Recent Advances in Two-Dimensional Transition Metal Dichalcogenide Nanocomposites Biosensors for Virus Detection before and during COVID-19 Outbreak
Abstract
:1. Introduction
1.1. Origin and Discovery of Viruses
1.2. The Pandemic
1.3. Conventional Diagnostic Tools on Viral Protein- and Nucleic Acid-Based Biomarkers
1.3.1. Cell Culture-Based Virus Diagnosis
1.3.2. Hemagglutination Assay
1.3.3. Electron Microscopy
1.3.4. Enzyme-Linked Immunosorbent Assay (ELISA)-Based Detection
1.3.5. PCR-Based Detection
1.3.6. Recently Advanced Biosensors as Alternative Viral Detection Methods
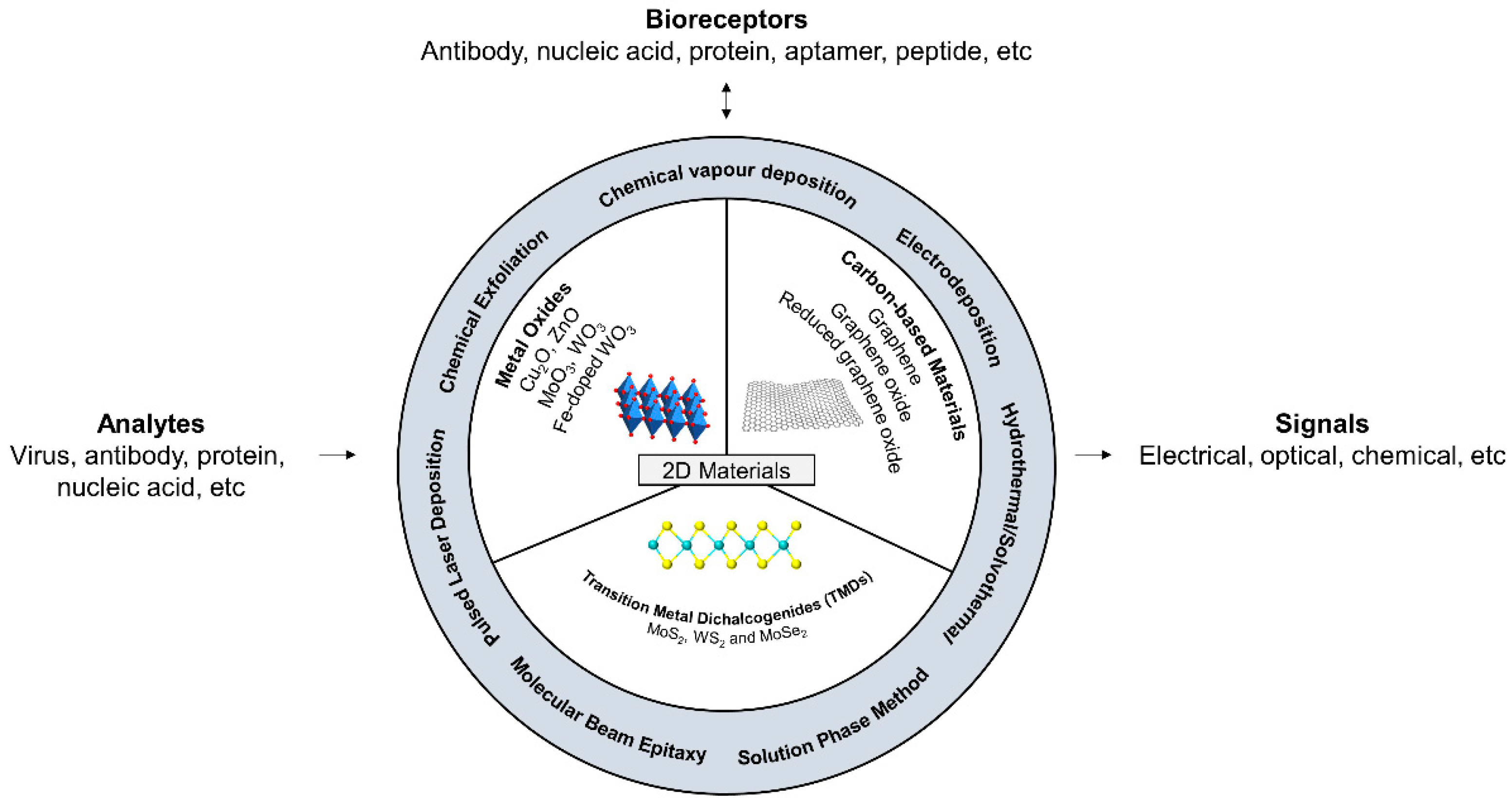
2. Two-Dimensional Nanocomposites
2.1. Transition Metal Dichalcogenides (TMDs) on Biosensing
2.1.1. Types of Elements as the Critical Parameters for Biocompatible Sensors
2.1.2. Polymorph Types and Morphologies of TMDs Alter Sensor Performance
2.1.3. TMDs-Based Disease-Related Proteins Biosensors
2.1.4. TMDs-Based Disease-Related Nucleic Acids Biosensors
2.1.5. Metal/Carbon-TMDs Nanocomposites Optimize Biosensor Performance
2.1.6. Summary
3. Conclusions and Perspective
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Bos, L. The embryonic beginning of virology: Unbiased thinking and dogmatic stagnation. Arch. Virol. 1995, 140, 613–619. [Google Scholar] [CrossRef] [PubMed]
- Bos, L. Beijerinck’s work on tobacco mosaic virus: Historical context and legacy. Philos. Trans. R. Soc. Lond. B Biol. Sci. 1999, 354, 675–685. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Prangishvili, D.; Forterre, P.; Garrett, R.A. Viruses of the Archaea: A unifying view. Nat. Rev. Microbiol. 2006, 4, 837–848. [Google Scholar] [CrossRef]
- Edwards, R.; Rohwer, F. Viral metagenomics. Nat. Rev. Genet. 2005, 3, 504–510. [Google Scholar] [CrossRef] [PubMed]
- Geraets, J.; Dykeman, E.C.; Stockley, P.; Ranson, N.; Twarock, R. Asymmetric Genome Organization in an RNA Virus Revealed via Graph-Theoretical Analysis of Tomographic Data. PLoS Comput. Biol. 2015, 11, e1004146. [Google Scholar] [CrossRef] [Green Version]
- Takizawa, T.; Matsukawa, S.; Higuchi, Y.; Nakamura, S.; Nakanishi, Y.; Fukuda, R. Induction of programmed cell death (apoptosis) by influenza virus infection in tissue culture cells. J. Gen. Virol. 1993, 74, 2347–2355. [Google Scholar] [CrossRef]
- Zhu, Z.; Lian, X.; Su, X.; Wu, W.; Marraro, G.A.; Zeng, Y. From SARS and MERS to COVID-19: A brief summary and comparison of severe acute respiratory infections caused by three highly pathogenic human coronaviruses. Respir. Res. 2020, 21, 224. [Google Scholar] [CrossRef]
- Yang, Y.; Peng, F.; Wang, R.; Guan, K.; Jiang, T.; Xu, G.; Sun, J.; Chang, C. The deadly coronaviruses: The 2003 SARS pandemic and the 2020 novel coronavirus epidemic in China. J. Autoimmun. 2020, 109, 102434. [Google Scholar] [CrossRef]
- Xu, R.-H.; He, J.-F.; Evans, M.R.; Peng, G.-W.; E Field, H.; Yu, D.-W.; Lee, C.-K.; Luo, H.-M.; Lin, W.-S.; Lin, P.; et al. Epidemiologic Clues to SARS Origin in China. Emerg. Infect. Dis. 2004, 10, 1030–1037. [Google Scholar] [CrossRef]
- Gumel, A.; Ruan, S.; Day, T.; Watmough, J.; Brauer, F.; Driessche, P.V.D.; Gabrielson, D.; Bowman, C.; Alexander, M.E.; Ardal, S.; et al. Modelling strategies for controlling SARS outbreaks. Proc. R. Soc. B Boil. Sci. 2004, 271, 2223–2232. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Perlman, S.; Azhar, E.; A Memish, Z.; Hui, D.S.; Zumla, P.S.A. Confronting the persisting threat of the Middle East respiratory syndrome to global health security. Lancet Infect. Dis. 2020, 20, 158–160. [Google Scholar] [CrossRef] [Green Version]
- Richter, C.D. European Patent Office grants controversial patent protecting virus: Lessons from the Middle East respiratory syndrome coronavirus outbreak. Nat. Biotechnol. 2021, 39, 287–290. [Google Scholar] [CrossRef] [PubMed]
- Wang, C.; Wang, Z.; Wang, G.; Lau, J.Y.-N.; Zhang, K.; Li, W. COVID-19 in early 2021: Current status and looking forward. Signal. Transduct. Target. Ther. 2021, 6, 1–14. [Google Scholar] [CrossRef] [PubMed]
- Plante, J.A.; Liu, Y.; Liu, J.; Xia, H.; Johnson, B.A.; Lokugamage, K.G.; Zhang, X.; Muruato, A.E.; Zou, J.; Fontes-Garfias, C.R.; et al. Spike mutation D614G alters SARS-CoV-2 fitness. Nature 2021, 592, 116–121. [Google Scholar] [CrossRef] [PubMed]
- Rees-Spear, C.; Muir, L.; Griffith, S.A.; Heaney, J.; Aldon, Y.; Snitselaar, J.L.; Thomas, P.; Graham, C.; Seow, J.; Lee, N.; et al. The effect of spike mutations on SARS-CoV-2 neutralization. Cell Rep. 2021, 34, 108890. [Google Scholar] [CrossRef] [PubMed]
- Cao, J.; Li, D. Searching for human oncoviruses: Histories, challenges, and opportunities. J. Cell. Biochem. 2018, 119, 4897–4906. [Google Scholar] [CrossRef]
- Douek, D.C.; Roederer, M.; Koup, R.A. Emerging Concepts in the Immunopathogenesis of AIDS. Annu. Rev. Med. 2009, 60, 471–484. [Google Scholar] [CrossRef] [Green Version]
- Sirohi, D.; Chen, Z.; Sun, L.; Klose, T.; Pierson, T.C.; Rossmann, M.G.; Kuhn, R.J. The 3.8 A resolution cryo-EM structure of Zika virus. Science 2016, 352, 467–470. [Google Scholar] [CrossRef] [Green Version]
- Jacob, S.T.; Crozier, I.; Wahl, V.; Griffiths, A.; Bollinger, L.; Kuhn, J.H.; Fischer, W.A., II; Hewlett, A.; Kraft, C.S.; De La Vega, M.-A.; et al. Ebola virus disease. Nat. Rev. Dis. Prim. 2020, 6, 13. [Google Scholar] [CrossRef] [Green Version]
- Hematian, A.; Sadeghifard, N.; Mohebi, R.; Taherikalani, M.; Nasrolahi, A.; Amraei, M.; Ghafourian, S. Traditional and Modern Cell Culture in Virus Diagnosis. Osong Public Health Res. Perspect. 2016, 7, 77–82. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Nisalak, A. Laboratory diagnosis of dengue virus infections. Southeast. Asian J. Trop. Med. Public Health 2015, 46, 55–76. [Google Scholar] [PubMed]
- Sridhar, S.; To, K.; Chan, J.F.-W.; Lau, S.K.P.; Woo, P.C.Y.; Yuen, K.-Y. A Systematic Approach to Novel Virus Discovery in Emerging Infectious Disease Outbreaks. J. Mol. Diagn. 2015, 17, 230–241. [Google Scholar] [CrossRef] [PubMed]
- Mokhtarzadeh, A.; Eivazzadeh-Keihan, R.; Pashazadeh, P.; Hejazi, M.; Gharaatifar, N.; Hasanzadeh, M.; Baradaran, B.; de la Guardia, M. Nanomaterial-based biosensors for detection of pathogenic virus. TrAC Trends Anal. Chem. 2017, 97, 445–457. [Google Scholar] [CrossRef] [PubMed]
- Balasuriya, U.B.R.; Crossley, B.M.; Timoney, P.J. A review of traditional and contemporary assays for direct and indirect detection of Equid herpesvirus 1 in clinical samples. J. Vet. Diagn. Investig. 2015, 27, 673–687. [Google Scholar] [CrossRef] [Green Version]
- Mathew, T. Cultivation and immunological studies of the pox group of viruses. I. Propagation of vaccinia virus in hamster kidney cell culture. Indian J. Pathol. Bacteriol. 1966, 9, 207–213. [Google Scholar] [PubMed]
- Brumback, B.G.; Wade, C.D. Simultaneous culture for adenovirus, cytomegalovirus, and herpes simplex virus in same shell vial by using three-color fluorescence. J. Clin. Microbiol. 1994, 32, 2289–2290. [Google Scholar] [CrossRef] [Green Version]
- Klespies, S.L.; E Cebula, D.; Kelley, C.L.; Galehouse, D.; Maurer, C.C. Detection of enteroviruses from clinical specimens by spin amplification shell vial culture and monoclonal antibody assay. J. Clin. Microbiol. 1996, 34, 1465–1467. [Google Scholar] [CrossRef] [Green Version]
- Hsiung, G.D. Diagnostic virology: From animals to automation. Yale J. Boil. Med. 1984, 57, 727–733. [Google Scholar]
- Hirst, G.K. The quantitative determination of influenza virus and antibodies by means of red cell agglutination. J. Exp. Med. 1942, 75, 49–64. [Google Scholar] [CrossRef]
- Haga, T.; Nagaki, D.; Iwasaki, K. Studies on bacterial hemagglutination. II. Hemagglutination inhibiting substance of stroma. Kitasato Arch. Exp. Med. 1951, 23, 105–109. [Google Scholar]
- Killian, M.L. Hemagglutination Assay for Influenza Virus. Methods Mol. Biol. 2014, 1161, 3–9. [Google Scholar] [CrossRef]
- Noah, D.L.; Hill, H.; Hines, D.; White, E.L.; Wolff, M.C. Qualification of the Hemagglutination Inhibition Assay in Support of Pandemic Influenza Vaccine Licensure. Clin. Vaccine Immunol. 2009, 16, 558–566. [Google Scholar] [CrossRef] [Green Version]
- Poirot, E.; Levine, M.Z.; Russell, K.; Stewart, R.J.; Pompey, J.M.; Chiu, S.; Fry, A.M.; Gross, L.; Havers, F.P.; Li, Z.-N.; et al. Detection of Avian Influenza A(H7N2) Virus Infection among Animal Shelter Workers Using a Novel Serological Approach—New York City, 2016–2017. J. Infect. Dis. 2019, 219, 1688–1696. [Google Scholar] [CrossRef]
- Müthing, J.; Unland, F. A comparative assessment of TLC overlay technique and microwell adsorption assay in the examination of influenza A and Sendai virus specificities towards oligosaccharides and sialic acid linkages of gangliosides. Glycoconj. J. 1994, 11, 486–492. [Google Scholar] [CrossRef]
- A Petrenko, V.; Sorokulova, I.B. Detection of biological threats. A challenge for directed molecular evolution. J. Microbiol. Methods 2004, 58, 147–168. [Google Scholar] [CrossRef]
- Hazelton, P.R.; Gelderblom, H.R. Electron microscopy for rapid diagnosis of infectious agents in emergent situations. Emerg. Infect. Dis. 2003, 9, 294–303. [Google Scholar] [CrossRef] [PubMed]
- Almeida, J.; Goffe, A. Antibody to wart virus in human sera demonstrated by electron microscopy and precipitin tests. Lancet 1965, 286, 1205–1207. [Google Scholar] [CrossRef]
- Briquet, S.; Vaquero, C. Immunolocalization Studies of an Antisense Protein in HIV-1-Infected Cells and Viral Particles. Virology 2002, 292, 177–184. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Fornera, S.; Walde, P. Spectrophotometric quantification of horseradish peroxidase with o-phenylenediamine. Anal. Biochem. 2010, 407, 293–295. [Google Scholar] [CrossRef] [PubMed]
- Lagunas-Rangel, F.A.; Chavez-Valencia, V. What do we know about the antibody responses to SARS-CoV-2? Immunobiology 2021, 226, 152054. [Google Scholar] [CrossRef]
- Dutta, N.K.; Mazumdar, K.; Gordy, J.T. The Nucleocapsid Protein of SARS-CoV-2: A Target for Vaccine Development. J. Virol. 2020, 94, e00647-20. [Google Scholar] [CrossRef]
- Zhang, L.; Guo, H. Biomarkers of COVID-19 and technologies to combat SARS-CoV-2. Adv. Biomark. Sci. Technol. 2020, 2, 1–23. [Google Scholar] [CrossRef]
- Jalandra, R.; Yadav, A.K.; Verma, D.; Dalal, N.; Sharma, M.; Singh, R.; Kumar, A.; Solanki, P.R. Strategies and perspectives to develop SARS-CoV-2 detection methods and diagnostics. Biomed. Pharm. 2020, 129, 110446. [Google Scholar] [CrossRef] [PubMed]
- Helmy, Y.A.; Fawzy, M.; Elaswad, A.; Sobieh, A.; Kenney, S.P.; Shehata, A.A. The COVID-19 Pandemic: A Comprehensive Review of Taxonomy, Genetics, Epidemiology, Diagnosis, Treatment, and Control. J. Clin. Med. 2020, 9, 1225. [Google Scholar] [CrossRef] [PubMed]
- Li, D.; Zhang, J.; Li, J. Primer design for quantitative real-time PCR for the emerging Coronavirus SARS-CoV-2. Theranostics 2020, 10, 7150–7162. [Google Scholar] [CrossRef]
- Michel, C.J.; Mayer, C.; Poch, O.; Thompson, J.D. Characterization of accessory genes in coronavirus genomes. Virol. J. 2020, 17, 131. [Google Scholar] [CrossRef] [PubMed]
- Park, M.; Won, J.; Choi, B.Y.; Lee, C.J. Optimization of primer sets and detection protocols for SARS-CoV-2 of coronavirus disease 2019 (COVID-19) using PCR and real-time PCR. Exp. Mol. Med. 2020, 52, 963–977. [Google Scholar] [CrossRef] [PubMed]
- Kim, S.; Lee, J.H.; Lee, S.; Shim, S.; Nguyen, T.T.; Hwang, J.; Kim, H.; Choi, Y.O.; Hong, J.; Bae, S.; et al. The Progression of SARS Coronavirus 2 (SARS-CoV2): Mutation in the Receptor Binding Domain of Spike Gene. Immune Netw. 2020, 20, e41. [Google Scholar] [CrossRef]
- Huang, W.E.; Lim, B.; Hsu, C.C.; Xiong, D.; Wu, W.; Yu, Y.J.; Jia, H.D.; Wang, Y.; Zeng, Y.D.; Ji, M.M.; et al. RT-LAMP for rapid diagnosis of coronavirus SARS-CoV-2. Microb. Biotechnol. 2020, 13, 950–961. [Google Scholar] [CrossRef] [Green Version]
- Chakraborty, D.; Kumar, S.; Chandrasekaran, N.; Mukherjee, A. Viral Diagnostics and Preventive Techniques in the Era of COVID-19: Role of Nanoparticles. Front. Nanotechnol. 2020, 2, 588795. [Google Scholar] [CrossRef]
- Carter, L.J.; Garner, L.V.; Smoot, J.W.; Li, Y.; Zhou, Q.; Saveson, C.J.; Sasso, J.M.; Gregg, A.C.; Soares, D.J.; Beskid, T.R.; et al. Assay Techniques and Test Development for COVID-19 Diagnosis. ACS Cent. Sci. 2020, 6, 591–605. [Google Scholar] [CrossRef] [PubMed]
- Thai, H.T.C.; Le, M.Q.; Vuong, C.D.; Parida, M.; Minekawa, H.; Notomi, T.; Hasebe, F.; Morita, K. Development and Evaluation of a Novel Loop-Mediated Isothermal Amplification Method for Rapid Detection of Severe Acute Respiratory Syndrome Coronavirus. J. Clin. Microbiol. 2004, 42, 1956–1961. [Google Scholar] [CrossRef] [Green Version]
- Broughton, J.P.; Deng, X.D.; Yu, G.X.; Fasching, C.L.; Servellita, V.; Singh, J.; Miao, X.; Streithorst, J.A.; Granados, A.; Sotomayor-Gonzalez, A.; et al. CRISPR-Cas12-based detection of SARS-CoV-2. Nat. Biotechnol. 2020, 38, 870–874. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hashtroudi, H.; Mackinnon, I.D.R.; Shafiei, M. Emerging 2D hybrid nanomaterials: Towards enhanced sensitive and selective conductometric gas sensors at room temperature. J. Mater. Chem. C 2020, 8, 13108–13126. [Google Scholar] [CrossRef]
- Gleason, A.M. Remote Monitoring of a Work-From-Home Employee to Identify Stress: A Case Report. Work. Health Saf. 2021. [Google Scholar] [CrossRef] [PubMed]
- Vermisoglou, E.; Panáček, D.; Jayaramulu, K.; Pykal, M.; Frébort, I.; Kolář, M.; Hajdúch, M.; Zbořil, R.; Otyepka, M. Human virus detection with graphene-based materials. Biosens. Bioelectron. 2020, 166, 112436. [Google Scholar] [CrossRef] [PubMed]
- Ehtesabi, H. Application of carbon nanomaterials in human virus detection. J. Sci. Adv. Mater. Devices 2020, 5, 436–450. [Google Scholar] [CrossRef]
- Chen, X.; Liu, C.; Mao, S. Environmental Analysis with 2D Transition-Metal Dichalcogenide-Based Field-Effect Transistors. Nano-Micro Lett. 2020, 12, 1–24. [Google Scholar] [CrossRef] [Green Version]
- Kannan, P.K.; Late, D.J.; Morgan, H.; Rout, C.S. Recent developments in 2D layered inorganic nanomaterials for sensing. Nanoscale 2015, 7, 13293–13312. [Google Scholar] [CrossRef]
- Wu, J.; Lv, W.; Yang, Q.; Li, H.; Li, F. Label-free homogeneous electrochemical detection of MicroRNA based on target-induced anti-shielding against the catalytic activity of two-dimension nanozyme. Biosens. Bioelectron. 2021, 171, 112707. [Google Scholar] [CrossRef]
- Li, H.; Lv, W.; Yang, Q.; Li, Q.; Li, F. Inorganic Recognizer-Assisted Homogeneous Electrochemiluminescence Determination of Organophosphorus Pesticides via Target-Controlled Emitter Release. J. Agric. Food Chem. 2021, 69, 6087–6095. [Google Scholar] [CrossRef] [PubMed]
- Wu, J.; Yang, Q.; Li, Q.; Li, H.; Li, F. Two-Dimensional MnO2 Nanozyme-Mediated Homogeneous Electrochemical Detection of Organophosphate Pesticides without the Interference of H2O2 and Color. Anal. Chem. 2021, 93, 4084–4091. [Google Scholar] [CrossRef] [PubMed]
- Védrine, J.C. Metal Oxides in Heterogeneous Oxidation Catalysis: State of the Art and Challenges for a More Sustainable World. ChemSusChem 2019, 12, 577–588. [Google Scholar] [CrossRef] [PubMed]
- Choi, W.; Choudhary, N.; Han, G.H.; Park, J.; Akinwande, D.; Lee, Y.H. Recent development of two-dimensional transition metal dichalcogenides and their applications. Mater. Today 2017, 20, 116–130. [Google Scholar] [CrossRef]
- Huy, V.H.; Ahn, Y.; Hur, J. Recent Advances in Transition Metal Dichalcogenide Cathode Materials for Aqueous Rechargeable Multivalent Metal-Ion Batteries. Nanomaterials 2021, 11, 1517. [Google Scholar] [CrossRef]
- Ahmed, S.; Yi, J. Two-Dimensional Transition Metal Dichalcogenides and Their Charge Carrier Mobilities in Field-Effect Transistors. Nano-Micro Lett. 2017, 9, 1–23. [Google Scholar] [CrossRef]
- Teo, W.Z.; Chng, E.L.K.; Sofer, Z.; Pumera, M. Cytotoxicity of Exfoliated Transition-Metal Dichalcogenides (MoS2, WS2, and WSe2) is Lower Than That of Graphene and its Analogues. Chem. A Eur. J. 2014, 20, 9627–9632. [Google Scholar] [CrossRef]
- Pessoa-Pureur, R.; Heimfarth, L.; Rocha, J.B. Signaling Mechanisms and Disrupted Cytoskeleton in the Diphenyl Ditelluride Neurotoxicity. Oxidative Med. Cell. Longev. 2014, 2014, 1–21. [Google Scholar] [CrossRef]
- Bhanu, U.; Islam, M.R.; Tetard, L.; Khondaker, S.I. Photoluminescence quenching in gold—MoS2 hybrid nanoflakes. Sci. Rep. 2014, 4, 5575. [Google Scholar] [CrossRef]
- Loo, A.H.; Bonanni, A.; Pumera, M. Strong dependence of fluorescence quenching on the transition metal in layered transition metal dichalcogenide nanoflakes for nucleic acid detection. Analyst. 2016, 141, 4654–4658. [Google Scholar] [CrossRef]
- Enyashin, A.N.; Yadgarov, L.; Houben, L.; Popov, I.; Weidenbach, M.; Tenne, R.; Bar-Sadan, M.; Seifert, G. New Route for Stabilization of 1T-WS2 and MoS2 Phases. J. Phy. Chem. C 2011, 115, 24586–24591. [Google Scholar] [CrossRef] [Green Version]
- Lan, L.; Chen, D.; Yao, Y.; Peng, X.; Wu, J.; Li, Y.; Ping, J.; Ying, Y. Phase-Dependent Fluorescence Quenching Efficiency of MoS2 Nanosheets and Their Applications in Multiplex Target Biosensing. ACS Appl. Mater. Interfaces 2018, 10, 42009–42017. [Google Scholar] [CrossRef] [PubMed]
- Barua, S.; Dutta, H.S.; Gogoi, S.; Devi, R.; Khan, R. Nanostructured MoS2-Based Advanced Biosensors: A Review. ACS Appl. Nano Mater. 2017, 1, 2–25. [Google Scholar] [CrossRef]
- Zhao, Z.-Y.; Liu, Q.-L. Study of the layer-dependent properties of MoS2 nanosheets with different crystal structures by DFT calculations. Catal. Sci. Technol. 2018, 8, 1867–1879. [Google Scholar] [CrossRef]
- Fan, Q.; Wang, L.; Xu, D.; Duo, Y.; Gao, J.; Zhang, L.; Wang, X.; Chen, X.; Li, J.; Zhang, H. Solution-gated transistors of two-dimensional materials for chemical and biological sensors: Status and challenges. Nanoscale 2020, 12, 11364–11394. [Google Scholar] [CrossRef] [PubMed]
- Wang, Y.; Xiao, J.; Zhu, H.; Li, Y.; Alsaid, Y.; Fong, K.Y.; Zhou, Y.; Wang, S.; Shi, W.; Wang, Y.; et al. Structural phase transition in monolayer MoTe2 driven by electrostatic doping. Nat. Cell Biol. 2017, 550, 487–491. [Google Scholar] [CrossRef] [PubMed]
- Song, S.; Keum, D.H.; Cho, S.; Perello, D.; Kim, Y.; Lee, Y.H. Room Temperature Semiconductor–Metal Transition of MoTe2 Thin Films Engineered by Strain. Nano Lett. 2016, 16, 188–193. [Google Scholar] [CrossRef] [PubMed]
- Sun, X.; Deng, H.; Zhu, W.; Yu, Z.; Wu, C.; Xie, Y. Interface Engineering in Two-Dimensional Heterostructures: Towards an Advanced Catalyst for Ullmann Couplings. Angew. Chem. Int. Ed. 2015, 55, 1704–1709. [Google Scholar] [CrossRef]
- Kufer, D.; Nikitskiy, I.; Lasanta, T.; Navickaite, G.; Koppens, F.; Konstantatos, G. Hybrid 2D-0D MoS2-PbS Quantum Dot Photodetectors. Adv. Mater. 2015, 27, 176–180. [Google Scholar] [CrossRef]
- Patolsky, F.; Zheng, G.; Hayden, O.; Lakadamyali, M.; Zhuang, X.; Lieber, C.M. Electrical detection of single viruses. Proc. Natl. Acad. Sci. USA 2004, 101, 14017–14022. [Google Scholar] [CrossRef] [Green Version]
- Mohanty, N.; Berry, V. Graphene-Based Single-Bacterium Resolution Biodevice and DNA Transistor: Interfacing Graphene Derivatives with Nanoscale and Microscale Biocomponents. Nano Lett. 2008, 8, 4469–4476. [Google Scholar] [CrossRef] [PubMed]
- Guo, X.; Small, J.P.; Klare, J.E.; Wang, Y.; Purewal, M.S.; Tam, I.W.; Hong, B.H.; Caldwell, R.; Huang, L.; Obrien, S.; et al. Covalently Bridging Gaps in Single-Walled Carbon Nanotubes with Conducting Molecules. Science 2006, 311, 356–359. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zhang, P.; Yang, S.; Pineda-Gomez, R.; Ibarlucea, B.; Ma, J.; Lohe, M.R.; Akbar, T.F.; Baraban, L.; Cuniberti, G.; Feng, X. Electrochemically Exfoliated High-Quality 2H-MoS2 for Multiflake Thin Film Flexible Biosensors. Small 2019, 15, e1901265. [Google Scholar] [CrossRef]
- Ge, J.; Ou, E.C.; Yu, R.Q.; Chu, X. A novel aptameric nanobiosensor based on the self-assembled DNA-MoS2 nanosheet architecture for biomolecule detection. J. Mater. Chem. B 2014, 2, 625–628. [Google Scholar] [CrossRef] [PubMed]
- Yoo, G.; Park, H.; Kim, M.; Song, W.G.; Jeong, S.; Kim, M.H.; Lee, H.; Lee, S.W.; Hong, Y.K.; Lee, M.G.; et al. Real-time electrical detection of epidermal skin MoS2 biosensor for point-of-care diagnostics. Nano Res. 2017, 10, 767–775. [Google Scholar] [CrossRef]
- Kukkar, M.; Tuteja, S.K.; Sharma, A.L.; Kumar, V.; Paul, A.K.; Kim, K.-H.; Sabherwal, P.; Deep, A. A New Electrolytic Synthesis Method for Few-Layered MoS2 Nanosheets and Their Robust Biointerfacing with Reduced Antibodies. ACS Appl. Mater. Interfaces 2016, 8, 16555–16563. [Google Scholar] [CrossRef] [PubMed]
- Park, H.; Lee, H.; Jeong, S.H.; Lee, E.; Lee, W.; Liu, N.; Yoon, D.S.; Kim, S.; Lee, S.W. MoS2 Field-Effect Transistor-Amyloid-beta1-42 Hybrid Device for Signal Amplified Detection of MMP-9. Anal. Chem. 2019, 91, 8252–8258. [Google Scholar] [CrossRef] [PubMed]
- Shi, J.; Lyu, J.; Tian, F.; Yang, M. A fluorescence turn-on biosensor based on graphene quantum dots (GQDs) and molybdenum disulfide (MoS2) nanosheets for epithelial cell adhesion molecule (EpCAM) detection. Biosens. Bioelectron. 2017, 93, 182–188. [Google Scholar] [CrossRef]
- Shin, M.; Yoon, J.; Yi, C.; Lee, T.; Choi, J.-W. Flexible HIV-1 Biosensor Based on the Au/MoS2 Nanoparticles/Au Nanolayer on the PET Substrate. Nanomaterials 2019, 9, 1076. [Google Scholar] [CrossRef] [Green Version]
- Li, F.; Li, Y.; Feng, J.; Gao, Z.; Lv, H.; Ren, X.; Wei, Q. Facile synthesis of MoS2@Cu2O-Pt nanohybrid as enzyme-mimetic label for the detection of the Hepatitis B surface antigen. Biosens. Bioelectron. 2018, 100, 512–518. [Google Scholar] [CrossRef]
- Gao, Z.; Li, Y.; Zhang, X.; Feng, J.; Kong, L.; Wang, P.; Chen, Z.; Dong, Y.; Wei, Q. Ultrasensitive electrochemical immunosensor for quantitative detection of HBeAg using Au@Pd/MoS2@MWCNTs nanocomposite as enzyme-mimetic labels. Biosens. Bioelectron. 2018, 102, 189–195. [Google Scholar] [CrossRef] [PubMed]
- Pathania, P.K.; Saini, J.K.; Vij, S.; Tewari, R.; Sabherwal, P.; Rishi, P.; Suri, C.R. Aptamer functionalized MoS2-rGO nanocomposite based biosensor for the detection of Vi antigen. Biosens. Bioelectron. 2018, 122, 121–126. [Google Scholar] [CrossRef]
- Solanki, S.; Soni, A.; Pandey, M.K.; Biradar, A.; Sumana, G. Langmuir-Blodgett Nanoassemblies of the MoS2-Au Composite at the Air-Water Interface for Dengue Detection. ACS Appl. Mater. Interfaces 2018, 10, 3020–3028. [Google Scholar] [CrossRef] [PubMed]
- Le, S.T.; Guros, N.B.; Bruce, R.C.; Cardone, A.; Amin, N.D.; Zhang, S.; Klauda, J.B.; Pant, H.C.; Richter, C.A.; Balijepalli, A. Quantum capacitance-limited MoS2 biosensors enable remote label-free enzyme measurements. Nanoscale 2019, 11, 15622–15632. [Google Scholar] [CrossRef] [Green Version]
- Song, Y.J.; Cao, K.H.; Li, W.J.; Ma, C.Y.; Qiao, X.W.; Li, H.L.; Hong, C.L. Optimal film thickness of rGO/MoS2 @ polyaniline nanosheets of 3D arrays for carcinoembryonic antigen high sensitivity detection. Microchem. J. 2020, 155, 104694. [Google Scholar] [CrossRef]
- Peng, X.; Zhou, Y.X.; Nie, K.X.; Zhou, F.F.; Yuan, Y.F.; Song, J.; Qu, J.L. Promising near-infrared plasmonic biosensor employed for specific detection of SARS-CoV-2 and its spike glycoprotein. New J. Phys. 2020, 22, 103046. [Google Scholar] [CrossRef]
- Vasudevan, M.; Tai, M.J.Y.; Perumal, V.; Gopinath, S.C.B.; Murthe, S.S.; Ovinis, M.; Mohamed, N.M.; Joshi, N. Cellulose acetate-MoS2 nanopetal hybrid: A highly sensitive and selective electrochemical aptasensor of Troponin I for the early diagnosis of Acute Myocardial Infarction. J. Taiwan Inst. Chem. Eng. 2021, 118, 245–253. [Google Scholar] [CrossRef]
- Zhang, X.; Lai, Z.C.; Tan, C.L.; Zhang, H. Solution-Processed Two-Dimensional MoS2 Nanosheets: Preparation, Hybridization, and Applications. Angew. Chem. Int. Ed. Engl. 2016, 55, 8816–8838. [Google Scholar] [CrossRef] [PubMed]
- Zeng, Z.; Yin, Z.; Huang, X.; Li, H.; He, Q.; Lu, G.; Boey, F.; Zhang, H. Single-Layer Semiconducting Nanosheets: High-Yield Preparation and Device Fabrication. Angew. Chem. Int. Ed. 2011, 50, 11093–11097. [Google Scholar] [CrossRef]
- Coleman, J.N.; Lotya, M.; O’Neill, A.; Bergin, S.D.; King, P.J.; Khan, U.; Young, K.; Gaucher, A.; De, S.; Smith, R.J.; et al. Two-Dimensional Nanosheets Produced by Liquid Exfoliation of Layered Materials. Science 2011, 331, 568–571. [Google Scholar] [CrossRef] [Green Version]
- Navani, N.K.; Mok, W.K.; Yingfu, L. In Vitro Selection of Protein-Binding DNA Aptamers as Ligands for Biosensing Applications. Methods Mol. Biol. 2009, 504, 399–415. [Google Scholar] [CrossRef] [PubMed]
- Johansson, M.K. Choosing Reporter-Quencher Pairs for Efficient Quenching Through Formation of Intramolecular Dimers. Fluoresc. Energy Transf. Nucleic Acid Probes 2006, 335, 17–30. [Google Scholar] [CrossRef]
- Wong, W.K.; Wong, S.H.D.; Bian, L. Long-Term Detection of Oncogenic MicroRNA in Living Human Cancer Cells by Gold@ Polydopamine–Shell Nanoprobe. ACS Biomater. Sci. Eng. 2020, 6, 3778–3783. [Google Scholar] [CrossRef] [PubMed]
- Lu, C.; Liu, Y.; Ying, Y.; Liu, J. Comparison of MoS2, WS2, and Graphene Oxide for DNA Adsorption and Sensing. Langmuir 2017, 33, 630–637. [Google Scholar] [CrossRef]
- Wang, L.; Dong, L.; Liu, G.; Shen, X.; Wang, J.; Zhu, C.; Ding, M.; Wen, Y. Fluorometric determination of HIV DNA using molybdenum disulfide nanosheets and exonuclease III-assisted amplification. Microchim. Acta 2019, 186, 286. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Y.; Zheng, B.; Zhu, C.; Zhang, X.; Tan, C.; Li, H.; Chen, B.; Yang, J.; Chen, J.; Wang, L.; et al. Single-Layer Transition Metal Dichalcogenide Nanosheet-Based Nanosensors for Rapid, Sensitive, and Multiplexed Detection of DNA. Adv. Mater. 2015, 27, 935–939. [Google Scholar] [CrossRef]
- Singhal, C.; Khanuja, M.; Chaudhary, N.; Pundir, C.S.; Narang, J. Detection of chikungunya virus DNA using two-dimensional MoS2 nanosheets based disposable biosensor. Sci. Rep. 2018, 8, 7734. [Google Scholar] [CrossRef]
- Huang, M.-B.; Xia, M.; Gao, Z.; Zhou, H.; Liu, M.; Huang, S.; Zhen, R.; Wu, J.Y.; Roth, W.W.; Bond, V.C.; et al. Characterization of Exosomes in Plasma of Patients with Breast, Ovarian, Prostate, Hepatic, Gastric, Colon, and Pancreatic Cancers. J. Cancer Ther. 2019, 10, 382–399. [Google Scholar] [CrossRef] [Green Version]
- Zhang, F.; Wang, S.; Feng, J.; Zou, R.; Xiang, L.; Cai, C. MoS2-loaded G-quadruplex molecular beacon probes for versatile detection of MicroRNA through hybridization chain reaction signal amplification. Talanta 2019, 202, 342–348. [Google Scholar] [CrossRef] [PubMed]
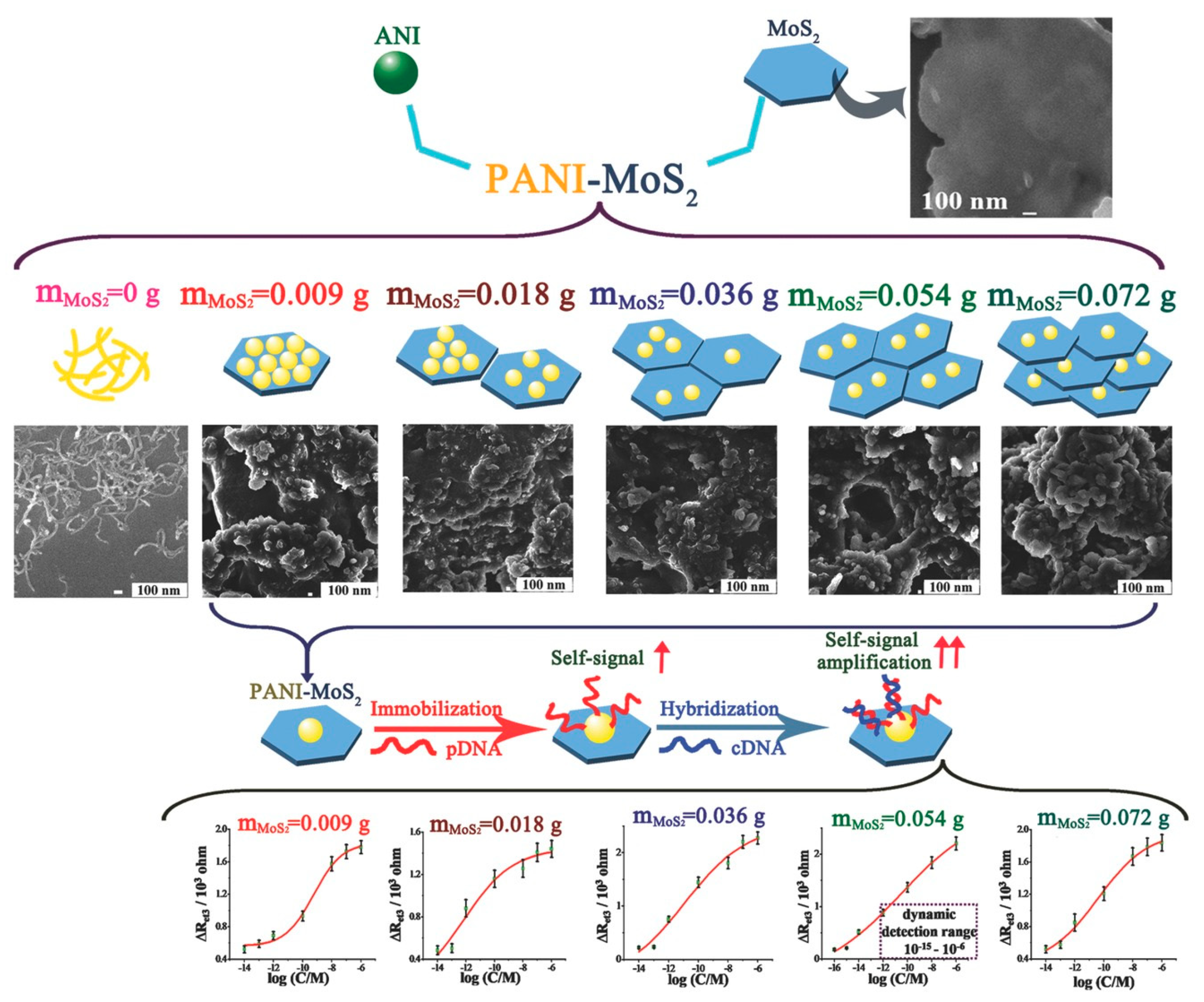
- Yang, T.; Meng, L.; Chen, H.; Luo, S.; Li, W.; Jiao, K. Synthesis of Thin-Layered Molybdenum Disulfide-Based Polyaniline Nanointerfaces for Enhanced Direct Electrochemical DNA Detection. Adv. Mater. Interfaces 2016, 3, 1500700. [Google Scholar] [CrossRef]
- Dutta, S.; Chowdhury, A.D.; Biswas, S.; Park, E.Y.; Agnihotri, N.; De, A.; De, S. Development of an effective electrochemical platform for highly sensitive DNA detection using MoS2—Polyaniline nanocomposites. Biochem. Eng. J. 2018, 140, 130–139. [Google Scholar] [CrossRef]
- Wang, T.; Zhu, R.; Zhuo, J.; Zhu, Z.; Shao, Y.; Li, M. Direct Detection of DNA below ppb Level Based on Thionin-Functionalized Layered MoS2Electrochemical Sensors. Anal. Chem. 2014, 86, 12064–12069. [Google Scholar] [CrossRef]
- Loan, P.T.K.; Zhang, W.; Lin, C.-T.; Wei, K.-H.; Li, L.-J.; Chen, C.-H. Graphene/MoS2Heterostructures for Ultrasensitive Detection of DNA Hybridisation. Adv. Mater. 2014, 26, 4838–4844. [Google Scholar] [CrossRef]
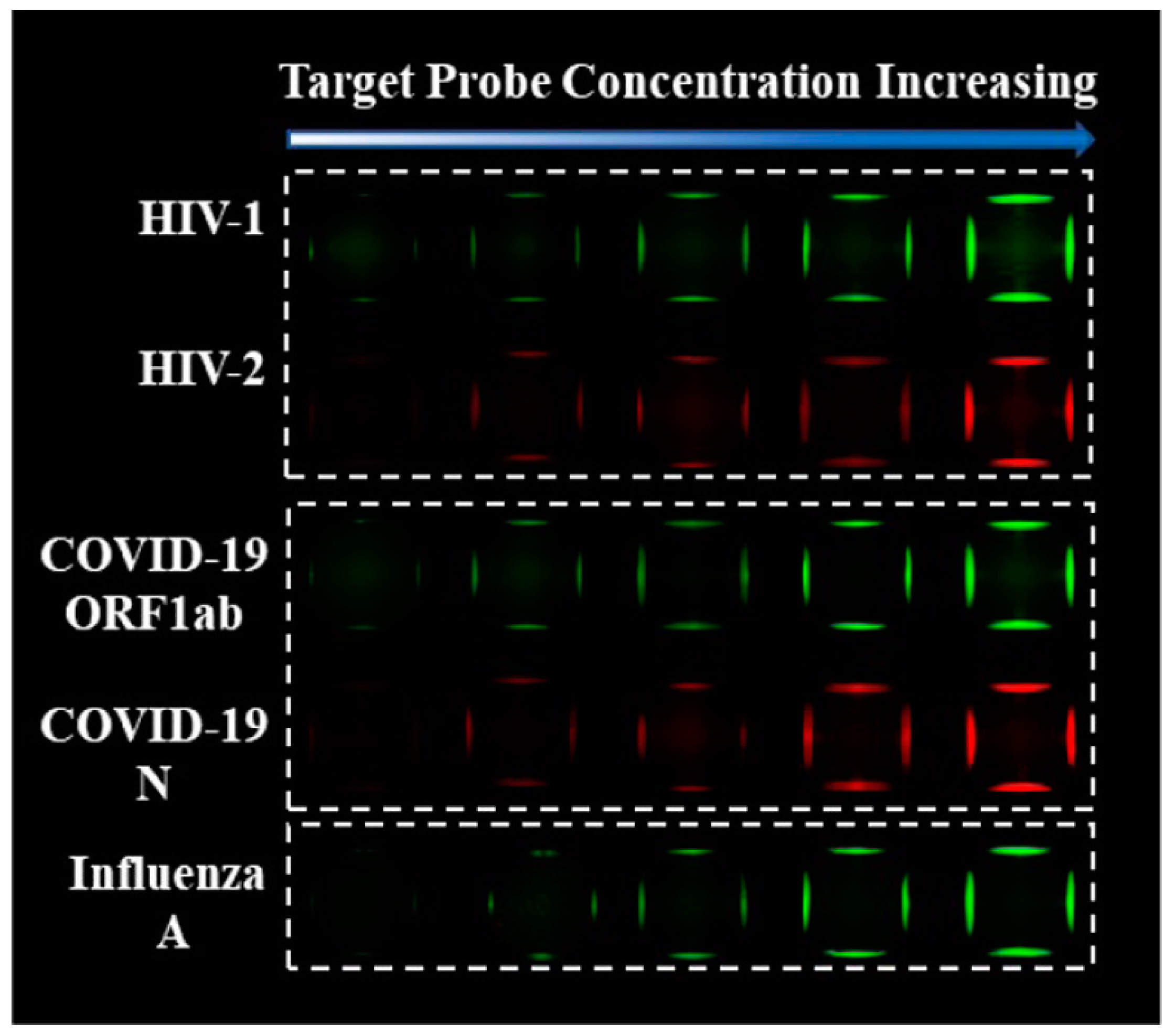
- Oudeng, G.; Benz, M.; Popova, A.A.; Zhang, Y.; Yi, C.; Levkin, P.A.; Yang, M. Droplet Microarray Based on Nanosensing Probe Patterns for Simultaneous Detection of Multiple HIV Retroviral Nucleic Acids. ACS Appl. Mater. Interfaces 2020, 12, 55614–55623. [Google Scholar] [CrossRef]
- Shariati, M.; Vaezjalali, M.; Sadeghi, M. Ultrasensitive and easily reproducible biosensor based on novel doped MoS2 nanowires field-effect transistor in label-free approach for detection of hepatitis B virus in blood serum. Anal. Chim. Acta 2021, 1156, 338360. [Google Scholar] [CrossRef] [PubMed]
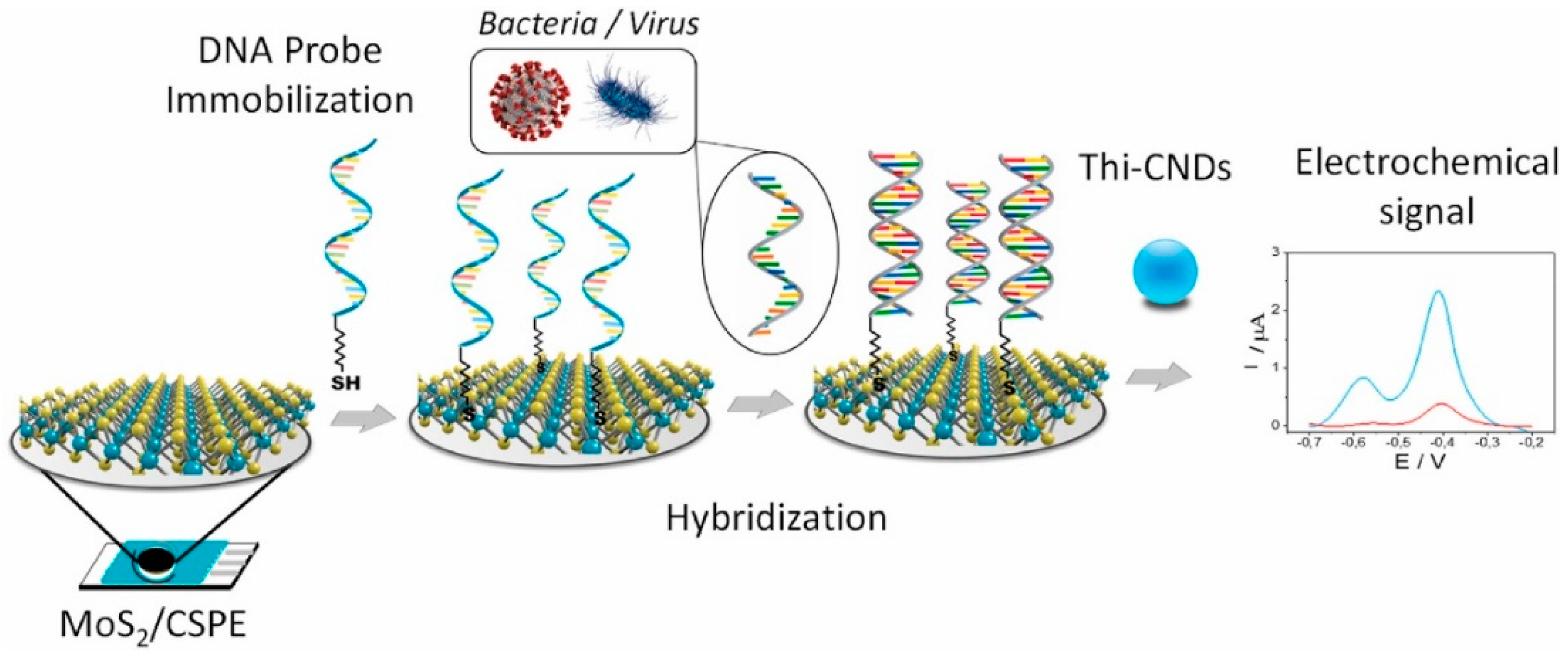
- Martínez-Periñán, E.; García-Mendiola, T.; Enebral-Romero, E.; del Caño, R.; Vera-Hidalgo, M.; Sulleiro, M.V.; Navío, C.; Pariente, F.; Pérez, E.M.; Lorenzo, E. A MoS2 platform and thionine-carbon nanodots for sensitive and selective detection of pathogens. Biosens. Bioelectron. 2021, 189, 113375. [Google Scholar] [CrossRef] [PubMed]
- Feng, T.; Chen, X.; Qiao, X.; Sun, Z.; Wang, H.; Qi, Y.; Hong, C. Graphene oxide supported rhombic dodecahedral Cu2O nanocrystals for the detection of carcinoembryonic antigen. Anal. Biochem. 2016, 494, 101–107. [Google Scholar] [CrossRef] [PubMed]
- Cho, I.-H.; Lee, J.; Kim, J.; Kang, M.-S.; Paik, J.K.; Ku, S.; Cho, H.-M.; Irudayaraj, J.; Kim, D.-H. Current Technologies of Electrochemical Immunosensors: Perspective on Signal Amplification. Sensors 2018, 18, 207. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Yin, B.; Wu, Y.; Ma, H.; Ma, X.; Fu, B.; Liu, J. Studies on the Asymmetric Catalytic Friedel-Crafts Alkylation of Indoles with Trifluoromethyl Pyruvate Catalyzed by Heteroarylidene-BOX-Cu Complexes. Chin. J. Org. Chem. 2015, 35, 2119–2124. [Google Scholar] [CrossRef]
- Wu, D.; Ma, H.; Zhang, Y.; Jia, H.; Yan, T.; Wei, Q. Corallite-like Magnetic Fe3O4@MnO2@Pt Nanocomposites as Multiple Signal Amplifiers for the Detection of Carcinoembryonic Antigen. ACS Appl. Mater. Interfaces 2015, 7, 18786–18793. [Google Scholar] [CrossRef] [PubMed]
- Huang, C.-L.; Weng, W.-L.; Huang, Y.-S.; Liao, C.-N. Enhanced photolysis stability of Cu2O grown on Cu nanowires with nanoscale twin boundaries. Nanoscale 2019, 11, 13709–13713. [Google Scholar] [CrossRef] [PubMed]
- Choi, J.W.; Yoon, J.; Lim, J.; Shin, M.; Lee, S.N. Graphene/MoS2 Nanohybrid for Biosensors. Materials 2021, 14, 518. [Google Scholar]
- Yin, B.; Ho, L.W.C.; Liu, S.; Hong, H.; Tian, X.Y.; Li, H.; Choi, C.H.J. Sub-10 nm Substrate Roughness Promotes the Cellular Uptake of Nanoparticles by Upregulating Endocytosis-Related Genes. Nano Lett. 2021, 21, 1839–1847. [Google Scholar] [CrossRef] [PubMed]
- Ho, L.W.C.; Yin, B.; Dai, G.; Choi, C.H.J. Effect of Surface Modification with Hydrocarbyl Groups on the Exocytosis of Nanoparticles. Biochemistry 2020, 60, 1019–1030. [Google Scholar] [CrossRef] [PubMed]
- Yang, H.; Yao, Y.; Li, H.; Ho, L.W.C.; Yin, B.; Yung, W.-Y.; Leung, K.C.-F.; Mak, A.F.-T.; Choi, C.H.J. Promoting intracellular delivery of sub-25 nm nanoparticles via defined levels of compression. Nanoscale 2018, 10, 15090–15102. [Google Scholar] [CrossRef]
- Wong, S.H.D.; Yin, B.; Yang, B.; Lin, S.; Li, R.; Feng, Q.; Yang, H.; Zhang, L.; Yang, Z.; Li, G.; et al. Anisotropic Nanoscale Presentation of Cell Adhesion Ligand Enhances the Recruitment of Diverse Integrins in Adhesion Structures and Mechanosensing-Dependent Differentiation of Stem Cells. Adv. Funct. Mater. 2019, 29, 1806822. [Google Scholar] [CrossRef]
- Yin, B.; Chan, C.K.W.; Liu, S.; Hong, H.; