0 | 1 | 1 | 0 | 1 | 1 | 0 | 1 | 1 |
| halite | 0 | 1 | 0 | 1 | 0 | 0 | 1 | 1 | 0 | 1 |
| microcline | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 1 | 0 |
| muscovite | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 |
| palygorskite | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 0 | 1 | 0 |
| quartz | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 |
| rutile | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| suhailite | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| takanelite | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 |
| RT | Identified Compound | Carbon No. |
|---|---|---|
| 9.63 | 2-methyl-1-pentanol | C6 |
| 9.82 | cycloheptanol | C7 |
| 10.06 | 1,2-dibromo-octane | C8 |
| 10.65 | 2,4,4-trimethyl-1-hexene | C9 |
| 11.08 | 1,2-dibromo-2-methyl-undecane | C12 |
| 12.41 | 1,2-dibromododecane | C12 |
| 13.17 | 3,7,11-trimethyl-1-dodecanol | C15 |
| 13.67 | 1-nonadecanol | C19 |
| 14.87 | hexadecanoic acid, (3-bromoprop-2-ynyl) ester | C19 |
| 15.39 | 1-bromoeicosane | C20 |
| 17.15 | 5,15-dimethylnonadecane | C21 |
| 18.12 | 2-nitro-1,3-bis(octyloxy)benzene | C22 |
| 19.08 | 7-hexyldocosane | C28 |
| 20.02 | 11-decyldocosane | C32 |
| 21.76 | tritriacontane | C33 |
| 22.06 | 1-hexadecylheptadecylcyclohexane | C39 |
| 22.58 | tetratetracontane | C44 |
| RT | Identified Compound | Carbon No. |
|---|---|---|
| 3.47 | 1,2-dibromo-2-methylundecane | C4 |
| 4.06 | 2-nitrocyclohexanone | C6 |
| 5.51 | 2,5-heptadecadione | C7 |
| 5.95 | 1,7-dichloroheptane | C7 |
| 6.49 | 2,2-dimethyl-3-pentanol | C7 |
| 6.84 | 1-chloro-heptane | C7 |
| 8.10 | N-methylcyclohexanamine | C7 |
| 9.64 | acetic acid, hexyl ester | C8 |
| 9.80 | 1,2-dibromo-octane | C8 |
| 11.06 | 1,2-dibromododecane | C12 |
| 12.42 | 1-chlorododecane | C12 |
| 13.18 | 1-nonadecanol | C19 |
| 14.88 | 1- eicosanol | C20 |
| 16.01 | 9-octadecenyl acetate | C20 |
| 17.14 | dimethylnonadecane | C21 |
| 18.11 | tetracosane | C24 |
| 19.09 | 7-hexyldocosane | C28 |
| 20.90 | 2-(1-decylundecyl)-1,4-dimethyl cyclohexane | C29 |
| 21.76 | 11-decyldocosane | C32 |
| 22.57 | tritriacontane | C33 |
© 2018 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Shibulal, B.; Al-Bahry, S.N.; Al-Wahaibi, Y.M.; Elshafie, A.E.; Al-Bemani, A.S.; Joshi, S.J. Microbial-Enhanced Heavy Oil Recovery under Laboratory Conditions by Bacillus firmus BG4 and Bacillus halodurans BG5 Isolated from Heavy Oil Fields. Colloids Interfaces 2018, 2, 1. https://doi.org/10.3390/colloids2010001
Shibulal B, Al-Bahry SN, Al-Wahaibi YM, Elshafie AE, Al-Bemani AS, Joshi SJ. Microbial-Enhanced Heavy Oil Recovery under Laboratory Conditions by Bacillus firmus BG4 and Bacillus halodurans BG5 Isolated from Heavy Oil Fields. Colloids and Interfaces. 2018; 2(1):1. https://doi.org/10.3390/colloids2010001
Chicago/Turabian StyleShibulal, Biji, Saif N. Al-Bahry, Yahya M. Al-Wahaibi, Abdulkadir E. Elshafie, Ali S. Al-Bemani, and Sanket J. Joshi. 2018. "Microbial-Enhanced Heavy Oil Recovery under Laboratory Conditions by Bacillus firmus BG4 and Bacillus halodurans BG5 Isolated from Heavy Oil Fields" Colloids and Interfaces 2, no. 1: 1. https://doi.org/10.3390/colloids2010001
APA StyleShibulal, B., Al-Bahry, S. N., Al-Wahaibi, Y. M., Elshafie, A. E., Al-Bemani, A. S., & Joshi, S. J. (2018). Microbial-Enhanced Heavy Oil Recovery under Laboratory Conditions by Bacillus firmus BG4 and Bacillus halodurans BG5 Isolated from Heavy Oil Fields. Colloids and Interfaces, 2(1), 1. https://doi.org/10.3390/colloids2010001






