SemiSeg-CAW: Semi-Supervised Segmentation of Ultrasound Images by Leveraging Class-Level Information and an Adaptive Multi-Loss Function
Abstract
1. Introduction
- Proposing SemiSeg-CAW, a semi-supervised segmentation and classification model that utilizes class-level information to compensate for the shortage of sufficient pixel-level annotated data.
- Proposing ClassElevateSeg, an auxiliary module to produce refined segmentation maps under multitask supervision, providing stable auxiliary features to enhance training.
- Proposing an adaptive weighting strategy to generate distinct, trainable weights for multiple loss functions, ensuring balanced and effective multitask learning.
2. Related Works
3. Materials and Methods
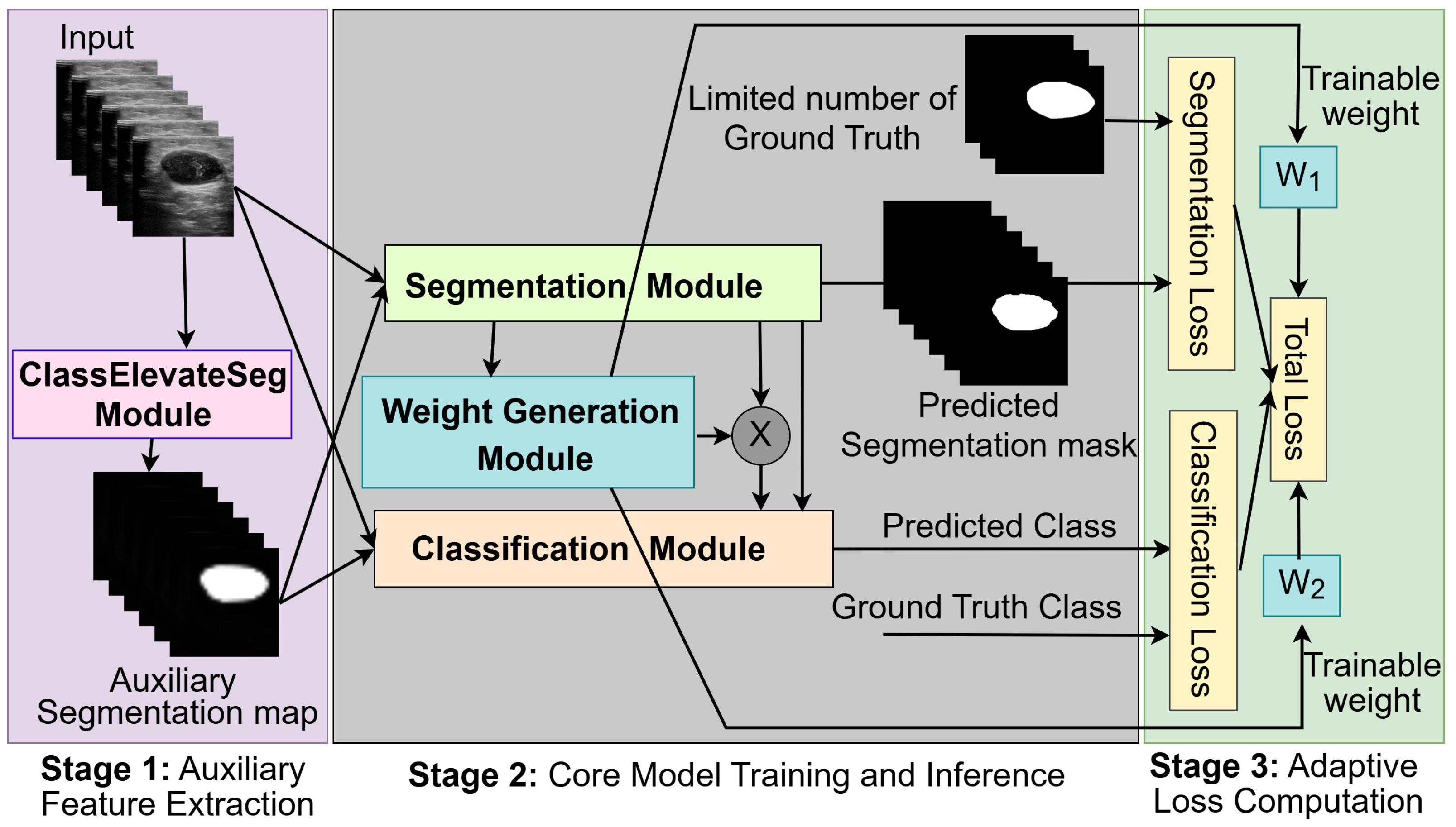
- Stage 1 (Auxiliary Feature Extraction): A pre-trained module, called ClassElevateSeg, processes input images to generate auxiliary segmentation maps. The auxiliary maps provide additional class-level guidance that compensates for the lack of reliable pixel-level annotations.
