Extracting Interpretable Knowledge from the Remote Monitoring of COVID-19 Patients
Abstract
1. Introduction
2. Materials and Methods
2.1. Overview of Data
- Age;
- Weight;
- Sex;
- Medical history, including the following:
- ○
- Comorbidities (asthma, hypertension, hyperlipidemia, diabetes, chronic obstructive pulmonary disease (COPD), and coronary heart disease);
- ○
- Smoking;
- ○
- Medications;
- COVID-19 history, including the following:
- ○
- Date of SARS-CoV-2 positive test;
- ○
- Onset of COVID-19 symptoms;
- ○
- Number of COVID-19 vaccine doses administered if vaccinated;
- ○
- Date of last COVID-19 vaccination dose;
- ○
- COVID-19 vaccine manufacturer;
- ○
- Initial symptoms;
- ○
- Previous SARS-CoV-2 infection.
- Heart rate;
- Blood pressure;
- Oxygen saturation level;
- Body temperature;
- Respiratory rate;
- Weight;
- Glucose (if appropriate, e.g., diabetic patients).
2.2. The Proposed Methodology
2.2.1. Data Preparation
- Classification of the mMRC Grade;
- Time-To-Event (TTE) analysis of fever remission.
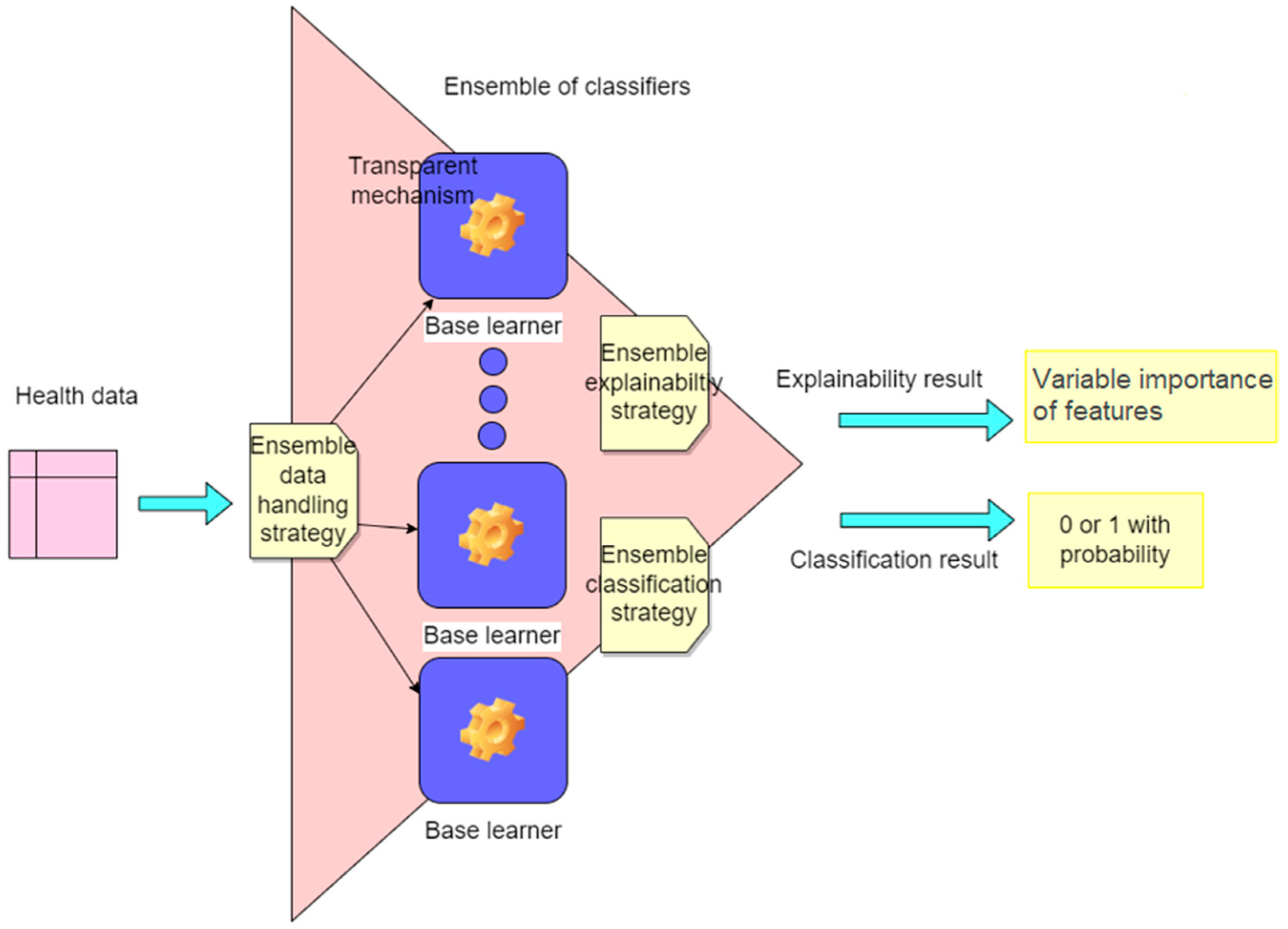
2.2.2. Classification and Interpretability Methodology
- Dataset preparation, where the data are partitioned into subgroups or assimilated as a whole by each underlying learning model according to the ensemble classifier strategy.
