1. Introduction
Cervical cancer remains one of the leading health threats for women worldwide, especially in low- and middle-income countries, where it is the second-most prevalent cancer among women [
1,
2]. Globally, over 600,000 new cases and over 340,000 deaths were reported in 2020, of which about 90% occurred in resource-limited regions [
3]. The high mortality in these areas largely stems from inadequate access to screening, human papillomavirus (HPV) vaccination, and timely medical intervention [
4]. HPV infection is recognized as the primary cause of cervical cancer, yet barriers to implementing preventive measures persist due to economic and healthcare disparities [
5]. Advances in diagnostic tools, including machine learning and artificial intelligence, show promise for enhancing early detection, which is critical given that early-stage intervention could reduce cervical cancer mortality by up to 60% [
6]. More broadly, AI and machine learning (ML) applications are increasingly transforming healthcare by improving diagnostics, optimizing treatments, and enhancing patient outcomes [
7,
8]
The integration of Deep Learning (DL) in cervical cancer diagnostics has advanced significantly in recent years, with applications that enhance automated image analysis and assist in early detection [
9,
10]. DL techniques now focus extensively on cervical cell classification, leveraging large datasets to train models, although publicly available data remains limited [
11]. For this reason, transfer learning has been widely employed to address data scarcity by adapting pre-trained models from large, general datasets [
12]. This method has shown particular promise in cervical cell detection tasks, allowing networks to achieve higher accuracy despite constrained data [
13]. As DL continues to evolve, its impact on cervical cancer detection grows, offering scalable solutions that aid in screening and reduce the diagnostic burden on pathologists.
Ensuring trustworthiness in AI models is essential, especially in healthcare, where biases in training data or model structures can disproportionately impact certain demographic groups. Recent research emphasizes the need for fairness-focused metrics and strategies to mitigate these biases, which are present in diverse applications such as environmental security and semantic segmentation, and now in clinical predictions as well [
14,
15,
16]. Despite advances in algorithmic fairness, the evaluation and reduction in biases within clinical models remains challenging due to the limited accessibility of representative health data and the complexity of high-dimensional medical information [
17]. In healthcare, these biases can lead to disparities in diagnostic outcomes, reinforcing existing inequities. Addressing these issues requires a comprehensive approach involving data quality enhancement, explicitly fair algorithms, and interdisciplinary collaboration to ensure ethical, equitable AI deployment in clinical settings [
18].
Transfer learning has emerged as a powerful technique to mitigate the effects of data scarcity and to address biases in medical image analysis by leveraging pre-trained models from tasks with varying dataset sizes, regardless of their relative scale [
19,
20]. In medical applications, transfer learning enables models to inherit feature representations from large, well-curated datasets, which can be fine-tuned to perform effectively even with limited domain-specific data [
21,
22]. This is particularly useful for complex tasks like cervical cell detection, where annotated data is often limited. Studies show that using domain-specific transfer learning can significantly improve model accuracy and reduce bias by aligning features more closely with the target medical context [
13,
22]. This approach not only enhances model robustness but also minimizes the need for extensive manual labeling, making it a viable solution to achieve fairer and more accurate predictions in clinical settings. Building on these foundations, this study introduces a novel dataset fusion methodology that further addresses the challenge of balancing domain-specific optimization with cross-domain adaptability.
The purpose of this work is to develop a methodology for creating more robust and Trustworthy Artificial Intelligence (TAI) models for health applications that mitigate bias effects and address the challenges described above. This objective will be achieved by developing a novel methodology, Interleaved Fusion Learning, that can be applied to a family of models, each specialized for a specific dataset, while also benefiting from shared knowledge across all datasets. By allowing models to share knowledge while maintaining specialization for their respective datasets, this methodology facilitates a synergistic fusion of diverse data sources, integrating heterogeneous data to enhance enhance predictive performance across a broader range of data. The proposed methodology will be evaluated using cervical cancer datasets to demonstrate its potential in improving screening solutions.
The organization of this paper is outlined as follows: initially,
Section 2 provides an overview of the materials and techniques essential for conducting this research. Subsequently,
Section 3 provides an explanation of the proposed approach for the development and evaluation of the interleaved fusion learned models. In
Section 4, we detail our implementation of the methodology and present a selection of the data obtained through our algorithm. Finally, in
Section 5 and
Section 6 , we engage in an analysis of the data and seek to draw some definitive conclusions.
2. Materials and Methods
2.1. Open Cervical Cancer Datasets
To develop robust models for cervical cell segmentation and classification, large and well-annotated datasets are essential. Among the most comprehensive publicly available resources, the APACC (Annotated PAp cell images and smear slices for Cell Classification) dataset and the CRIC (Center for Recognition and Inspection of Cells) Cervix collection stand out for their extensive cell annotations and segmentation data, which make them highly suitable for DL applications [
23,
24]. Both datasets offer the detailed, large-scale data necessary for effective model training and evaluation in the context of cervical cancer screening, supporting a range of tasks from detection to classification. Given their quality and scope, APACC dataset and CRIC Cervix collection provide an invaluable foundation for advancing automated cytological analysis.
In contrast, other commonly used datasets in cervical cell research, such as SIPaKMeD, Herlev and Mendeley, are less suited to our objectives. SIPaKMeD and Herlev datasets focus on isolated cells and are primarily used for image-based classification rather than full-image detection and segmentation, limiting their application for models requiring comprehensive smear data [
25,
26,
27]. On the other hand, the Mendeley dataset includes images with pointed-out cells but lacks full cell labeling, which restricts its effectiveness for segmentation-focused tasks [
27,
28].
2.1.1. CRIC Cervix Collection
The CRIC Cervix collection is a robust dataset, specifically curated to support automated analysis and detection in cervical cytology. Created as part of the CRIC initiative, this dataset includes 400 high-resolution RGB images (1376 × 1020 pixels), each containing manually classified cells. With a total of over 11,000 annotated cells, the CRIC Cervix collection offers the high-quality labels needed for machine learning models focused on cytopathological tasks [
24].
The CRIC Cervix collection uses the Bethesda System, which is the standardized terminology most widely adopted worldwide for cervical cytopathology, ensuring uniformity and reproducibility across laboratories and pathologists [
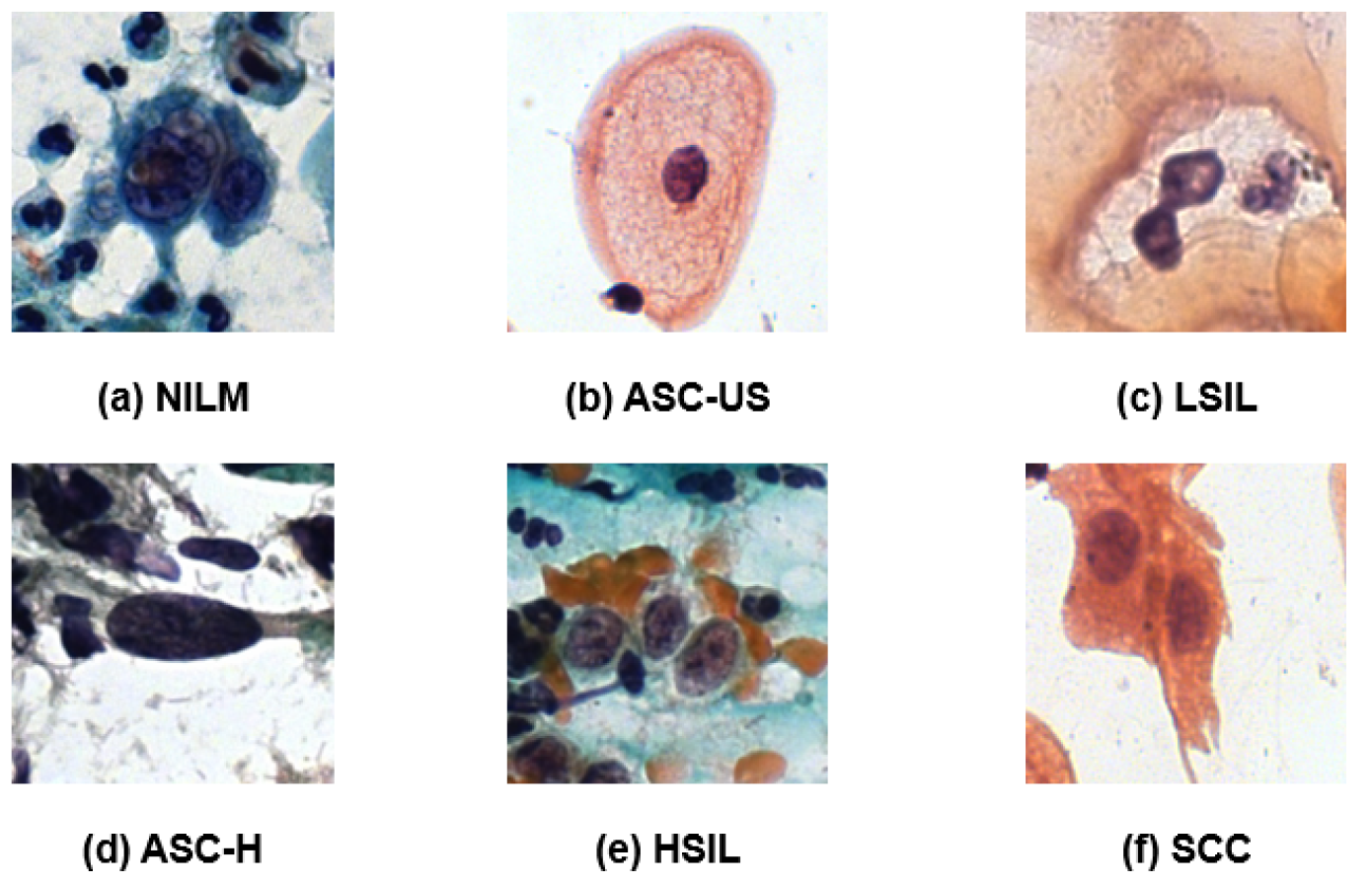
29]. This dataset classifies cells into six categories based on Bethesda nomenclature: (1) negative for intraepithelial lesion or malignancy (NILM), (2) atypical squamous cells of undetermined significance, possibly non-neoplastic (ASC-US), (3) low-grade squamous intraepithelial lesion (LSIL), (4) atypical squamous cells that cannot exclude a high-grade lesion (ASC-H), (5) high-grade squamous intraepithelial lesion (HSIL), and (6) squamous cell carcinoma (SCC). To streamline model development and enhance classification homogeneity with the APACC database, we have unified these categories into a binary classification system. The NILM category is maintained as is, while all other categories are combined and labeled as “Positive”.