- Stage 2 (Core Model Training and Inference): The main network integrates segmentation and classification modules, which share a significant portion of their structure. Segmentation is the main objective, and classification provides complementary global information. Both the segmentation and classification tasks are guided by the auxiliary maps from Stage 1.
- Stage 3 (Adaptive Loss Computation): The weight generation module (WGM) dynamically assigns trainable weights to segmentation and classification losses during training to balance the contributions of the tasks without manual tuning.
3.1. Auxiliary Feature Extraction (ClassElevateSeg Module)
3.2. Core Model Training and Inference
3.2.1. Segmentation
3.2.2. Classification
3.2.3. Weight Generation Module (WGM)
3.3. Adaptive Loss Computation
4. Experimental Results
4.1. Dataset
4.2. Implementation Details
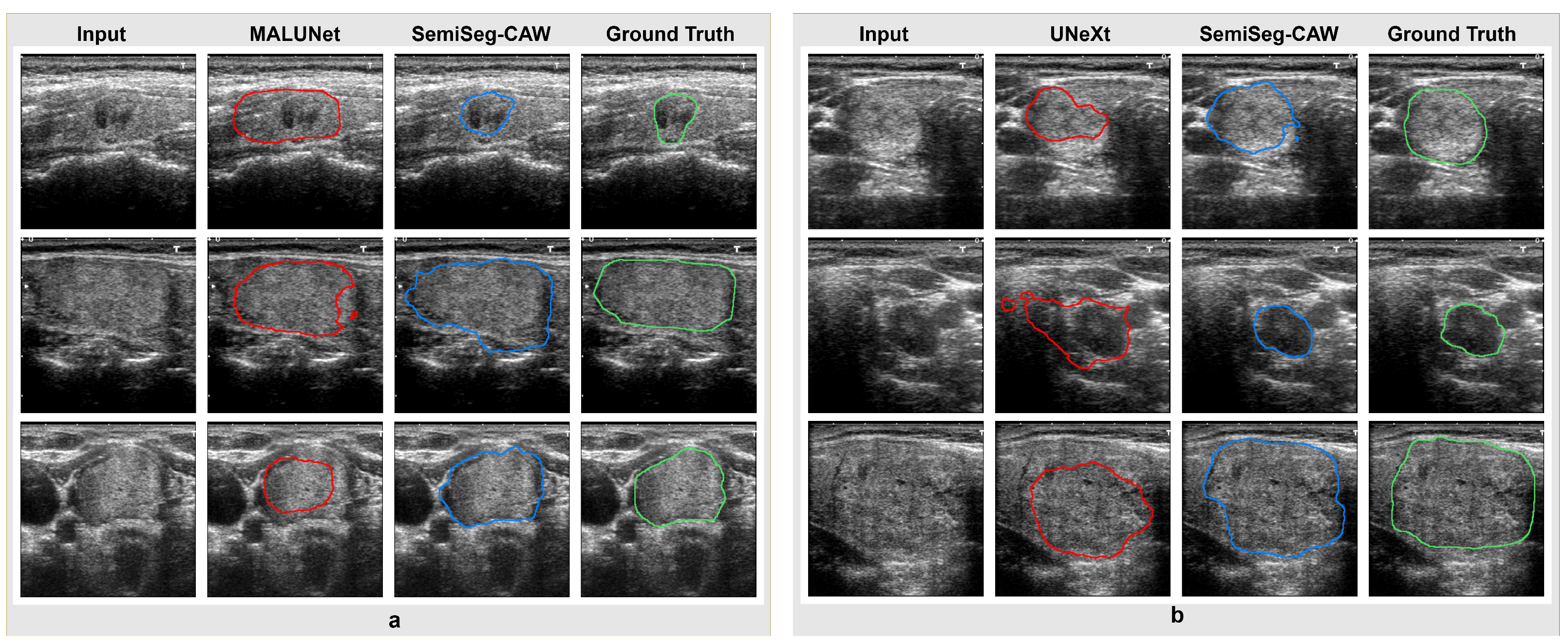
4.3. Segmentation Results
5. Conclusions
Author Contributions
Funding
Data Availability Statement
Conflicts of Interest
Abbreviations
| SemiSeg-CAW | Semi-supervised Segmentation and Classification with Adaptive Weighting |
| CAM | Class Activation Map |
| WGM | Weight Generation Module |
| IoU | Intersection over Union |
| Dice | Dice Coefficient |
| BCE | Binary Cross-Entropy |
| BCEwithLogits | Binary Cross-Entropy with Logits |
| Ftv | Focal Tversky |
| CE | Cross-Entropy |
| AdamW | Adaptive Moment Estimation with Weight Decay Optimizer |
| LR | Learning Rate |
| CosineAnnealingLR | Cosine Annealing Learning Rate Scheduler |
| BN | Batch Normalization |
| ReLU | Rectified Linear Unit |
| BUSI | Breast Ultrasound Images Dataset |
| DDTI | Digital Database of Thyroid Ultrasound Images |
| UNet | U-shaped Convolutional Neural Network |
Appendix A
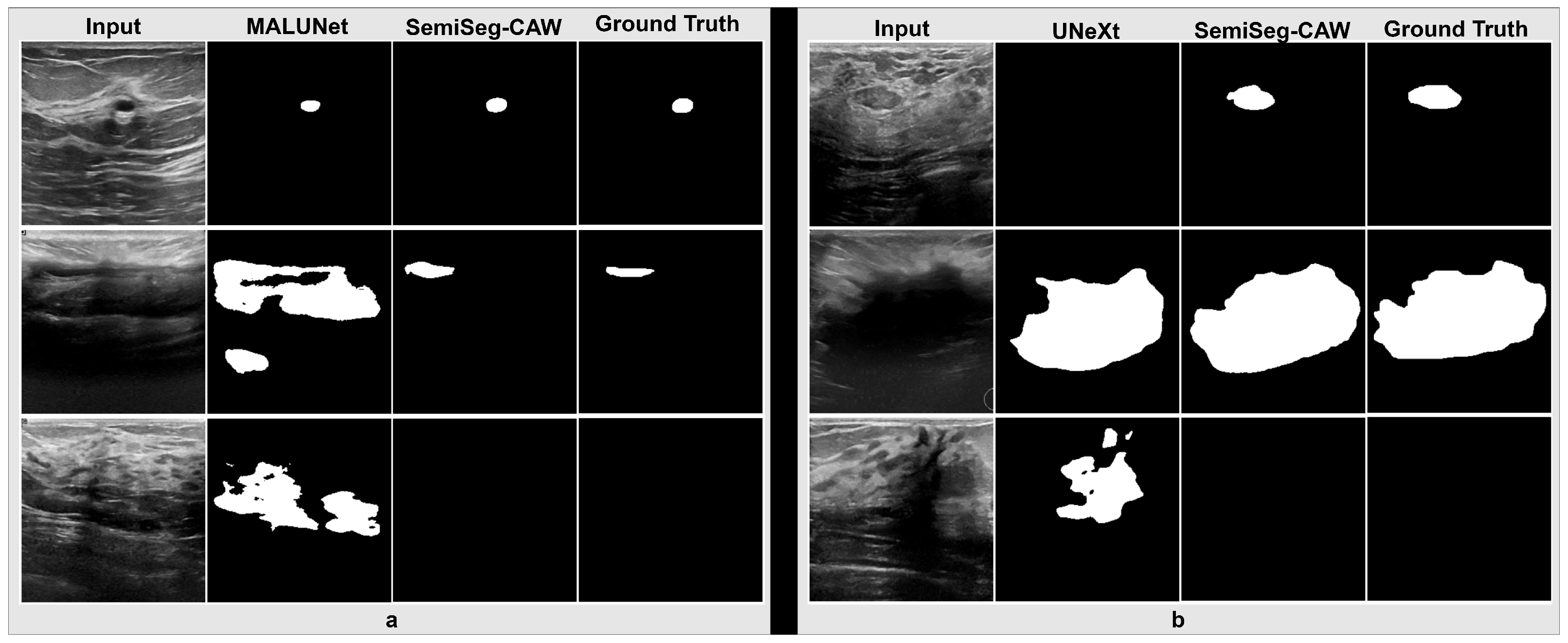
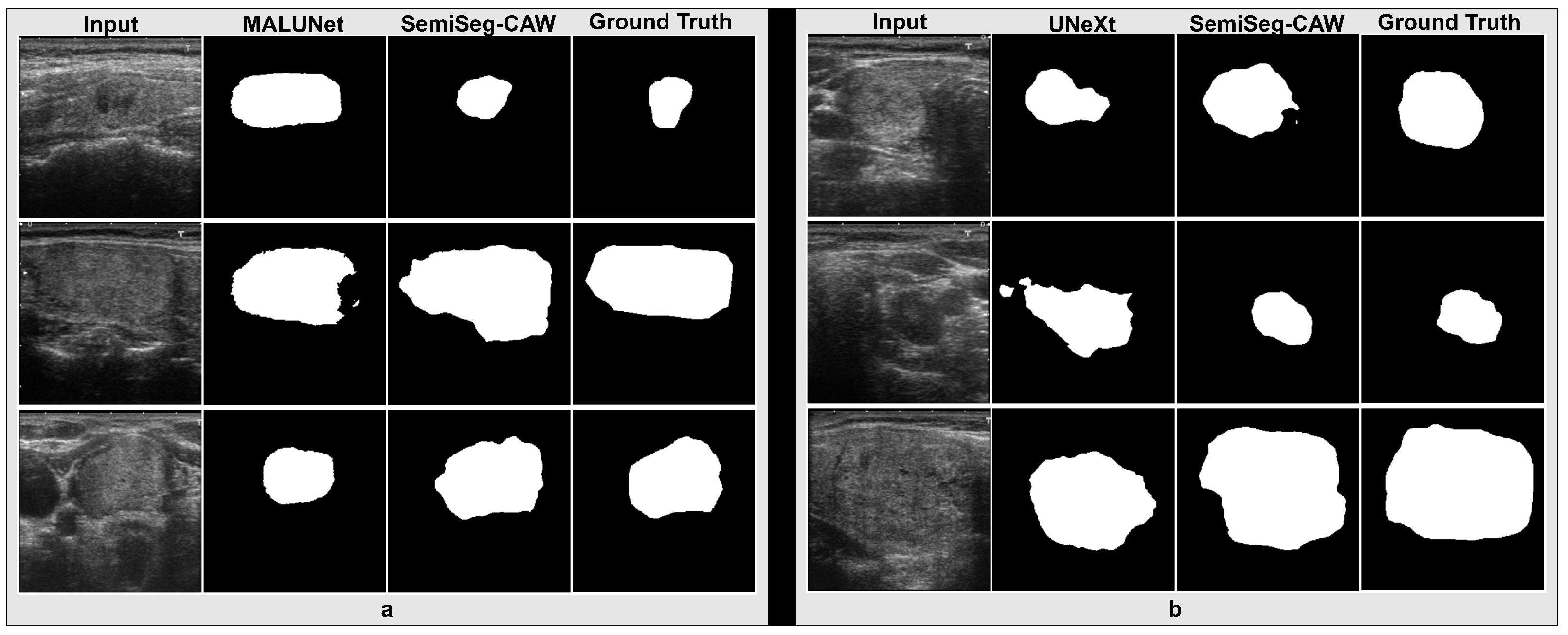
References
- Liu, X.; Song, L.; Liu, S.; Zhang, Y. A review of deep-learning-based medical image segmentation methods. Sustainability 2021, 13, 1224. [Google Scholar] [CrossRef]
- Lyu, Y.; Xu, Y.; Jiang, X.; Liu, J.; Zhao, X.; Zhu, X. AMS-PAN: Breast ultrasound image segmentation model combining attention mechanism and multi-scale features. Biomed. Signal Process. Control 2023, 81, 104425. [Google Scholar] [CrossRef]
- Ronneberger, O.; Fischer, P.; Brox, T. U-net: Convolutional networks for biomedical image segmentation. In Proceedings of the Medical Image Computing and Computer-Assisted Intervention–MICCAI 2015: 18th International Conference, Munich, Germany, 5–9 October 2015; Proceedings, Part III 18. Springer: Berlin/Heidelberg, Germany, 2015; pp. 234–241. [Google Scholar]
