Systematic Analysis of Retrieval-Augmented Generation-Based LLMs for Medical Chatbot Applications
Abstract
1. Introduction
- A comprehensive analysis of three open-source LLMs (Flan-T5-Large, LLaMA-2-7B, Mistral-7B) and four datasets (MedQA, MedMCQA, Meadow-MedQA, Comprehensive Medical Q&A) examines their performance in medical chatbot applications. Experimental results comparing base models with RAG and fine-tuned models with and without RAG reveal that RAG and Fine-tuning are key for best results.
- A systematic analysis of subjective questions and multiple-choice questions (MCQs). The MCQs are further divided into two methodologies: Type A (similar to NLP question-answering tasks) and Type B (similar to language generation tasks). While Type A assesses model accuracy through exact matches between predicted options and reference answers, Type B generates text based on the question and context retrieval, measures similarity with all candidate answers, and calculates a probability confidence score to select the best possible answer for performance evaluation. While Type A methodology has shown superior results in our experiments, there is room for further development of the Type B methodology to bridge the performance gap.
- A comparative analysis of fine-tuned LLMs for medical data in resource-constrained environments using techniques such as quantization, PEFT, and LoRA. This involves evaluating their effectiveness in improving model performance and efficiency. These techniques significantly reduce memory usage and inference time, with fine-tuned models using only 5 to 8 GB of GPU RAM compared to higher memory requirements of unoptimised models, highlighting their potential for future work.
2. Literature Review
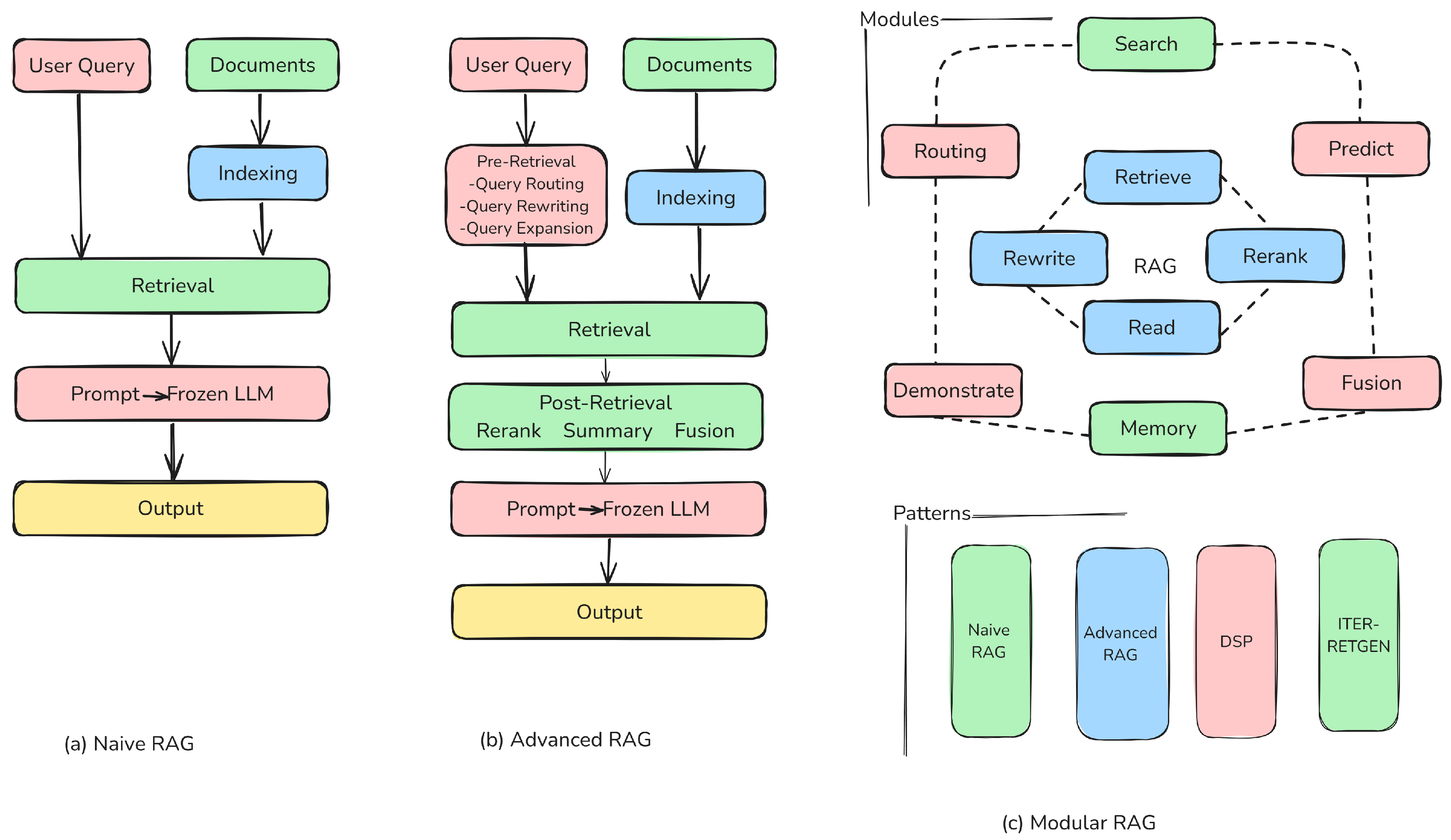
3. Theoretical Background
3.1. Embedding Model
3.2. LLMs
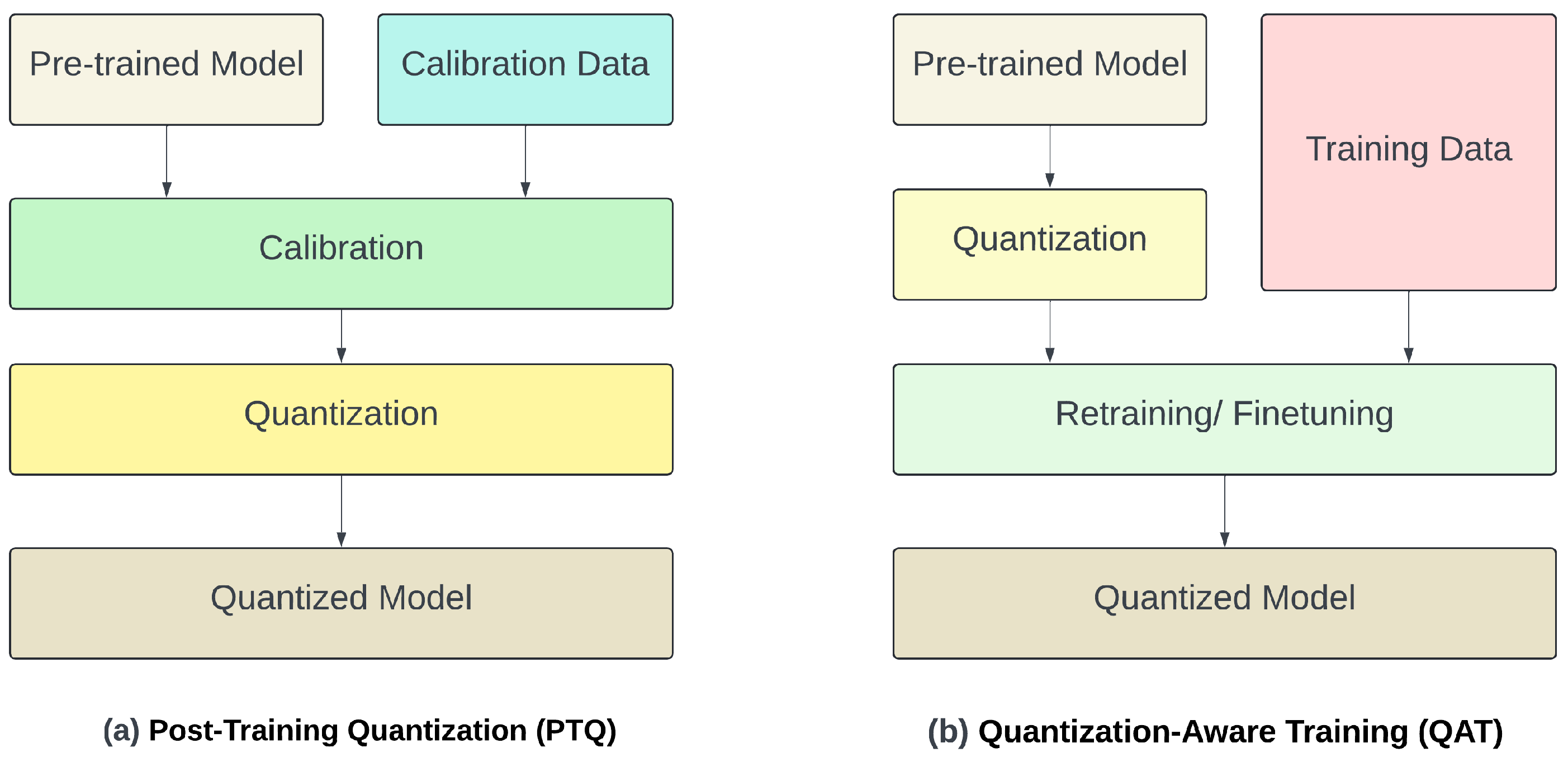
3.3. Quantization of LLMs
3.4. PEFT and LoRA
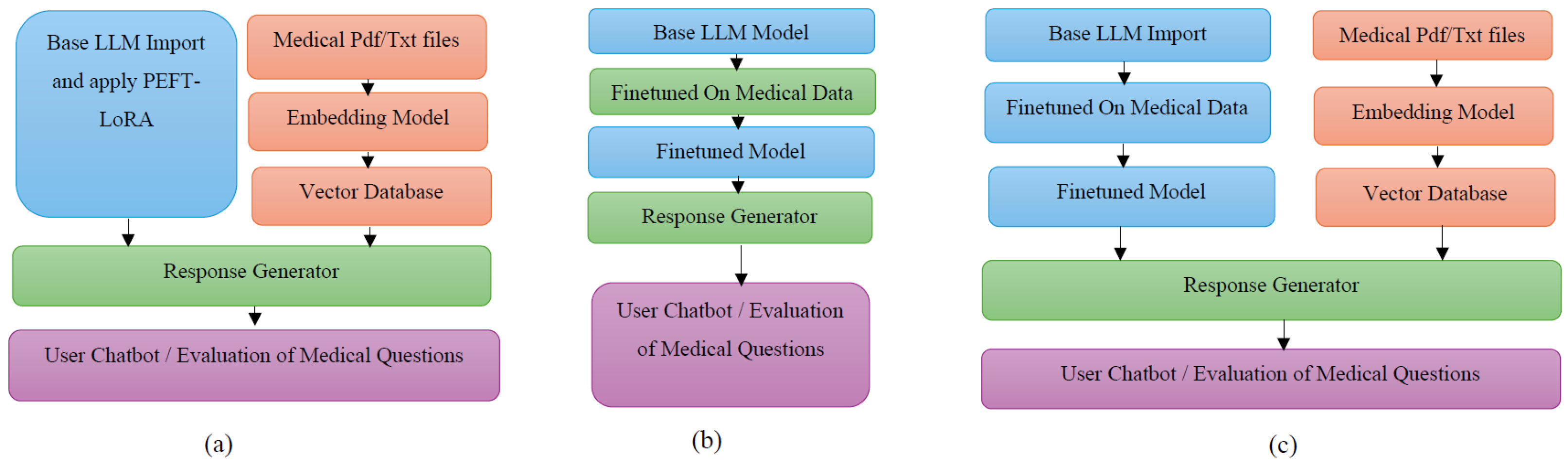
4. Methodology
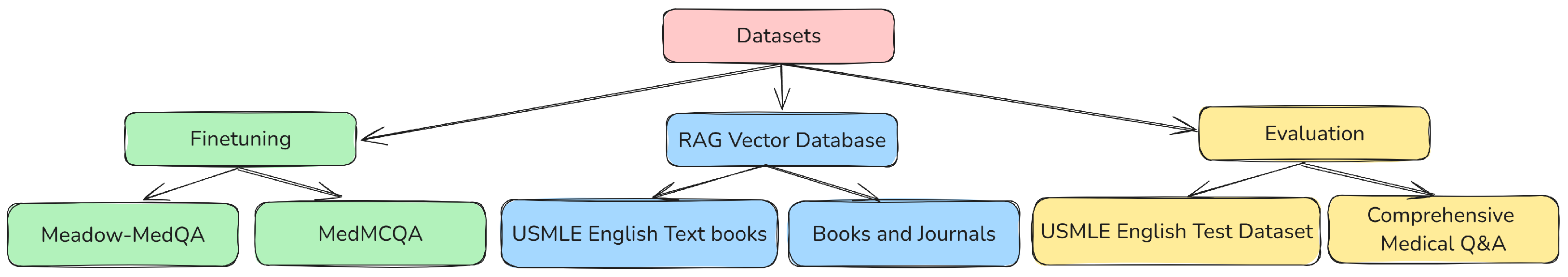
4.1. Medical Data Retrieval for Fine-Tuning, RAG Database and Evaluation
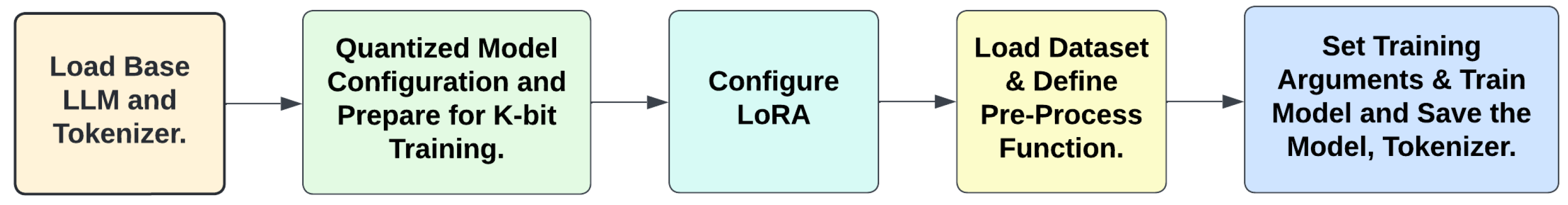
4.2. Fine-Tuning LLMs
4.3. Vector Database for RAG
- Document Loading: The documents are first loaded into the system.