The reference images for each category are shown in
Figure 1, with subfigures (a) through (f) illustrating representative cells from each class.
2.1.2. APACC Dataset
The APACC dataset is one of the most recent and comprehensive publicly available resources for cervical cell analysis. This dataset includes 103,675 annotated cell images, extracted from 107 whole Pap smears, and divided into over 21,000 sub-regions to support finer analysis [
23]. This sub-regions are RGB images with a resolution of 1984 × 1984 pixels.
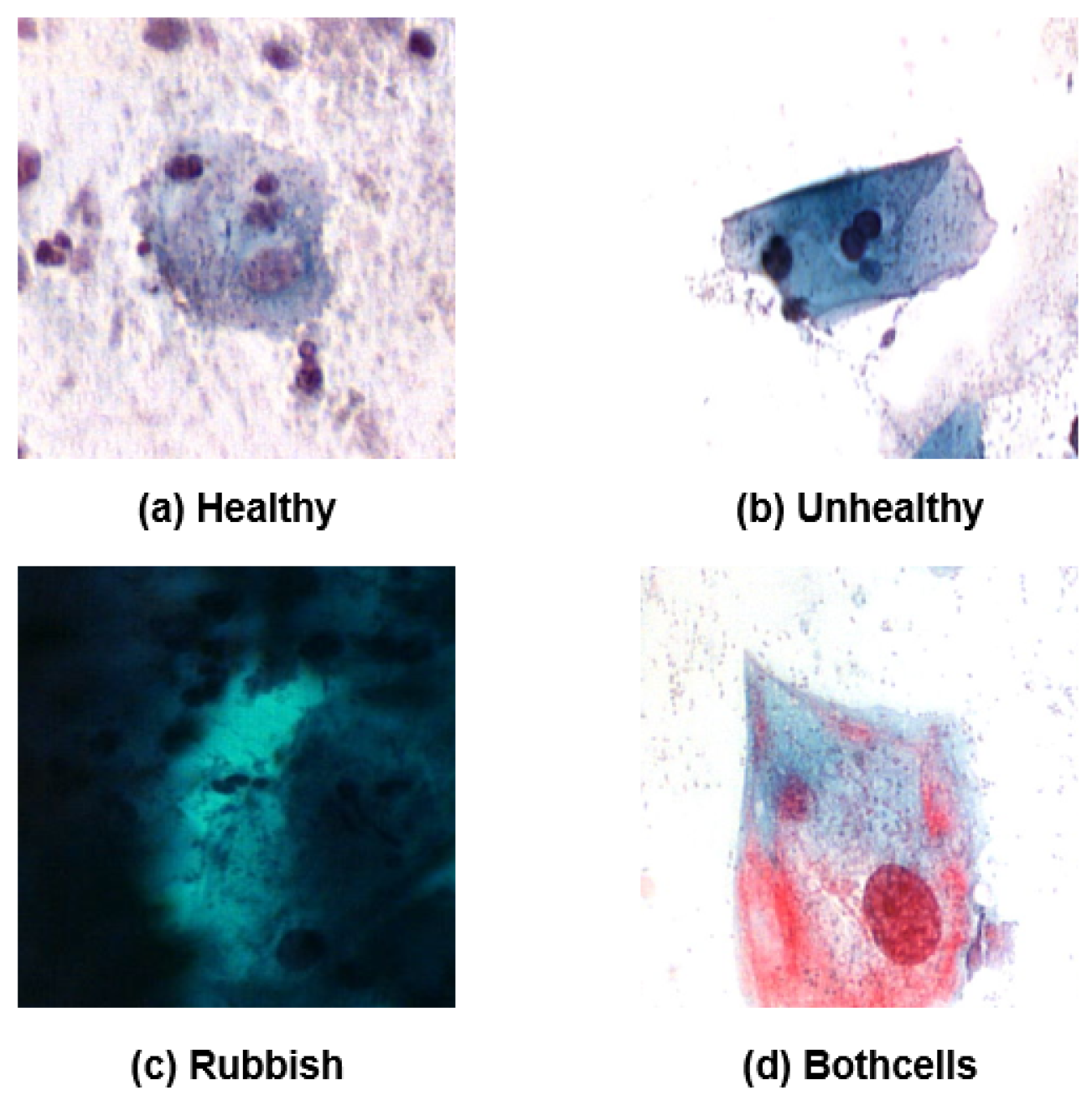
The APACC dataset categorizes cells into four classes: healthy (normal), unhealthy (abnormal), rubbish (not valid), and bothcells (a mixture of healthy and unhealthy cells). These classes loosely map onto Bethesda categories, where “healthy” corresponds to the NILM category, “unhealthy” represents cells from the broader Epithelial cell abnormality category (though without subdivisions like ASC, LSIL, or HSIL), “rubbish” aligns with Unsatisfactory for evaluation, and “bothcells” also falls within Epithelial cell abnormality as it includes malignant cells intermingled with normal cells. For our analysis, we have simplified the dataset by applying the same binary classification approach as in the CRIC database: NILM cells remain as “Negative,” while both “unhealthy” and “bothcells” are consolidated under a “Positive” label. The “rubbish” class has been excluded to ensure data relevance and consistency.
Representative examples of each original class from APACC are shown in
Figure 2, with subfigures (a) through (d) illustrating these types.
2.2. Deep Learning Architectures for Object Recognition: YOLOv8
YOLOv8 (You Only Look Once, Version 8), developed by Ultralytics, is one of the most advanced and efficient architectures for real-time object detection in medical imaging [
30,
31]. This DL model combines high accuracy with speed, making it ideal for applications requiring rapid identification of abnormalities, such as cervical cancer screening. With its optimized structure, YOLOv8 is particularly effective in identifying abnormal cells within complex, high-resolution images, which is crucial for early detection of precancerous and malignant lesions.
Its flexibility also allows it to be adapted to clinical environments where both diagnostic precision and processing speed are essential. Supporting both detection and classification tasks, YOLOv8 enhances the efficiency of cytological analysis, contributing to faster and more reliable early cancer screening workflows.