- Liu, S.; Wang, Y.; Yang, X.; Lei, B.; Liu, L.; Li, S.X.; Ni, D.; Wang, T. Deep learning in medical ultrasound analysis: A review. Engineering 2019, 5, 261–275. [Google Scholar] [CrossRef]
- Su, J.; Luo, Z.; Lian, S.; Lin, D.; Li, S. Mutual learning with reliable pseudo label for semi-supervised medical image segmentation. Med. Image Anal. 2024, 94, 103111. [Google Scholar] [CrossRef] [PubMed]
- Li, X.; Yu, L.; Chen, H.; Fu, C.W.; Xing, L.; Heng, P.A. Transformation-consistent self-ensembling model for semisupervised medical image segmentation. IEEE Trans. Neural Netw. Learn. Syst. 2020, 32, 523–534. [Google Scholar] [CrossRef] [PubMed]
- Barzegar, S.; Khan, N. Skin lesion segmentation using a semi-supervised U-NetSC model with an adaptive loss function. In Proceedings of the 2022 44th Annual International Conference of the IEEE Engineering in Medicine & Biology Society (EMBC), Glasgow, Scotland, 11–15 July 2022; IEEE: Piscataway, NJ, USA, 2022; pp. 3776–3780. [Google Scholar]
- Yuan, Y. Automatic skin lesion segmentation with fully convolutional-deconvolutional networks. arXiv 2017, arXiv:1703.05165. [Google Scholar] [CrossRef]
- Xie, Y.; Zhang, J.; Xia, Y.; Shen, C. A mutual bootstrapping model for automated skin lesion segmentation and classification. IEEE Trans. Med. Imaging 2020, 39, 2482–2493. [Google Scholar] [CrossRef] [PubMed]