- Text Chunking: The text is then divided into smaller, manageable chunks.
- Encoding: These chunks are passed through the embedding model, where they are transformed into vector data.
- Indexing: The generated vector data are stored as indexes in the vector database.
4.4. Testing Framework for Evaluation
4.4.1. Performance Metrics
4.4.2. Evaluation of Subjective Questions
4.4.3. Evaluation of Multiple-Choice Question Answering
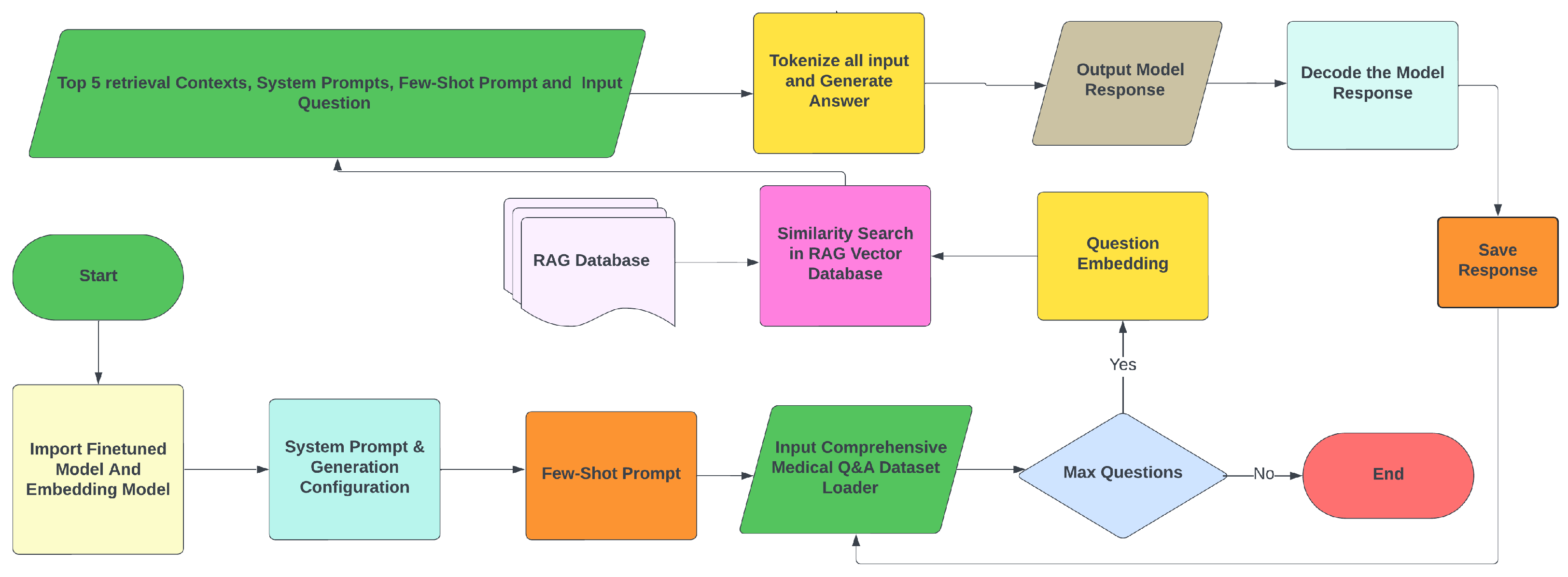
- Type A Methodology: Answers are generated according to an NLP question-answering task based on the context retrieval, system prompt, few-shot prompt, question, and reference options (candidate answers). For few-shot prompting, the model is provided with a small number of examples related to the input prompt. In this way, the Few-Shot prompt is provided to enhance the model’s efficiency in answer selection and to avoid unnecessary text generation. Figure 7 illustrates this methodology. For computational reasons, only the first 100 MCQs from the USMLE Test Data are used for the evaluation. The process generates answers for these questions, saving responses, questions, ground truth, exact matches of answers, and generation times in a text file. Typically, LLMs generate text one token at a time, and for each token, they calculate a probability distribution. In our methodology, since the model takes first the input question, system prompt, and retrieved context, the probability of that sequence can be expressed as follows:where is the probability of token given all previous tokens , Y is the output word sequence, and X includes all inputs. When candidate answers (options) are provided, the model generates scores for each option, selecting the one with the highest score [42] according towhere represents the option’s log probability.