- Ensemble classification, where base classifiers are trained in parallel or serially, and their predictions are aggregated to produce the output of the ensemble classifier [36].
- Ensemble interpretability, where base classifiers return an importance value for each individual input feature in the final result, and the importance values for each feature of the base classifiers are summed following the ensemble model logic.
2.2.3. Time-to-Event Analysis Methodology
3. Experimental Results
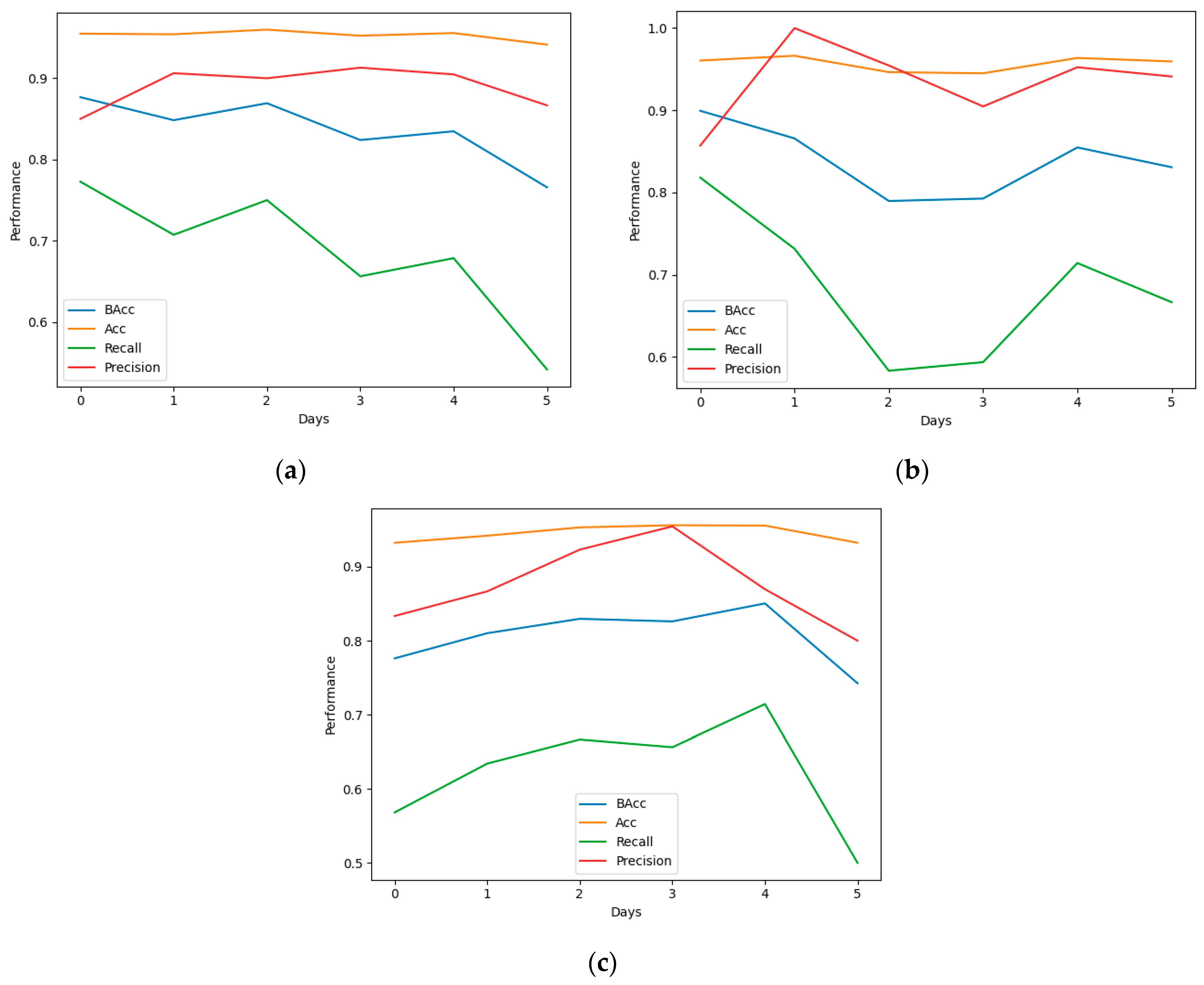
3.1. Classification
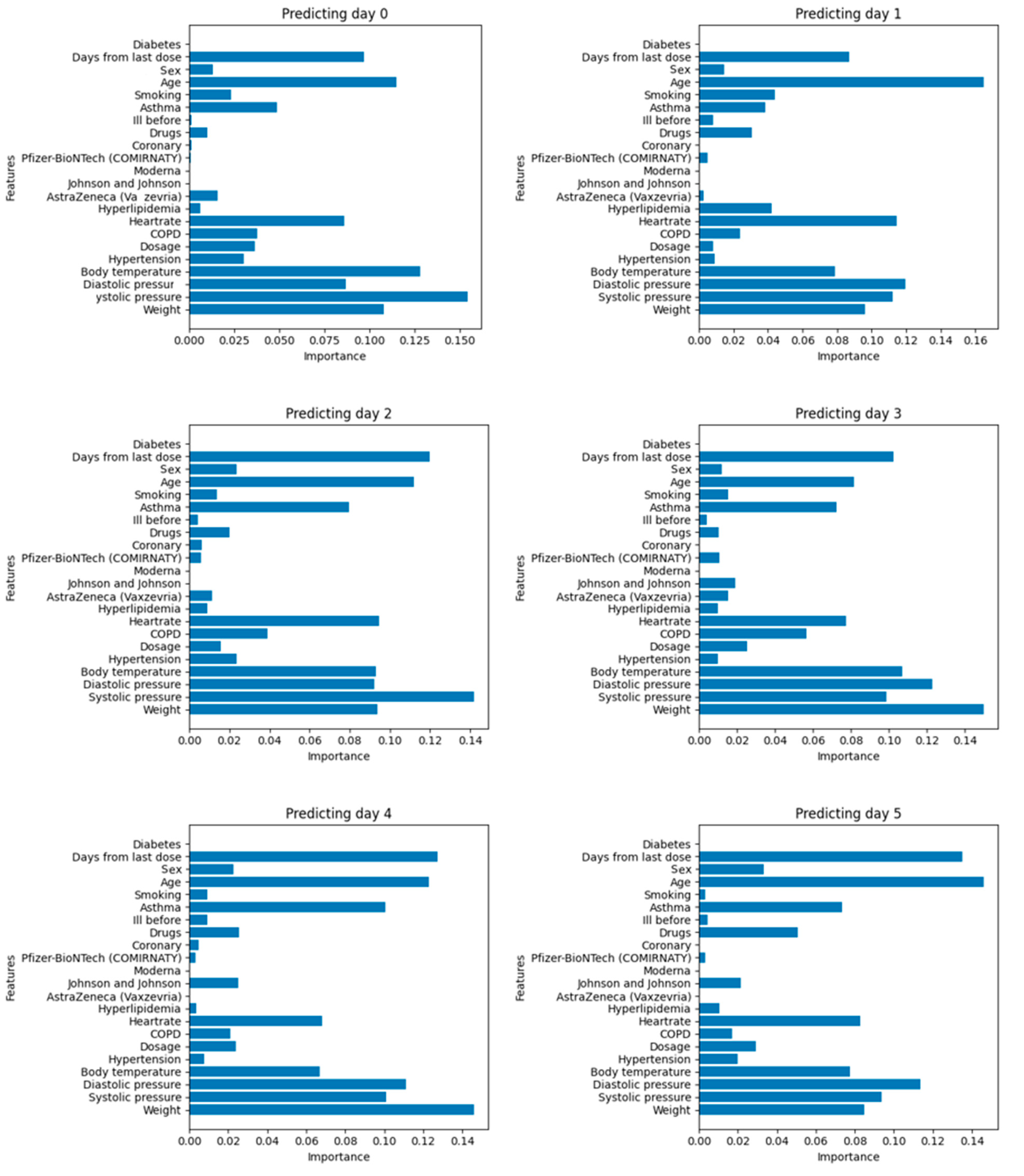
3.2. Interpretability
3.3. Improving Models through Feature Variable Selection
- Days since the last dose;
- Age;
- Asthma;
- Heart rate;
- Diastolic pressure;
- Systolic pressure;
- Body temperature;
- Weight.

3.4. Time-to-Event Analysis for Fever Remission
4. The System in Practice
4.1. Platform for Data Collection
4.2. Platform for Data Analysis
| GET/daybyday_inference?&cls_type=RF&sex_flag=1.0&days_ahead=3&age_lower_flag=50&age_upper_flag=80 |
| GET/cox_regression/?follow_up_period_flag=2&sex_flag=1&age_lower_flag=20&initial_positive_test_date_lower_flag=2020-12-03&initial_positive_test_date_upper_flag=2022-04-30 |
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Spänig, S.; Emberger-Klein, A.; Sowa, J.; Canbay, A.; Menrad, K.; Heider, D. The Virtual Doctor: An Interactive Artificial Intelligence based on Deep Learning for Non-Invasive Prediction of Diabetes. Artif. Intell. Med. 2019, 100, 101706. [Google Scholar] [CrossRef] [PubMed]
- Gu, D.; Zhao, W.; Xie, Y.; Wang, X.; Su, K.; Zolotarev, O. A Personalized Medical Decision Support System Based on Explainable Machine Learning Algorithms and ECC Features: Data from the Real World. Diagnostics 2021, 11, 1677. [Google Scholar] [CrossRef] [PubMed]
- Qin, F.; Lv, Z.; Wang, D.; Hu, B.; Wu, C. Health status prediction for the elderly based on machine learning. Arch. Gerontol. Geriatr. 2020, 90, 104121. [Google Scholar] [CrossRef] [PubMed]
- Habib, M.; Wang, Z.; Qiu, S.; Zhao, H.; Murthy, A.S. Machine Learning Based Healthcare System for Investigating the Association Between Depression and Quality of Life. IEEE J. Biomed. Health Inform. 2022, 26, 2008–2019. [Google Scholar] [CrossRef] [PubMed]
- Latif, S.; Qadir, J.; Farooq, S.; Imran, M.A. How 5G Wireless (and Concomitant Technologies) Will Revolutionize Healthcare? Future Internet 2017, 9, 93. [Google Scholar] [CrossRef]
- Iqbal, S.M.; Mahgoub, I.; Du, E.; Leavitt, M.A.; Asghar, W. Advances in healthcare wearable devices. NPJ Flex. Electron. 2021, 5, 1–14. [Google Scholar] [CrossRef]
- Pradhan, B.K.; Bhattacharyya, S.; Pal, K. IoT-Based Applications in Healthcare Devices. J. Healthc. Eng. 2021, 2021, 6632599. [Google Scholar] [CrossRef]
- Chen, P.; Lin, C.; Wu, W. Big data management in healthcare: Adoption challenges and implications. Int. J. Inf. Manag. 2020, 53, 102078. [Google Scholar] [CrossRef]
- Vellido, A. The importance of interpretability and visualization in machine learning for applications in medicine and health care. Neural Comput. Appl. 2019, 32, 18069–18083. [Google Scholar] [CrossRef]
- Ranjan, Y.; Rashid, Z.; Stewart, C.L.; Conde, P.; Begale, M.; Verbeeck, D.; Boettcher, S.; Dobson, R.J.; Folarin, A.A. RADAR-Base: Open Source Mobile Health Platform for Collecting, Monitoring, and Analyzing Data Using Sensors, Wearables, and Mobile Devices. JMIR mHealth uHealth 2019, 7, e11734. [Google Scholar] [CrossRef]
- Bhat, G.; Deb, R.; Ogras, U.Y. OpenHealth: Open-Source Platform for Wearable Health Monitoring. IEEE Des. Test 2019, 36, 27–34. [Google Scholar] [CrossRef]
- Bahmani, A.; Alavi, A.; Buergel, T.; Upadhyayula, S.; Wang, Q.; Ananthakrishnan, S.K.; Alavi, A.H.; Celis, D.; Gillespie, D.; Young, G.; et al. A scalable, secure, and interoperable platform for deep data-driven health management. Nat. Commun. 2021, 12, 5757. [Google Scholar] [CrossRef] [PubMed]
- Hermes, S.; Riasanow, T.; Clemons, E.K.; Böhm, M.; Krcmar, H. The digital transformation of the healthcare industry: Exploring the rise of emerging platform ecosystems and their influence on the role of patients. Bus. Res. 2020, 13, 1033–1069. [Google Scholar] [CrossRef]
- Najim, A.H.; Elkhediri, S.; Alrashidi, M.; Nasri, N. The Impact of using IoT for Elderly and Disabled Peoples Healthcare: An Overview. In Proceedings of the 2022 2nd International Conference on Computing and Information Technology (ICCIT), Tabuk, Saudi Arabia, 25–27 January 2022; pp. 394–398. [Google Scholar]
- Menychtas, A.; Tsanakas, P.; Maglogiannis, I. Knowledge discovery on IoT-enabled mHealth applications. In GeNeDis 2018: Computational Biology and Bioinformatics; Springer International Publishing: Berlin/Heidelberg, Germany, 2020; pp. 181–191. [Google Scholar]
- Kyriazis, D.; Autexier, S.; Boniface, M.; Engen, V.; Jimenez-Peris, R.; Jordan, B.; Jurak, G.; Kiourtis, A.; Kosmidis, T.; Lustrek, M.; et al. The CrowdHEALTH project and the hollistic health records: Collective wisdom driving public health policies. Acta Inform. Medica 2019, 27, 369. [Google Scholar] [CrossRef]
- Dinh-Le, C.; Chuang, R.; Chokshi, S.K.; Mann, D.M. Wearable Health Technology and Electronic Health Record Integration: Scoping Review and Future Directions. JMIR mHealth uHealth 2019, 7, e12861. [Google Scholar] [CrossRef]
- Rawajbeh, M.A.; Band, S.S.; Mosavi, A.H.; Kumar, R.L.; Khan, F.; Din, S.; Ibeke, E. Recurrent Neural Network and Reinforcement Learning Model for COVID-19 Prediction. Front. Public Health 2021, 9, 744100. [Google Scholar]
- Kallipolitis, A.; Gallos, P.; Menychtas, A.; Tsanakas, P.; Maglogiannis, I. Medical Knowledge Extraction from Graph-Based Modeling of Electronic Health Records. In Proceedings of the IFIP International Conference on Artificial Intelligence Applications and Innovations, León, Spain, 14–17 June 2023; Springer: Cham, Switzerland, 2023. [Google Scholar]
- Yuan, Z.; Ding, H.; Chao, G.; Song, M.; Wang, L.; Ding, W.; Chu, D. A Diabetes Prediction System Based on Incomplete Fused Data Sources. Mach. Learn. Knowl. Extr. 2023, 5, 384–399. [Google Scholar] [CrossRef]
- Cutillo, C.M.; Sharma, K.R.; Foschini, L.; Kundu, S.; Mackintosh, M.; Mandl, K.D.; MI in Healthcare Workshop Working Group. Machine intelligence in healthcare—perspectives on trustworthiness, explainability, usability, and transparency. NPJ Digital Medicine 2020, 3, 47. [Google Scholar] [CrossRef]
- Rueda, J.; Rodríguez, J.D.; Jounou, I.P.; Hortal-Carmona, J.; Ausín, T.; Rodríguez-Arias, D. “Just” accuracy? Procedural fairness demands Intepretability in AI-based medical resource allocations. Ai Soc. 2022, 1–12. [Google Scholar]
- Rai, A. Explainable AI: From black box to glass box. J. Acad. Mark. Sci. 2019, 48, 137–141. [Google Scholar] [CrossRef]
- Tziomaka, M.; Kallipolitis, A.; Tsanakas, P.; Maglogiannis, I. Evaluating Mental Patients Utilizing Video Analysis of Facial Expressions. In Proceedings of the IFIP International Conference on Artificial Intelligence Applications and Innovations, Crete, Greece, 25–27 June 2021; Springer International Publishing: Cham, Switzerland, 2021; pp. 182–193. [Google Scholar]
- Reza Soroushmehr, S.M.; Najarian, K. Transforming big data into computational models for personalized medicine and health care. Dialogues Clin. Neurosci. 2016, 18, 339–343. [Google Scholar] [CrossRef]
- Fröhlich, H.; Balling, R.; Beerenwinkel, N.; Kohlbacher, O.; Kumar, S.; Lengauer, T.; Maathuis, M.H.; Moreau, Y.; Murphy, S.A.; Przytycka, T.M.; et al. From hype to reality: Data science enabling personalized medicine. BMC Med. 2018, 16, 150. [Google Scholar] [CrossRef]
- Lonergan, M.; Senn, S.J.; McNamee, C.; Daly, A.K.; Sutton, R.; Hattersley, A.T.; Pearson, E.R.; Pirmohamed, M. Defining drug response for stratified medicine. Drug Discov. Today 2017, 22, 173–179. [Google Scholar] [CrossRef]
- Evans, S.S.; Repasky, E.A.; Fisher, D.T. Fever and the thermal regulation of immunity: The immune system feels the heat. Nat. Rev. Immunol. 2015, 15, 335–349. [Google Scholar] [CrossRef]
- Launey, Y.; Nesseler, N.; Mallédant, Y.; Seguin, P. Clinical review: Fever in septic ICU patients–friend or foe? Crit Care 2011, 15, 222. [Google Scholar] [CrossRef]
- Huang, T.; Guo, Y. Application and effects of fever screening system in the prevention of nosocomial infection in the only designated hospital of coronavirus disease 2019 (COVID-19) in Shenzhen, China. Infect. Control Hosp. Epidemiol. 2020, 41, 978–981. [Google Scholar] [CrossRef]
- Mahler, D.A.; Wells, C.K. Evaluation of clinical methods for rating dyspnea. Chest 1988, 93, 580–586. [Google Scholar] [CrossRef]
- Silverman, B.W.; Jones, M.C.E. Fix and J.L. Hodges (1951): An Important Contribution to Nonparametric Discriminant Analysis and Density Estimation: Commentary on Fix and Hodges (1951). Int. Stat. Rev./Rev. Int. De Stat. 1951, 57, 233–238. [Google Scholar] [CrossRef]
- Chen, T.; Guestrin, C. XGBoost: A Scalable Tree Boosting System. In Proceedings of the 22nd ACM SIGKDD International Conference on Knowledge Discovery and Data Mining, San Francisco, CA, USA, 13–17 August 2016. [Google Scholar]
- Freund, Y.; Schapire, R.E. A decision-theoretic generalization of on-line learning and an application to boosting. J. Comput. Syst. Sci. 1997, 55, 119–139. [Google Scholar] [CrossRef]
- Breiman, L. Random Forests. Machine Learning 2001, 45, 5–32. [Google Scholar] [CrossRef]
- Alshayeji, M.H. Early Thyroid Risk Prediction by Data Mining and Ensemble Classifiers. Mach. Learn. Knowl. Extr. 2023, 5, 1195–1213. [Google Scholar] [CrossRef]
- Cox, D.R. Regression Models and Lifetables. J. R. Stat. Soc. Ser. B 1972, 34, 187–202. [Google Scholar] [CrossRef]
- Emmert-Streib, F.; Dehmer, M. Introduction to Survival Analysis in Practice. Mach. Learn. Knowl. Extr. 2019, 1, 1013–1038. [Google Scholar] [CrossRef]
- Xue, X.; Kim, M.Y.; Shore, R.E. Cox regression analysis in presence of collinearity: An application to assessment of health risks associated with occupational radiation exposure. Lifetime Data Anal 2007, 13, 333–350. [Google Scholar] [CrossRef]





| Demographic and Clinical Characteristics of Patients | |
|---|---|
| Variable | Data |
| Patients, N | 162 |
| Age, years (median, IQR) | 51 (42–60) |
| Sex, N (%) | |
| Male | 72 (44.4) |
| Female | 90 (55.6) |
| Smoking status, N (%) | |
| Yes | 87 (53.7) |
| No | 75 (46.3) |
| Comorbidities, N (%) | 35 (21.6) |
| Hypertension | 33 |
| Hyperlipidemia | 33 |
| Coronary artery disease | 5 |
| Diabetes | 4 |
| Thyroid disease | 4 |
| Asthma | 4 |
| COPD | 2 |
| Weight, kg (mean ± SD) | 70.57 ± 11.94 |
| Vaccination status, N (%) | |
| 4 doses | 1 (0.6) |
| 3 doses | 113 (69.8) |
| 2 doses | 37 (22.8) |
| 1 dose | 5 (3.1) |
| Unvaccinated | 6 (3.7) |
| Vaccine type, N (%) | |
| Pfizer | 142 (87.6) |
| Moderna | 5 (3.1) |
| Johnson & Johnson | 5 (3.1) |
| AstraZeneca | 4 (2.5) |
| Days since the last vaccine dose (median, IQR) | 120 (88–160) |
| Previous infection, N (%) | 6 (3.7) |
| Days from positive test prior to enrolment (median, IQR) | 3 (1–11) |
| Days of symptoms prior to enrolment (median, IQR) | 3 (2–5) |
| Days of monitoring (median, IQR) | 14 (13–15) |
| mMRC Scale | |
|---|---|
| Grade | Description |
| 1 | No shortness of breath or shortness of breath only during strenuous work. |
| 2 | Shortness of breath when walking quickly on level ground or a slight incline. |
| 3 | You walk more slowly than people of the same age on level ground because of shortness of breath or stop for breath if you walk alone. |
| 4 | Stopping for breath after walking for about 100 m or after a few minutes of walking on a level surface. |
| 5 | Too breathless to leave the house or breathless when dressing/undressing. |
| Day | mMRC Grade 1 | mMRC Grade 2 |
|---|---|---|
| 0 | 1018 | 146 |
| 1 | 938 | 134 |
| 2 | 861 | 120 |
| 3 | 791 | 105 |
| 4 | 721 | 92 |
| 5 | 647 | 79 |
| 6 | 574 | 68 |
| 7 | 520 | 60 |
| 8 | 430 | 48 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Tziomaka, M.; Kallipolitis, A.; Menychtas, A.; Gallos, P.; Panagopoulos, C.; Vassiliou, A.G.; Jahaj, E.; Dimopoulou, I.; Kotanidou, A.; Maglogiannis, I. Extracting Interpretable Knowledge from the Remote Monitoring of COVID-19 Patients. Mach. Learn. Knowl. Extr. 2024, 6, 1323-1342. https://doi.org/10.3390/make6020062
Tziomaka M, Kallipolitis A, Menychtas A, Gallos P, Panagopoulos C, Vassiliou AG, Jahaj E, Dimopoulou I, Kotanidou A, Maglogiannis I. Extracting Interpretable Knowledge from the Remote Monitoring of COVID-19 Patients. Machine Learning and Knowledge Extraction. 2024; 6(2):1323-1342. https://doi.org/10.3390/make6020062
Chicago/Turabian StyleTziomaka, Melina, Athanasios Kallipolitis, Andreas Menychtas, Parisis Gallos, Christos Panagopoulos, Alice Georgia Vassiliou, Edison Jahaj, Ioanna Dimopoulou, Anastasia Kotanidou, and Ilias Maglogiannis. 2024. "Extracting Interpretable Knowledge from the Remote Monitoring of COVID-19 Patients" Machine Learning and Knowledge Extraction 6, no. 2: 1323-1342. https://doi.org/10.3390/make6020062
APA StyleTziomaka, M., Kallipolitis, A., Menychtas, A., Gallos, P., Panagopoulos, C., Vassiliou, A. G., Jahaj, E., Dimopoulou, I., Kotanidou, A., & Maglogiannis, I. (2024). Extracting Interpretable Knowledge from the Remote Monitoring of COVID-19 Patients. Machine Learning and Knowledge Extraction, 6(2), 1323-1342. https://doi.org/10.3390/make6020062