- Zhou, B.; Khosla, A.; Lapedriza, A.; Oliva, A.; Torralba, A. Learning deep features for discriminative localization. In Proceedings of the IEEE Conference on Computer Vision and Pattern Recognition, Las Vegas, NV, USA, 27–30 June 2016; pp. 2921–2929. [Google Scholar]
- Feng, J.; Li, C.; Wang, J. CAM-TMIL: A Weakly-Supervised Segmentation Framework for Histopathology based on CAMs and MIL. In Proceedings of the 4th International Conference on Computing and Data Science, Macau, China, 16–25 July 2022; Volume 2547, p. 012014. [Google Scholar]
- Al-Dhabyani, W.; Gomaa, M.; Khaled, H.; Fahmy, A. Dataset of breast ultrasound images. Data Brief 2020, 28, 104863. [Google Scholar] [CrossRef] [PubMed]
- Pedraza, L.; Vargas, C.; Narváez, F.; Durán, O.; Muñoz, E.; Romero, E. An open access thyroid ultrasound image database. In Proceedings of the 10th International Symposium on Medical Information Processing and Analysis, Cartagena, Colombia, 14–16 October 2014; SPIE: Bellingham, WA, USA, 2015; Volume 9287, pp. 188–193. [Google Scholar]
- Eiraoi. Thyroid Ultrasound Data. Kaggle, 2024. [Online]. Available online: https://www.kaggle.com/datasets/eiraoi/thyroidultrasound/data (accessed on 17 June 2024).