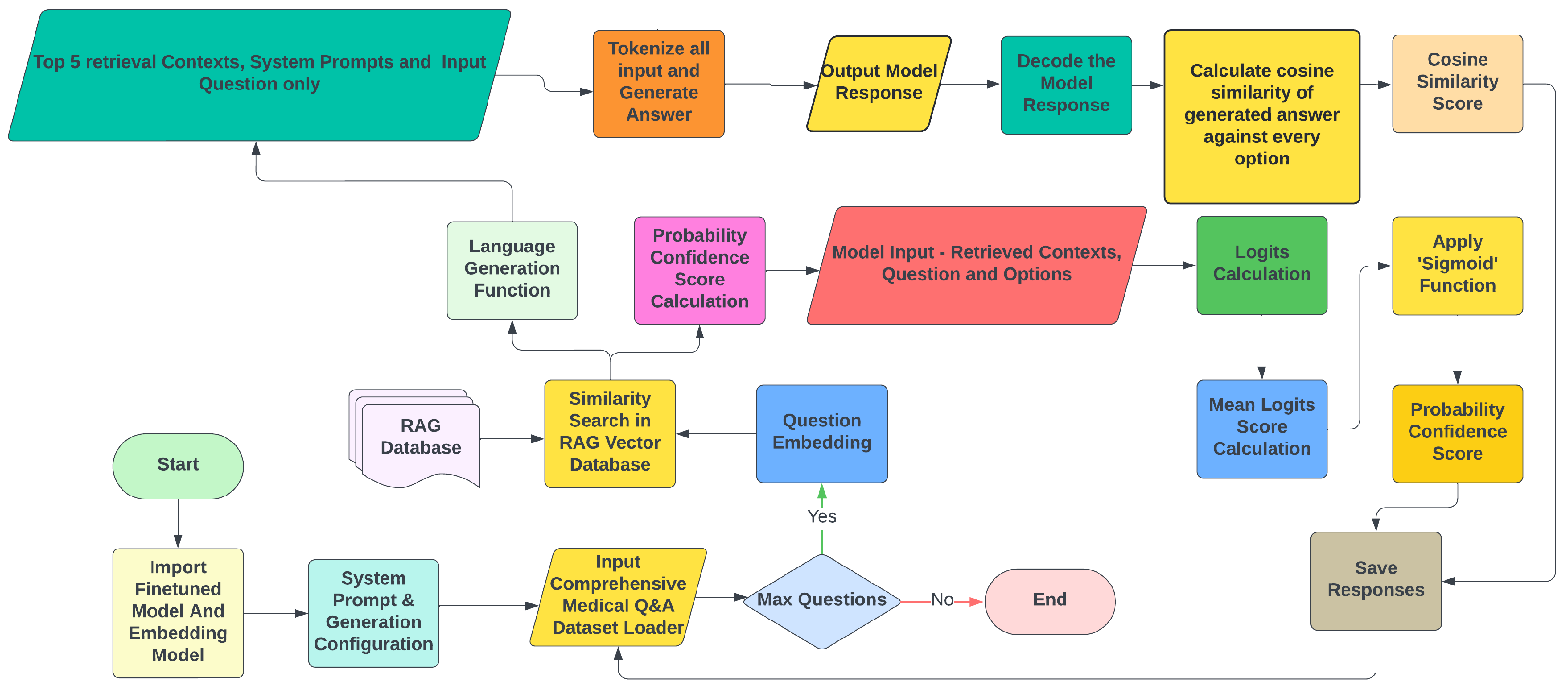
- Type B Methodology: It is designed to experiment with textual answer generation, while simultaneously calculating confidence scores for each option. This method generates text based on the input question and retrieved context without any few-shot prompts or candidate options. A function converts the generated answer and all reference options into vector representations using an embedding model, calculating the cosine similarity between each option and the generated answer aswhere is the vector representation of the generated answer, and is the vector representation of option i.Logits and probability are distinct mathematical concepts. While probabilities represent the likelihood of an event occurring, logits are raw, unnormalised scores that indicate how confident the model is in one outcome over another. In this methodology illustrated in Figure 8, for any given question, the mean logits score for every option is calculated. Then, mean logits are converted into probabilities using the ’Sigmoid’ function: , where is the mean logit score.Last but not least, mean reciprocal rank (MRR) is calculated according to , and Lenient Accuracy with Cutoff at and (LaCC) according to , where Q is the total number of questions, and k is the cutoff rank, set to 1 and 3.
4.5. Implementation
5. Results and Discussion
5.1. Subjective Question Evaluation
5.2. MCQ Evaluation
5.2.1. Type A Methodology
5.2.2. Type B Methodology
5.3. Discussion
6. Summary and Conclusions
Author Contributions
Funding
Data Availability Statement
Conflicts of Interest
Abbreviations
| AI | Artificial Intelligence |
| NLP | Natural Language Processing |
| LLM | Large Language Model |
| BERT | Bidirectional Encoder Representations from Transformers |
| RAG | Retrieval-Augmented Generation |
| ICL | In-Context Learning |
| CoT | Chain-of-Thought |
| PEFT | Parameter Efficient Fine-Tuning |
| LoRA | Low-Rank Adaptation |
| PTQ | Post-Training quantization |
| QAT | quantization-Aware Training |
| MCQ | Multi-Choice Questions |
| MRR | Mean Reciprocal Rank |
| LaCC | Lenient Accuracy |
Appendix A
Appendix A.1. System Requirements for the Research
- A Hugging Face account to access models, datasets, and integrations.
- Hardware specifications:
- −
- Minimum 15 GB of RAM to ensure smooth operation of deep learning models and data processing.
- −
- At least 200 GB of free storage on an SSD or HDD for storing datasets, model checkpoints, and other necessary files.
- −
- An NVIDIA GeForce RTX 3060 or a higher GPU with at least 12 GB of GPU memory for efficient model training and inference.
Appendix A.2. Overview of Required Python Libraries
Appendix A.3. Example of Subjective Question Evaluation
- Question: Who is at risk for Lymphocytic Choriomeningitis (LCM)?
- Ground Truth: Individuals of all ages who come into contact with urine, faeces, saliva, or blood of wild mice are potentially at risk for infection. Owners of pet mice or hamsters may be at risk for infection if these animals originate from colonies that were contaminated with LCMV, or if their animals are infected from other wild mice. Human fetuses are at risk of acquiring infection vertically from an infected mother. Laboratory workers who work with the virus or handle infected animals are also at risk. However, this risk can be minimised by utilising animals from sources that regularly test for the virus, wearing proper protective laboratory gear, and following appropriate safety precautions.
| Model | Responses | Performance Metrics |
|---|---|---|
| Flan-T5-Large | LCMV infections can occur after exposure to fresh urine, droppings, saliva, or nesting materials from infected rodents. | BERT Score F1: 0.872 ROUGE-1 Score: 0.125 ROUGE-L Score: 0.078 BLEU Score: 0.0012 METEOR Score: 0.0393 Perplexity Score: 83.20 |
| Llama-2-7B | LCMV infections can occur after exposure to fresh urine, droppings, saliva, or nesting materials from infected rodents. Transmission may also occur when these materials are directly introduced into broken skin, the nose, the eyes, or the mouth, or presumably, via the bite of an infected rodent. Person-to-person transmission has not been reported, with the exception of vertical transmission from infected mother to fetus. | BERT Score F1: 0.863 ROUGE-1 Score: 0.295 ROUGE-L Score: 0.1477 BLEU Score: 0.0346 METEOR Score: 0.140 Perplexity Score: 34.628 |
| Mistral-7B | LCMV infections can occur after exposure to fresh urine, droppings, saliva, or nesting materials from infected rodents. Transmission may also occur when these materials are directly introduced into broken skin, the nose, the eyes, or the mouth, or presumably, via the bite of an infected rodent. Person-to-person transmission has not been reported, with the exception of vertical transmission from infected mother to fetus, and rarely, through organ transplantation. LCMV infections can also occur after exposure to infected farm animals, including cattle, sheep, goats, and pigs. | BERT Score F1: 0.863 ROUGE-1 Score: 0.3030 ROUGE-L Score: 0.1717 BLEU Score: 0.0412 METEOR Score: 0.1337 Perplexity Score: 23.0744 |
Appendix A.4. Example of MCQ Evaluation
- Question: A 35-year-old man comes to the physician because of itchy, watery eyes for the past week. He has also been sneezing multiple times a day during this period. He had a similar episode 1 year ago around springtime. He has iron deficiency anaemia and ankylosing spondylitis. Current medications include ferrous sulfate, artificial tear drops, and indomethacin. He works as an elementary school teacher. His vital signs are within normal limits. Visual acuity is 20/20 without correction. Physical examination shows bilateral conjunctival injection with watery discharge. The pupils are 3 mm, equal, and reactive to light. Examination of the anterior chamber of the eye is unremarkable. Which of the following is the most appropriate treatment?