YOLOv8 provides several adjustable parameters that allow fine-tuning for specific tasks and datasets. The initial learning rate. controls the speed of weight updates during training, while the optimizer manages how the model minimizes the loss function. Early stopping prevents overfitting by halting training when validation metrics cease to improve. Data augmentation enhances generalization by applying random transformations to the training data. The batch size defines the number of samples processed before updating model weights, and the image size determines the resolution at which images are resized for input, balancing accuracy and computational efficiency.
2.3. Transfer Learning
Transfer learning is a DL approach that enables models to leverage knowledge acquired from one task (source task) to improve performance on a related task (target task), especially useful when the target domain has limited labeled data [
32].
Formally, a domain is defined as , where is a feature space, is a label space, and the function ensures that each feature has a corresponding label , such that . Given a specific domain D, we define a task , where is a subset of the feature space. For a specific task T, any function learned based on the relationships determined by the image of is referred to as a predictive function.
In transfer learning, the objective is to enhance a predictive function in a target domain with a corresponding target task , by leveraging knowledge from a source predictive function learned from a source domain and a source task . This process is valid under the assumption that either or .
The condition implies that the feature spaces, label spaces, or mapping functions differ between the source and target domains, i.e., , , or . Conversely, implies that the tasks differ, either in the subset of the feature space or in the label space .
Enhancing Transfer Learning
Two primary strategies for improving transfer learning outcomes are fine-tuning and weight initialization. Fine-tuning involves initializing a new model with pre-trained weights from the source domain and adapting specific layers to target domain data. This can involve adjusting all layers, or selectively fine-tuning only the last layers tailored to the target task. Weight initialization, by contrast, freezes certain pre-trained layers to retain general feature representations while adapting to the new domain through the remaining layers [
19].
Let represent the learned parameters of the source function . In fine-tuning, we initialize the target model with parameters and proceed to optimize a subset or the entirety of using labeled data from to better fit . On the other hand, weight initialization can be expressed by partitioning into two parameter sets, and , such that , where only is optimized with the target domain data, preserving the general representations from for use in .
3. Method
In this section we present Interleaved Fusion Learning (IFL), a new methodology designed to enhance model robustness and bias mitigation. The core concept behind this approach is to develop a sequence of models that effectively perform in their source datasets while adding essential knowledge from other datasets in the sequence. To determine whether this objective is achieved, a comprehensive evaluation framework will be established to assess and compare results across an arbitrary number n of datasets.
In
Figure 3, we present a schematic representation of the methodology pipeline. The diagram illustrates the IFL process: initially, each dataset
is employed to obtain a specific model
trained only on this dataset. Furthermore, a global model
f is trained on the combined dataset
. The specific models
then undergo the IFL pipeline, to produce a set of final models
, each model
specialized in
. Each final model
is evaluated against the global model
f on the dataset
, and against its corresponding initial model
on the remaining datasets
, with
.
To optimize performance across datasets while ensuring robustness and fairness, we start by formally defining the problem, which serves as the foundation for the proposed methodology.
3.1. Problem Definition
Building on the concepts introduced in
Section 2.3, transfer learning offers several key advantages for developing TAI systems. By leveraging transfer learning, models can be designed with enhanced robustness and fairness, mitigating biases inherent in training data while preserving performance specific to the target domain.
One approach to enhancing trustworthiness is through integrating a primary, potentially biased dataset , which represents the model’s main objective, with a secondary, less biased or unbiased dataset . Transfer learning between these datasets can adjust parameters in a way that preserves key knowledge from while mitigating bias through fine-tuning with . Formally, let and denote the learned parameters from and , respectively. Let be a partition of , and we can then optimize such that only is fine-tuned using samples from , adding robustness against bias while retaining essential information from .
The principles outlined above set the foundation for applying sequential transfer learning, where each dataset contributes to refining the model parameters progressively.
3.2. Sequential Transfer Learning
The methodology we propose defines a domain for each dataset, where is the set of features in the elements of the dataset, is the set of their possible labels, and d is the optimal function that correctly classifies the features with their corresponding labels. We also define the task , where X represents the labeled features of the dataset, i.e., the for which the image of is known. Specifically, each dataset can be represented as , where defines the domain with feature space , label space , and mapping function , and represents the unique task associated with the labeled subset and label space . Each trained model on a dataset functions as a predictive function , and its learned parameters correspond to the weights of .
The process is repeated sequentially for every
, where
n is the total number of available datasets. To begin, we define a baseline model
, trained from scratch using only the source dataset
, resulting in initial weights
. We then construct a sequence by choosing a subsequent index
j defined by
where
represents the step within the sequence. For each
j in this sequence, a model
is obtained by training a model in the dataset
, initializing it with the weights from training on the previous dataset in the sequence, denoted as
, where
This sequence continues iteratively through each dataset until we reach
, obtaining a final model
that has been adapted across all datasets in
. After completing this sequence for a given
i, the process is repeated for each
, resulting in a set of
n models
, each initialized with a different dataset. A schematic representation of this method is shown in
Figure 4.
Let represent a model trained from scratch on the combined dataset . Our goal for each transfer-learned model is to outperform when evaluated on its initial dataset . Additionally, each should yield better results on any other dataset (where ) compared to the baseline model , which is trained solely on . This criterion aims to demonstrate the advantage of transfer learning in enhancing performance consistency and fairness across diverse domains.
Learning Rate Selection for Transfer Learning
For each new dataset
, this methodology aims to set a learning rate that reflects the model’s performance on
. A high learning rate can reduce bias but may cause the model to lose specificity for the source datasets. To balance this trade-off, the new learning rate for transfer learning from
to
is determined as
where
with
controlling the maximum learning rate, and
representing the label corresponding each
in the dataset
.
The choice of this learning rate definition is motivated by several factors. Primarily, we aim for
, as is typical in transfer learning. This holds because the sum
is an integer in
, so dividing by
normalizes it to
. Multiplying by
, where
, ensures
Since is the minimum of this product and , both in , it follows that . The parameter controls the maximum learning rate, setting an upper bound of . By using in the other term of the minimum function, the learning rate is modulated by even when the error measure is low. This setup allows flexibility to adjust based on specific model requirements, enabling either a higher or lower learning rate as needed.
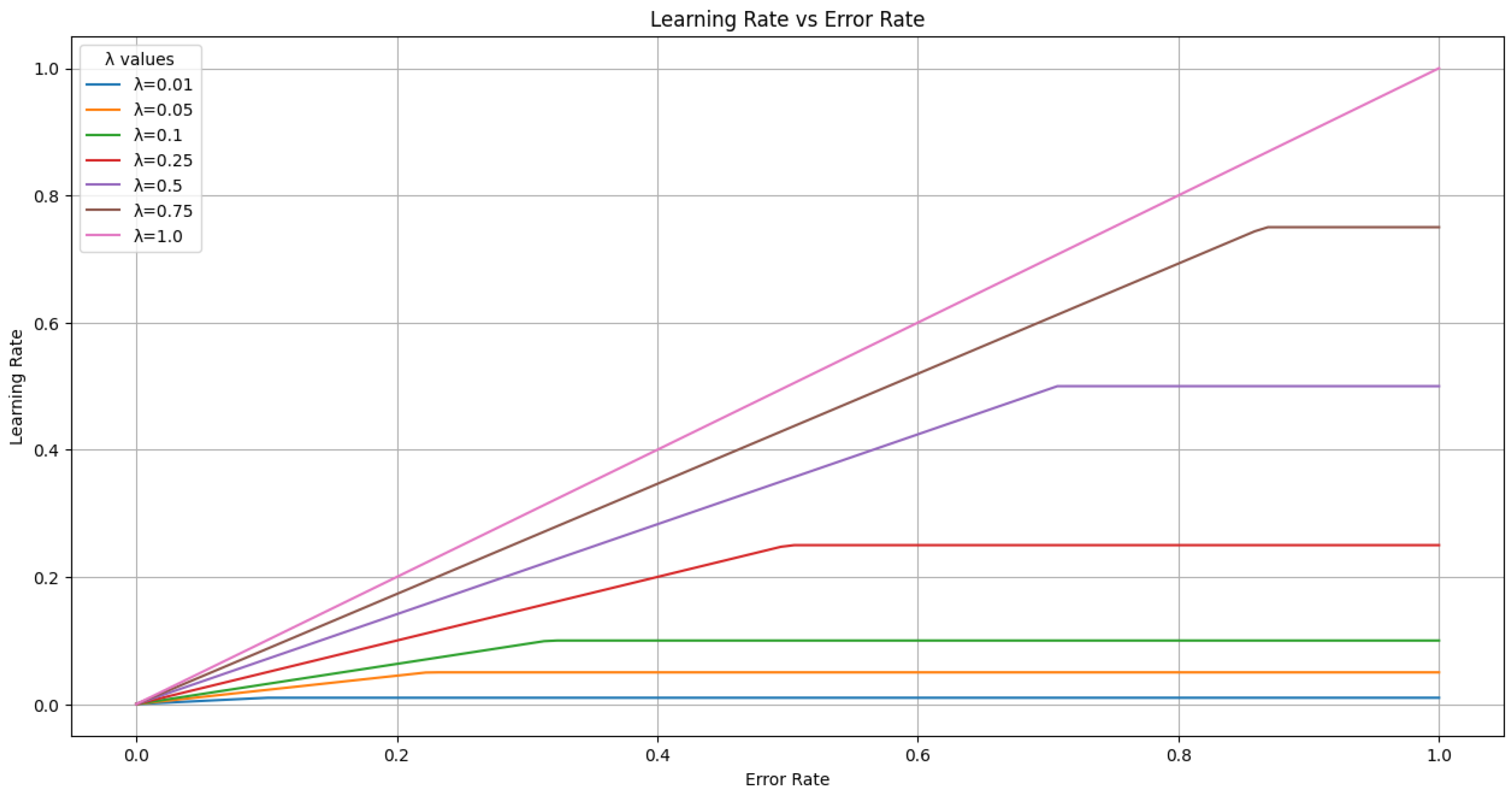
In
Figure 5, we present a graph to illustrate the possible values of
based on the error rate on the new dataset,
, and the chosen value of
.
3.3. Evaluation
Once
is trained, it is evaluated through two comparisons. First,
is compared to the model
f on the initial dataset
. Second,
is compared to
on datasets different from
.
Figure 6 illustrates these comparisons.
To comprehensively evaluate the performance of our methodology, we designed an evaluation protocol that focuses on three key aspects: (1) measuring cross-domain generalization through a structured performance matrix, (2) analyzing performance deviations to assess consistency and robustness, and (3) benchmarking IFL models against baseline and global models to validate their effectiveness.
3.3.1. Cross-Domain Performance Matrix
To systematically evaluate the IFL models, we propose a cross-domain performance matrix
M, where each entry
represents the performance of model
when evaluated on dataset
using a chosen evaluation metric. The matrix is constructed as follows:
where Metric is a user-specified evaluation metric, such as Accuracy, F1-score, or AUC-ROC. The diagonal entries
quantify the model’s performance on its source dataset using the selected metric, while off-diagonal entries
(for
) measure its generalization to other datasets.
3.3.2. Performance Deviation Analysis
We define two aggregate metric vectors to summarize cross-domain performance using the elements of the Cross-Domain Performance Matrix ():
Cross-Domain Generalization Score (CDGS): The vector whose elements are the average performance of
on all datasets other than its source, computed as
where Metric determines the range of
. For normalized metrics such as accuracy or F1-score,
. Higher values indicate better generalization across datasets.
Performance Variance (PV): The vector whose elements are the variance of each
across all datasets, reflecting the consistency of the model’s performance:
where
is the mean performance of
across all datasets. Lower PV values indicate more consistent performance, while higher values suggest variability or bias. Again, for normalized metrics, we have
, and consequently
, with smaller values indicating greater consistency across datasets in this case.
3.3.3. Benchmark Comparison Matrix
To validate the effectiveness of our methodology, we construct a benchmark comparison matrix
B, which directly mirrors the structure of the cross-domain performance matrix
M. The entries of
B are defined as follows:
The diagonal entries represent the performance of the global model on dataset , while the off-diagonal entries (for ) capture the performance of the baseline model on the other datasets .
By directly comparing the cross-domain performance matrix M with the benchmark comparison matrix B, we can evaluate the improvements introduced by the IFL models . Specifically, this comparison enables the following:
Assessment of Cross-Domain Generalization: Comparing against (for ) reveals whether the IFL model generalizes better to other datasets than the baseline model .
Evaluation of Source Dataset Performance: Comparing with indicates whether the IFL model outperforms the global model on its original dataset .
This benchmarking approach allows for a straightforward comparison, as each element in both matrices represents a single value of the chosen metric. This simplicity ensures that performance differences can be directly interpreted while preserving the properties of the utilized metric, such as normalization or scale consistency.
5. Discussion
The results of this study offer valuable insights into the application of the novel IFL method, specifically in the context of cervical cytology image analysis. The methodology presented is versatile and holds significant potential for broader applications across various domains. However, several important considerations arise when interpreting these findings.
In our experiment, 2 out of the 16 models do not improve their specific benchmark models. Specifically, the model did not surpass the global model f on dataset , and the model has the same accuracy as the global model f on dataset , even though the rest of the models either matched or exceeded the anticipated performance. This discrepancy highlights the challenges in ensuring consistent improvement across all models and datasets.
The APACC and CRIC Cervix datasets, while extensive and well-annotated, are the only publicly available datasets that meet the requirements for this study, particularly the detailed labeling and high resolution needed for both detection and classification tasks. Splitting these datasets allows for multidimensional analysis and IFL experimentation, but it may also reduce the diversity and variability within each subset. Future work could address this limitation by incorporating additional datasets, either from the domain of cervical cancer or from other medical imaging fields, to test the generalizability of the methodology across entirely different domains. This expansion could further validate the robustness of the proposed approach and explore its applicability to other pathologies. We also have to note that the selected order of the experiment could have varied. We chose the assignments , but there are possible permutations, and we can choose different permutations with different cycle order. Each permutation could potentially yield specific results due to differences in training order influencing the observed outcomes. Future research could explore the impact of such permutations systematically. This analysis could provide additional insights into the stability and optimality of the training process for transfer learning and multidimensional experiments.
Another critical factor in the methodology is the choice of the parameter
, which governs the maximum learning rate during transfer learning. As shown in Section Learning Rate Selection for Transfer Learning,
directly influences the adaptability of the model to new datasets. A higher
allows for faster adaptation but risks overfitting to the target dataset, potentially causing the model to lose important information from its source domain. Conversely, a lower
enforces more conservative updates, which can preserve knowledge from the source dataset but might limit the model’s ability to effectively learn features specific to the new domain. Optimizing
for each transfer step is therefore crucial, and future studies could explore adaptive or data-driven methods for determining this parameter to achieve an optimal balance between source retention and target adaptation. As illustrated in
Figure 5, the learning rate cap
mainly scales the error-dependent update. Preliminary checks showed that values in the range
produced near-identical accuracy, with fluctuations comparable to the variance observed across random seeds. While this indicates that
has limited influence under the present conditions, we acknowledge its potential relevance in larger or more heterogeneous settings and identify it as a promising direction for future work. In addition, Equation (3) offers a more stable and interpretable alternative to validation loss or gradient-based schedulers, which can fluctuate under class imbalance or noisy labels.
Preliminary checks indicated that variations in interleaving, learning rate schedule (fixed vs. dynamic), dataset order, and freezing policy produced only marginal changes, generally within the variance observed across random seeds. While this suggests that these design factors are not decisive in our current datasets, we acknowledge that they may become more influential in larger or more heterogeneous settings. For this reason, we did not include a full ablation grid in the main results, but we explicitly note this as a limitation and highlight it as a relevant direction for future research.
YOLOv8 was selected as it integrates detection and classification in a single pipeline and represents the state of the art in many recent medical imaging works, making it especially suitable for cytology where abnormal cells must be first localized. Alternative backbones such as ViT, Swin, or ConvNeXt achieve strong results on isolated cell crops, but do not address whole-slide detection. Importantly, our Interleaved Fusion Learning framework is architecture-agnostic: the sequential transfer and dynamic learning rate adaptation can be applied to any network supporting fine-tuning, and could in future be explored with CNN or transformer backbones.
For the evaluation, in
Section 3.3, by explicitly incorporating Metric into the definitions of
,
, and
, this framework remains adaptable to different evaluation needs. For example, if fairness is a key concern, one might prioritize metrics such as the F1-score or balanced accuracy, as they are particularly suited to scenarios with class imbalances or where equitable performance across classes is critical. We acknowledge that the absolute accuracies remain modest. However, the focus here is not on achieving state-of-the-art single-dataset results, but on showing consistent improvements over global and local baselines, indicating that residual knowledge can indeed be preserved and transferred. Direct comparison with alternative approaches was intentionally avoided, as their objectives and setups differ substantially. Depending on the chosen metric, these methods could appear either superior or inferior, leading to potentially misleading conclusions. Instead, the most meaningful baselines for assessing IFL are the global and local training strategies across datasets, which directly reflect its goal of preserving and transferring residual knowledge between domains.
The structured methodology relies on curated binary class mappings and dynamic learning rate adjustments. While these choices have been effective for the current datasets, they may require modifications for datasets with more complex label distributions or larger imbalances. Investigating methods to handle multi-class problems or severe label imbalances within this IFL framework could expand its utility and improve its fairness in real-world applications.
In
Section 2.3, a new notation for domains in transfer learning was introduced to address inconsistencies identified in previous literature. Although these studies provide initial domain definitions, their practical application often involves inconsistent alterations, resulting in discrepancies between the theoretical framework and its implementation. The revised notation seeks to promote a consistent and coherent use of domain definitions, improving the clarity and reproducibility of transfer learning methodologies.
Although the present study employed binary mappings to simplify the evaluation, it is important to acknowledge that many medical applications, including cervical cytology, are inherently multi-class in nature, with diagnostic categories such as NILM, ASC-US, LSIL, HSIL, and SCC. Extending the IFL framework to these settings is not trivial, as class overlap, hierarchical relationships, and unequal misclassification costs become critical factors. Furthermore, severe class imbalance remains a pervasive challenge in medical data, where minority categories, although clinically decisive, are often underrepresented. While balanced accuracy and F1-score partially mitigate these effects, future work should explore the integration of imbalance-aware strategies such as cost-sensitive losses, resampling techniques, or hierarchical classification schemes within the IFL pipeline. Addressing these two aspects—multi-class complexity and class imbalance—is essential to ensure that performance improvements are equitably distributed across categories.
In conclusion, while the IFL methodology demonstrates clear advantages in improving performance and robustness, the study also underscores the need for broader dataset diversity, careful parameter selection, and enhanced interpretability. These factors should guide future research to maximize the impact and applicability of IFL in medical imaging and beyond.
6. Conclusions
The results, summarized in
Section 4, highlight key advantages of the IFL models (
) over both the baseline models (
) trained solely on their respective source datasets and the combined dataset model (
) trained on the entire dataset
.
This work not only advances the field by proposing a novel IFL methodology but also provides a comprehensive set of practical tools to support future research. These tools include methods for the informed and systematic selection of learning rates, offering a structured optimization process based on our proposed dynamic learning rate for transfer steps. In the method, we provide a novel evaluation stage designed to accommodate diverse performance metrics, such as fairness, robustness, or domain-specific criteria. In addition, we address longstanding inconsistencies in transfer learning notation by presenting an enhanced and standardized framework. Collectively, these contributions establish a solid foundation for further developments in IFL and transfer learning methodologies.
In the studied cervix datasets, the IFL models exhibit consistent improvements in cross-domain generalization, as evidenced by values presented in
Table 1. These models generally achieve higher accuracy on datasets other than their sources compared to the baseline models, demonstrating their ability to retain critical knowledge from the source dataset while adapting to new domains. The low PV values further support the robustness of the IFL models, indicating a relatively consistent performance across datasets.
These findings underscore the potential of IFL to enhance the robustness and fairness of DL models in complex multi-domain settings. Future work will focus on expanding the methodology to incorporate additional metrics, such as fairness measures, and exploring its applicability to other medical imaging domains. Furthermore, integrating explainability techniques could provide additional insights into the decision-making processes of the IFL models, facilitating their adoption in clinical practice.