- Singh, V.K.; Rashwan, H.A.; Romani, S.; Akram, F.; Pandey, N.; Sarker, M.M.K.; Saleh, A.; Arenas, M.; Arquez, M.; Puig, D.; et al. Breast tumor segmentation and shape classification in mammograms using generative adversarial and convolutional neural network. Expert Syst. Appl. 2020, 139, 112855. [Google Scholar] [CrossRef]
- Feng, X.; Lin, J.; Feng, C.M.; Lu, G. GAN inversion-based semi-supervised learning for medical image segmentation. Biomed. Signal Process. Control 2024, 88, 105536. [Google Scholar] [CrossRef]
- Chen, Z.; Tian, Z.; Zhu, J.; Li, C.; Du, S. C-cam: Causal cam for weakly supervised semantic segmentation on medical image. In Proceedings of the IEEE/CVF Conference on Computer Vision and Pattern Recognition, New Orleans, LA, USA, 18–24 June 2022; pp. 11676–11685. [Google Scholar]
- Wang, J.; Liu, B.; Li, Y.; Li, J.; Pei, Y. CAM-CycleGAN: A Weakly Supervised Segmentation Method for Medical Images Based on Cycle Consistensy Generative Adversarial Networks. In Proceedings of the 2024 IEEE 6th Advanced Information Management, Communicates, Electronic and Automation Control Conference (IMCEC), Chongqing, China, 24–26 May 2024; IEEE: Piscataway, NJ, USA, 2024; Volume 6, pp. 1293–1296. [Google Scholar]
- Yang, C.L.; Harjoseputro, Y.; Chen, Y.Y. A hybrid approach of simultaneous segmentation and classification for medical image analysis. Multimed. Tools Appl. 2024, 84, 21805–21827. [Google Scholar]
- Ling, Y.; Wang, Y.; Dai, W.; Yu, J.; Liang, P.; Kong, D. Mtanet: Multi-task attention network for automatic medical image segmentation and classification. IEEE Trans. Med. Imaging 2023, 43, 674–685. [Google Scholar] [CrossRef] [PubMed]
- Liu, L.; Li, Y.; Kuang, Z.; Xue, J.; Chen, Y.; Yang, W.; Liao, Q.; Zhang, W. Towards impartial multi-task learning. In Proceedings of the ICLR 2021 International Conference on Learning Representations, Vienna, Austria, 4 May 2021. [Google Scholar]
- Ruan, J.; Xiang, S.; Xie, M.; Liu, T.; Fu, Y. MALUNet: A multi-attention and light-weight unet for skin lesion segmentation. In Proceedings of the 2022 IEEE International Conference on Bioinformatics and Biomedicine (BIBM), Las Vegas, NV, USA, 6–8 December 2022; IEEE: Piscataway, NJ, USA, 2022; pp. 1150–1156. [Google Scholar]
- He, K.; Zhang, X.; Ren, S.; Sun, J. Deep Residual Learning for Image Recognition. In Proceedings of the 2016 IEEE Conference on Computer Vision and Pattern Recognition (CVPR), Las Vegas, NV, USA, 27–30 June 2016; pp. 770–778. [Google Scholar]
- Valanarasu, J.M.J.; Patel, V.M. Unext: Mlp-based rapid medical image segmentation network. In Proceedings of the International Conference on Medical Image Computing and Computer-Assisted Intervention, Singapore, 18–22 September 2022; Springer: Berlin/Heidelberg, Germany, 2022; pp. 23–33. [Google Scholar]
- Ruano, J. MALUNet: A Multi-Attention and Light-Weight UNet for Skin Lesion Segmentation. 2023. Available online: https://github.com/JCruan519/MALUNet (accessed on 1 November 2023).
- Valanarasu, J.M.J. UNeXt–Pytorch Implementation. 2022. Available online: https://github.com/jeya-maria-jose/UNeXt-pytorch (accessed on 16 November 2024).
- Jiang, P.T.; Zhang, C.B.; Hou, Q.; Cheng, M.M.; Wei, Y. Layercam: Exploring hierarchical class activation maps for localization. IEEE Trans. Image Process. 2021, 30, 5875–5888. [Google Scholar] [CrossRef] [PubMed]
- Yazıcı, Z.A.; Öksüz, İ.; Ekenel, H.K. GLIMS: Attention-guided lightweight multi-scale hybrid network for volumetric semantic segmentation. Image Vis. Comput. 2024, 146, 105055. [Google Scholar] [CrossRef]
- Ma, J.; He, Y.; Li, F.; Han, L.; You, C.; Wang, B. Segment anything in medical images. Nat. Commun. 2024, 15, 654. [Google Scholar] [CrossRef] [PubMed]

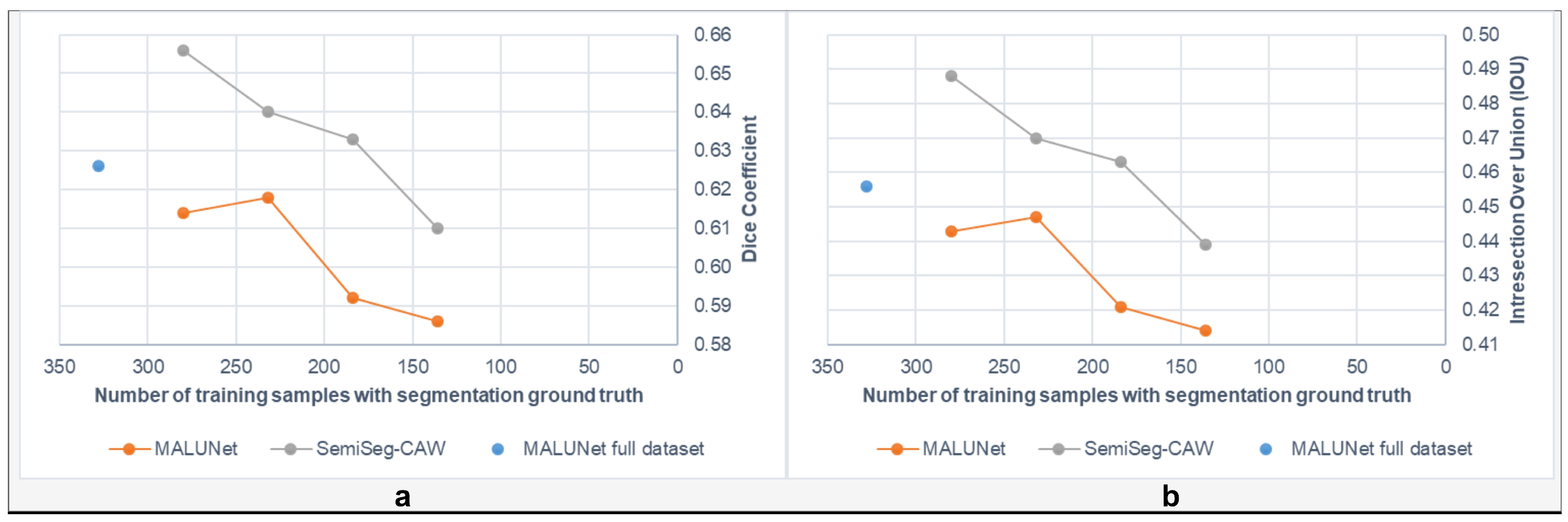


| Dataset | Method | Total | Class Only | Both Labels | Dice | IOU | Recall | Precision | Specificity | Accuracy |
|---|---|---|---|---|---|---|---|---|---|---|
| BUSI | MALUNet—Full data | 572 | - | 572 | 0.672 | 0.506 | 0.671 | 0.673 | 0.970 | 0.944 |
| MALUNet—Reduced | 472 | - | 472 | 0.662 | 0.494 | 0.737 | 0.601 | 0.954 | 0.936 | |
| SemiSeg-CAW | 568 | 100 | 468 | 0.684 | 0.520 | 0.659 | 0.712 | 0.975 | 0.948 | |
| DDTI | MALUNet—Full data | 328 | - | 328 | 0.628 | 0.457 | 0.831 | 0.504 | 0.905 | 0.897 |
| MALUNet—Reduced | 280 | - | 280 | 0.614 | 0.443 | 0.770 | 0.511 | 0.914 | 0.899 | |
| SemiSeg-CAW | 328 | 48 | 280 | 0.656 | 0.488 | 0.792 | 0.560 | 0.928 | 0.913 | |
| SemiSeg-CAW | 328 | 96 | 232 | 0.640 | 0.470 | 0.795 | 0.535 | 0.920 | 0.907 | |
| SemiSeg-CAW | 328 | 144 | 184 | 0.633 | 0.463 | 0.817 | 0.517 | 0.911 | 0.901 |
| Changes to SemiSeg-CAW | Dice | IOU | Training Time/Epoch [s] |
|---|---|---|---|
| Removing ClassElevateSeg | 0.571 | 0.400 | |
| Static weights instead of WGM | 0.572 | 0.401 | |
| Strategy of [21] instead of WGM | 0.537 | 0.367 | |
| Removing ClassElevateSeg and WGM [7] | 0.601 | 0.429 | |
| LayerCAM [27] instead of ClassElevateSeg | 0.583 | 0.411 | |
| SemiSeg-CAW (Full model) | 0.610 | 0.439 |
| Backbone | Model | Params [M] | Training/Epoch [s] | Inference [ms/img] | ||
|---|---|---|---|---|---|---|
| BUSI | DDTI | BUSI | DDTI | |||
| MALUNet | MALUNet (base) | 0.18 | 2.96 | 2.84 | ||
| SemiSeg–CAW | 14.50 | 5.26 | 4.98 | |||
| UNeXt | UNeXt (base) | 1.47 | 1.57 | 2.28 | ||
| SemiSeg–CAW | 15.85 | 3.75 | 4.41 | |||
| Dataset | Method | Dice | IOU | Recall | Precision |
|---|---|---|---|---|---|
| BUSI | UNeXt | 0.710 | 0.595 | 0.688 | 0.722 |
| SemiSeg-CAW | 0.813 | 0.704 | 0.673 | 0.752 | |
| DDTI | UNeXt | 0.678 | 0.519 | 0.813 | 0.598 |
| SemiSeg-CAW | 0.710 | 0.552 | 0.737 | 0.699 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Barzegar, S.; Khan, N. SemiSeg-CAW: Semi-Supervised Segmentation of Ultrasound Images by Leveraging Class-Level Information and an Adaptive Multi-Loss Function. Mach. Learn. Knowl. Extr. 2025, 7, 124. https://doi.org/10.3390/make7040124
Barzegar S, Khan N. SemiSeg-CAW: Semi-Supervised Segmentation of Ultrasound Images by Leveraging Class-Level Information and an Adaptive Multi-Loss Function. Machine Learning and Knowledge Extraction. 2025; 7(4):124. https://doi.org/10.3390/make7040124
Chicago/Turabian StyleBarzegar, Somayeh, and Naimul Khan. 2025. "SemiSeg-CAW: Semi-Supervised Segmentation of Ultrasound Images by Leveraging Class-Level Information and an Adaptive Multi-Loss Function" Machine Learning and Knowledge Extraction 7, no. 4: 124. https://doi.org/10.3390/make7040124
APA StyleBarzegar, S., & Khan, N. (2025). SemiSeg-CAW: Semi-Supervised Segmentation of Ultrasound Images by Leveraging Class-Level Information and an Adaptive Multi-Loss Function. Machine Learning and Knowledge Extraction, 7(4), 124. https://doi.org/10.3390/make7040124