- Options: ‘A’: ‘Erythromycin ointment’, ‘B’: ‘Ketotifen eye drops’, ‘C’: ‘Warm compresses’, ‘D’: ‘Fluorometholone eye drops’, ‘E’: ‘Latanoprost eye drops’
- Ground Truth: Ketotifen eye drops
| Methodology | Output Response Format | Performance Metrics |
|---|---|---|
| Type A | Generated Answer: B’: Ketotifen eye drops | Cosine Similarity = 1.0 Exact Match |
| Type B | Options and Confidence scores: Option A:: Erythromycin ointment - Confidence: 0.4048, Similarity: 0.5229 Option B:: Ketotifen eye drops - Confidence: 0.4423, Similarity: 0.5074 Option C:: Warm compresses - Confidence: 0.2088, Similarity: 0.5516 Option D:: Fluorometholone eye drops - Confidence: 0.3739, Similarity: 0.5061 Option E:: Latanoprost eye drops - Confidence: 0.2895, Similarity: 0.5345 Generated Answer: The physician should prescribe topical antihistamines to relieve symptoms. Selected Answer: Option B:: Ketotifen eye drops Ground Truth: Ketotifen eye drops | MRR = 1.0 LaCC (k = 1) = 100% LaCC (k = 3) = 100% |
References
- Silvera-Tawil, D. Robotics in Healthcare: A Survey. SN Comput. Sci. 2024, 5, 189. [Google Scholar] [CrossRef]
- Topol, E.J. High-performance medicine: The convergence of human and artificial intelligence. Nat. Med. 2019, 25, 44–56. [Google Scholar] [CrossRef] [PubMed]
- Toukmaji, C.; Tee, A. Retrieval-Augmented Generation and LLM Agents for Biomimicry Design Solutions. In Proceedings of the AAAI Spring Symposium Series (SSS-24), Stanford, CA, USA, 25–27 March 2024. [Google Scholar]
- Zeng, F.; Gan, W.; Wang, Y.; Liu, N.; Yu, P.S. Large Language Models for Robotics: A Survey. arXiv 2023, arXiv:2311.07226. [Google Scholar]
- Vaswani, A. Attention Is All You Need. In Proceedings of the 31st International Conference on Neural Information Processing Systems (NIPS), Long Beach, CA, USA, 4–9 December 2017. [Google Scholar]
- Jiang, A.Q.; Sablayrolles, A.; Mensch, A.; Bamford, C.; Chaplot, D.S.; de las Casas, D.; Bressand, F.; Lengyel, G.; Lample, G.; Saulnier, L.; et al. Mistral 7B. arXiv 2023, arXiv:2310.06825. [Google Scholar]
- Ni, J.; Qu, C.; Lu, J.; Dai, Z.; Ábrego, G.H.; Ma, J.; Zhao, V.Y.; Luan, Y.; Hall, K.B.; Chang, M.-W.; et al. Large Dual Encoders Are Generalizable Retrievers. arXiv 2021, arXiv:2112.07899. [Google Scholar]
- Reimers, N.; Gurevych, I. Sentence-BERT: Sentence Embeddings using Siamese BERT-Networks. arXiv 2019, arXiv:1908.10084. [Google Scholar]
- Zhang, T.; Kishore, V.; Wu, F.; Weinberger, K.Q.; Artzi, Y. BERTScore: Evaluating Text Generation with BERT. arXiv 2020, arXiv:1904.09675. [Google Scholar]
- Wolfe, C.R. LLaMA-2 from the Ground Up. 2023. Available online: https://cameronrwolfe.substack.com/p/llama-2-from-the-ground-up (accessed on 7 June 2024).
- Driess, D.; Xia, F.; Sajjadi, M.S.M.; Lynch, C.; Chowdhery, A.; Ichter, B.; Wahid, A.; Tompson, J.; Vuong, Q.; Yu, T.; et al. PaLM-E: An Embodied Multimodal Language Model. In Proceedings of the 40th International Conference on Machine Learning (ICML’23), Honolulu, HI, USA, 23–29 July 2023; Volume 202, pp. 8469–8488. [Google Scholar]
- Wei, J.; Wang, X.; Schuurmans, D.; Bosma, M.; Ichter, B.; Xia, F.; Chi, E.; Le, Q.; Zhou, D. Chain-of-Thought Prompting Elicits Reasoning in Large Language Models. arXiv 2023, arXiv:2201.11903. [Google Scholar]
- Béchard, P.; Ayala, O.M. Reducing hallucination in structured outputs via Retrieval-Augmented Generation. arXiv 2024, arXiv:2404.08189. [Google Scholar]
- Banerjee, S.; Agarwal, A.; Singla, S. LLMs Will Always Hallucinate, and We Need to Live with This. arXiv 2024, arXiv:2409.05746. [Google Scholar]
- Gao, Y.; Xiong, Y.; Gao, X.; Jia, K.; Pan, J.; Bi, Y.; Dai, Y.; Sun, J.; Wang, M.; Wang, H. Retrieval-Augmented Generation for Large Language Models: A Survey. arXiv 2024, arXiv:2312.10997. [Google Scholar]
- Lewis, P.; Perez, E.; Piktus, A.; Petroni, F.; Karpukhin, V.; Goyal, N.; Küttler, H.; Lewis, M.; Yih, W.-t.; Rocktäschel, T.; et al. Retrieval-Augmented Generation for Knowledge-Intensive NLP Tasks. arXiv 2021, arXiv:2005.11401. [Google Scholar]
- Gholami, A.; Kim, S.; Dong, Z.; Yao, Z.; Mahoney, M.W.; Keutzer, K. A Survey of Quantization Methods for Efficient Neural Network Inference. arXiv 2021, arXiv:2103.13630. [Google Scholar]
- Bajwa, J.; Munir, U.; Nori, A.; Williams, B. Artificial intelligence in healthcare: Transforming the practice of medicine. Future Healthc. J. 2021, 8, e188–e194. [Google Scholar] [CrossRef] [PubMed] [PubMed Central]
- Pal, A.; Umapathi, L.K.; Sankarasubbu, M. MedMCQA: A Large-Scale Multi-Subject Multi-Choice Dataset for Medical Domain Question Answering. arXiv 2022, arXiv:2203.14371. [Google Scholar]
- Gu, Y.; Tinn, R.; Cheng, H.; Lucas, M.; Usuyama, N.; Liu, X.; Naumann, T.; Gao, J.; Poon, H. Domain-Specific Language Model Pretraining for Biomedical Natural Language Processing. ACM Trans. Health Inform. 2022, 3, 1–23. [Google Scholar] [CrossRef]
- Bedi, S.; Liu, Y.; Orr-Ewing, L.; Dash, D.; Koyejo, S.; Callahan, A.; Fries, J.A.; Wornow, M.; Swaminathan, A.; Lehmann, L.S.; et al. A Systematic Review of Testing and Evaluation of Healthcare Applications of Large Language Models (LLMs). medRxiv 2024, 2024.04.15.24305869. [Google Scholar] [CrossRef]
- Ge, J.; Sun, S.; Owens, J.; Galvez, V.; Gologorskaya, O.; Lai, J.C.; Pletcher, M.J.; Lai, K. Development of a Liver Disease-Specific Large Language Model Chat Interface Using Retrieval Augmented Generation. medRxiv 2023, 2023.11.10.23298364. [Google Scholar] [CrossRef]
- Ramjee, P.; Sachdeva, B.; Golechha, S.; Kulkarni, S.; Fulari, G.; Murali, K.; Jain, M. CataractBot: An LLM-Powered Expert-in-the-Loop Chatbot for Cataract Patients. arXiv 2024, arXiv:2402.04620. [Google Scholar]
- Liévin, V.; Hother, C.E.; Motzfeldt, A.G.; Winther, O. Can large language models reason about medical questions? Patterns 2024, 5, 100943. [Google Scholar] [CrossRef]
- Jovanović, M.; Baez, M.; Casati, F. Chatbots as Conversational Healthcare Services. IEEE Internet Comput. 2021, 25, 44–51. [Google Scholar] [CrossRef]
- Zhou, H.; Liu, F.; Gu, B.; Zou, X.; Huang, J.; Wu, J.; Li, Y.; Chen, S.S.; Zhou, P.; Liu, J.; et al. A Survey of Large Language Models in Medicine: Progress, Application, and Challenge. arXiv 2024, arXiv:2311.05112. [Google Scholar]
- Raffel, C.; Shazeer, N.; Roberts, A.; Lee, K.; Narang, S.; Matena, M.; Zhou, Y.; Li, W.; Liu, P.J. Exploring the Limits of Transfer Learning with a Unified Text-to-Text Transformer. arXiv 2023, arXiv:1910.10683. [Google Scholar]
- Chung, H.W.; Hou, L.; Longpre, S.; Zoph, B.; Tay, Y.; Fedus, W.; Li, Y.; Wang, X.; Dehghani, M.; Brahma, S.; et al. Scaling Instruction-Finetuned Language Models. arXiv 2022, arXiv:2210.11416. [Google Scholar]
- Touvron, H.; Martin, L.; Stone, K.; Albert, P.; Almahairi, A.; Babaei, Y.; Bashlykov, N.; Batra, S.; Bhargava, P.; Bhosale, S.; et al. Llama 2: Open Foundation and Fine-Tuned Chat Models. arXiv 2023, arXiv:2307.09288. [Google Scholar]
- Gao, Y.; Liu, Y.; Zhang, H.; Li, Z.; Zhu, Y.; Lin, H.; Yang, M. Estimating GPU Memory Consumption of Deep Learning Models. In Proceedings of the ACM, Virtual, 8–13 November 2020. [Google Scholar]
- Jeon, H.; Kim, Y.; Kim, J.-J. L4Q: Parameter Efficient Quantization-Aware Fine-Tuning on Large Language Models. arXiv 2024, arXiv:2402.04902. [Google Scholar]
- Dettmers, T.; Pagnoni, A.; Holtzman, A.; Zettlemoyer, L. QLoRA: Efficient finetuning of quantized LLMs. arXiv 2023, arXiv:2305.14314. [Google Scholar]
- Xu, Y.; Xie, L.; Gu, X.; Chen, X.; Chang, H.; Zhang, H.; Chen, Z.; Zhang, X.; Tian, Q. QA-LoRA: Quantization-Aware Low-Rank Adaptation of Large Language Models. arXiv 2023, arXiv:2309.14717. [Google Scholar]
- Christophe, C.; Kanithi, P.K.; Munjal, P.; Raha, T.; Hayat, N.; Rajan, R.; Al-Mahrooqi, A.; Gupta, A.; Salman, M.U.; Gosal, G.; et al. Med42—Evaluating Fine-Tuning Strategies for Medical LLMs: Full-Parameter vs. Parameter-Efficient Approaches. arXiv 2024, arXiv:2404.14779v1. [Google Scholar]
- Han, T.; Adams, L.C.; Papaioannou, J.-M.; Grundmann, P.; Oberhauser, T.; Löser, A.; Truhn, D.; Bressem, K.K. MedAlpaca—An Open-Source Collection of Medical Conversational AI Models and Training Data. arXiv 2023, arXiv:2304.08247. [Google Scholar]
- Jin, Q.; Dhingra, B.; Liu, Z.; Cohen, W.W.; Lu, X. PubMedQA: A Dataset for Biomedical Research Question Answering. arXiv 2019, arXiv:1909.06146. [Google Scholar]
- Abacha, A.B.; Demner-Fushman, D. A Question-Entailment Approach to Question Answering. BMC Bioinform. 2019, 20, 511. [Google Scholar]
- Hu, T.; Zhou, X.-H. Unveiling LLM Evaluation Focused on Metrics: Challenges and Solutions. arXiv 2024, arXiv:2404.09135. [Google Scholar]
- Papineni, K.; Roukos, S.; Ward, T.; Zhu, W.-J. BLEU: A Method for Automatic Evaluation of Machine Translation. In Proceedings of the 40th Annual Meeting of the Association for Computational Linguistics, Philadelphia, PA, USA, 6–12 July 2002; Association for Computational Linguistics: Philadelphia, PA, USA, 2002; pp. 311–318. [Google Scholar]
- Lin, C.-Y. ROUGE: A Package for Automatic Evaluation of Summaries. In Text Summarization Branches Out; Association for Computational Linguistics: Barcelona, Spain, 2004; pp. 74–81. [Google Scholar]
- Banerjee, S.; Lavie, A. METEOR: An Automatic Metric for MT Evaluation with Improved Correlation with Human Judgments. In Proceedings of the ACL Workshop on Intrinsic and Extrinsic Evaluation Measures for Machine Translation and/or Summarization, Ann Arbor, MI, USA, 29 June 2005; pp. 65–72. [Google Scholar]
- Zhou, J. QOG: Question and Options Generation based on Language Model. arXiv 2024, arXiv:2406.12381. [Google Scholar]
- Wu, J.; Zhu, J.; Qi, Y. Medical Graph RAG: Towards Safe Medical Large Language Model via Graph Retrieval-Augmented Generation. arXiv 2024, arXiv:2408.04187. [Google Scholar]
- Singhal, K.; Tu, T.; Gottweis, J.; Sayres, R.; Wulczyn, E.; Hou, L.; Clark, K.; Pfohl, S.; Cole-Lewis, H.; Neal, D.; et al. Towards Expert-Level Medical Question Answering with Large Language Models. arXiv 2023, arXiv:2305.09617. [Google Scholar]
- Zhao, W.X.; Zhou, K.; Li, J.; Tang, T.; Wang, X.; Hou, Y.; Min, Y.; Zhang, B.; Zhang, J.; Dong, Z.; et al. A Survey of Large Language Models. arXiv 2023, arXiv:2303.18223. [Google Scholar]
- Mhatre, A.; Warhade, S.R.; Pawar, O.; Kokate, S.; Jain, S.; Emmanuel, M. Leveraging LLM: Implementing an Advanced AI Chatbot for Healthcare. Int. J. Innov. Sci. Res. Technol. 2024, 9. [Google Scholar] [CrossRef]
- Singhal, K.; Azizi, S.; Tu, T.; Mahdavi, S.S.; Wei, J.; Chung, H.W.; Scales, N.; Tanwani, A.; Cole-Lewis, H.; Pfohl, S.; et al. Large language models encode clinical knowledge. Nature 2023, 620, 172–180. [Google Scholar] [CrossRef]

| Feature | Flan-T5-Large | LLaMA2-7B | Mistral-7B |
|---|---|---|---|
| Architecture | Transformer (Encoder-Decoder) | Transformer (Decoder) | Transformer (Decoder) |
| Model Parameters | 780 million | 7 billion | 7 billion |
| Training Data | Massive text corpus | Publicly Available Data | Publicly Available Data |
| Training Objective | Text-to-Text | Autoregressive Language Modelling | Autoregressive Language Modelling |
| Use Cases | Translation, Summarisation, Q&A | Text Generation, Summarisation, Code Generation | Text Generation, Summarisation |
| Inference Speed | Fast | Moderate to High | Moderate to High |
| Memory Requirements | Moderate to High | High | High |
| Open-Source | Yes | Yes | Yes |
| Quantization | Yes | Yes | Yes |
| Licensing | Apache 2.0 | Apache 2.0 | Apache 2.0 |
| Metrics | Classification | Flan-T5-Large | Llama-2-7B | Mistral-7B |
|---|---|---|---|---|
| BERT F1 | Base with RAG | 0.071 | 0.068 | 0.181 |
| FT without RAG | 0.063 | 0.16 | 0.17 | |
| FT with RAG | 0.066 | 0.233 | 0.221 | |
| ROUGE-1 | Base with RAG | 0.162 | 0.232 | 0.3456 |
| FT without RAG | 0.153 | 0.339 | 0.339 | |
| FT with RAG | 0.136 | 0.284 | 0.308 | |
| ROUGE-L | Base with RAG | 0.133 | 0.168 | 0.2512 |
| FT without RAG | 0.129 | 0.254 | 0.246 | |
| FT with RAG | 0.113 | 0.23 | 0.221 | |
| BLEU | Base with RAG | 0.01 | 0.05 | 0.127 |
| FT without RAG | 0.02 | 0.119 | 0.103 | |
| FT with RAG | 0.019 | 0.095 | 0.091 | |
| METEOR | Base with RAG | 0.086 | 0.16 | 0.271 |
| FT without RAG | 0.084 | 0.259 | 0.238 | |
| FT with RAG | 0.077 | 0.219 | 0.288 | |
| Perplexity | Base with RAG | 4.0188 | 7.967 | 6.4691 |
| FT without RAG | 7.822 | 8.393 | 6.9795 | |
| FT with RAG | 12.592 | 7.797 | 4.84 | |
| Avg. Time p/Q (s) | Base with RAG | 17.6975 | 8.128 | 78.5243 |
| FT without RAG | 6.8529 | 33.7098 | 56.7355 | |
| FT with RAG | 13.0491 | 116.372 | 150.658 |
| Metrics | Classification | Flan-T5-Large | Llama-2-7B | Mistral-7B |
|---|---|---|---|---|
| BERT F1 | Base with RAG | 0.071 | 0.068 | 0.181 |
| FT without RAG | 0.072 | 0.039 | 0.113 | |
| FT with RAG | 0.074 | 0.152 | 0.073 | |
| ROUGE-1 | Base with RAG | 0.162 | 0.232 | 0.3456 |
| FT without RAG | 0.166 | 0.204 | 0.292 | |
| FT with RAG | 0.159 | 0.295 | 0.31 | |
| ROUGE-L | Base with RAG | 0.133 | 0.168 | 0.2512 |
| FT without RAG | 0.1376 | 0.15 | 0.222 | |
| FT with RAG | 0.13 | 0.217 | 0.181 | |
| BLEU | Base with RAG | 0.01 | 0.05 | 0.127 |
| FT without RAG | 0.011 | 0.049 | 0.091 | |
| FT with RAG | 0.01 | 0.085 | 0.05 | |
| METEOR | Base with RAG | 0.086 | 0.16 | 0.271 |
| FT without RAG | 0.09 | 0.146 | 0.21 | |
| FT with RAG | 0.09 | 0.208 | 0.278 | |
| Perplexity | Base with RAG | 4.0188 | 7.967 | 6.4691 |
| FT without RAG | 5.823 | 6.99 | 5.95 | |
| FT with RAG | 8.67 | 7.088 | 6.253 | |
| Avg. Time p/Q (s) | Base with RAG | 17.6975 | 8.128 | 78.5243 |
| FT without RAG | 9.4703 | 151.56 | 81.871 | |
| FT with RAG | 9.8312 | 123.47 | 54.715 |
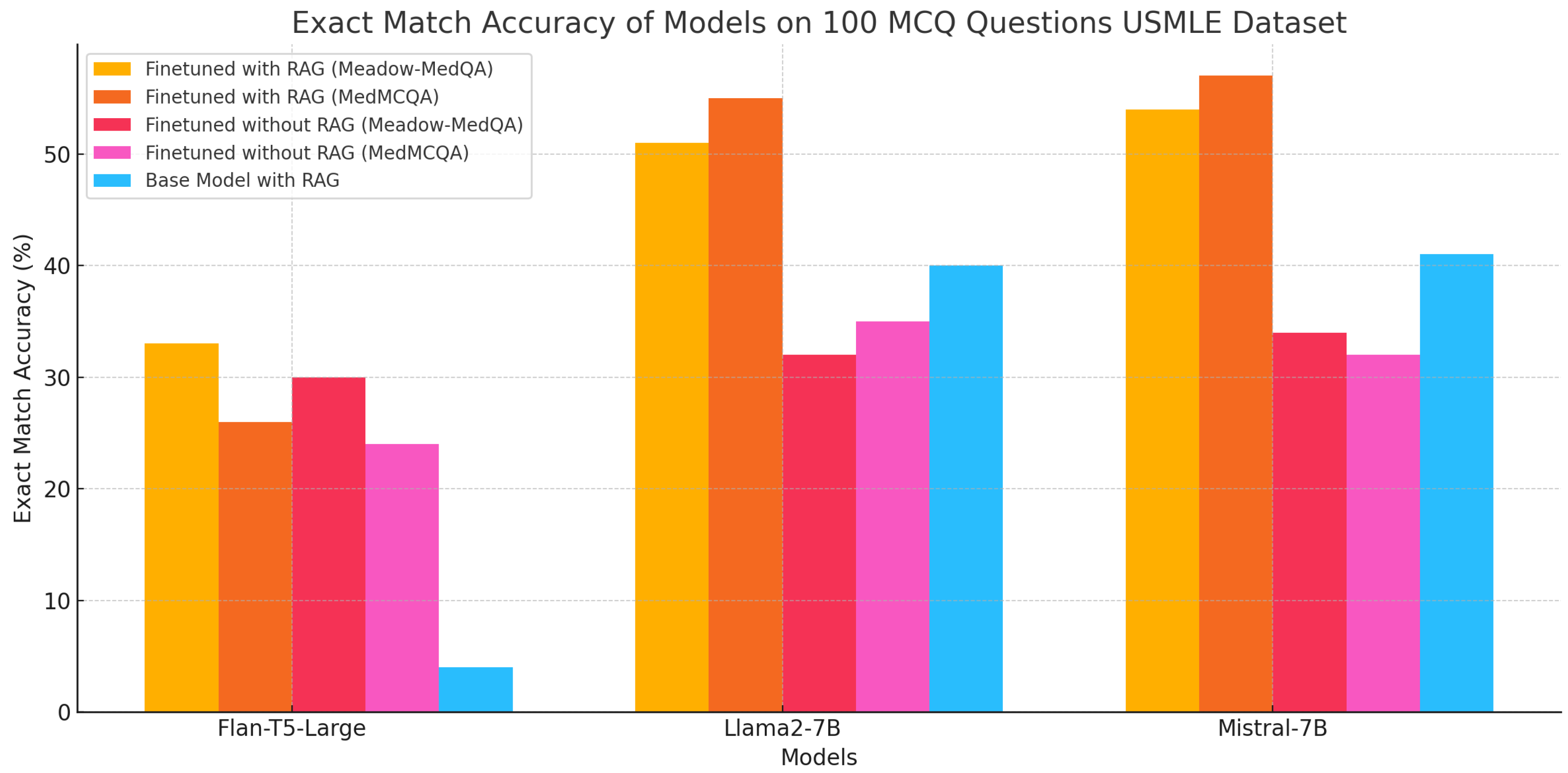
| Type | Fine-Tuning Dataset | Model | Exact Match Accuracy | Avg. Time p/Question (In Seconds) |
|---|---|---|---|---|
| Base with RAG | - | Flan-T5-Large | 4% | 3.0446 |
| - | Llama-2-7B | 40% | 13.4805 | |
| - | Mistral-7B | 41% | 18.5270 | |
| FT without RAG | Meadow-MedQA | Flan-T5-Large | 30% | 0.6309 |
| MedMCQA | Flan-T5-Large | 24% | 0.2713 | |
| Meadow-MedQA | Llama-2-7B | 32% | 10.4805 | |
| MedMCQA | Llama-2-7B | 35% | 10.5743 | |
| Meadow-MedQA | Mistral-7B | 34% | 15.3815 | |
| MedMCQA | Mistral-7B | 32% | 11.1937 | |
| FT with RAG | Meadow-MedQA | Flan-T5-Large | 33% | 4.4675 |
| MedMCQA | Flan-T5-Large | 26% | 0.3956 | |
| Meadow-MedQA | Llama-2-7B | 51% | 14.1219 | |
| MedMCQA | Llama-2-7B | 55% | 14.2405 | |
| Meadow-MedQA | Mistral-7B | 54% | 14.1282 | |
| MedMCQA | Mistral-7B | 57% | 12.2159 |
| Model | Fine-tuning Training Dataset | Mean Reciprocal Rank (MRR) | LaCC (K = 1) | LaCC (k = 3) |
|---|---|---|---|---|
| Flan-T5-Large | Meadow-MedQA | 0.44 | 21.42% | 61.05% |
| MedMCQA | 0.43 | 18.6% | 56.84% | |
| Llama-2-7B | Meadow-MedQA | 0.44 | 22.8% | 58.90% |
| MedMCQA | 0.47 | 26.2% | 61.80% | |
| Mistral-7B | Meadow-MedQA | 0.51 | 30.7% | 67.74% |
| MedMCQA | 0.53 | 32.8% | 70.83% |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Bora, A.; Cuayáhuitl, H. Systematic Analysis of Retrieval-Augmented Generation-Based LLMs for Medical Chatbot Applications. Mach. Learn. Knowl. Extr. 2024, 6, 2355-2374. https://doi.org/10.3390/make6040116
Bora A, Cuayáhuitl H. Systematic Analysis of Retrieval-Augmented Generation-Based LLMs for Medical Chatbot Applications. Machine Learning and Knowledge Extraction. 2024; 6(4):2355-2374. https://doi.org/10.3390/make6040116
Chicago/Turabian StyleBora, Arunabh, and Heriberto Cuayáhuitl. 2024. "Systematic Analysis of Retrieval-Augmented Generation-Based LLMs for Medical Chatbot Applications" Machine Learning and Knowledge Extraction 6, no. 4: 2355-2374. https://doi.org/10.3390/make6040116
APA StyleBora, A., & Cuayáhuitl, H. (2024). Systematic Analysis of Retrieval-Augmented Generation-Based LLMs for Medical Chatbot Applications. Machine Learning and Knowledge Extraction, 6(4), 2355-2374. https://doi.org/10.3390/make6040116






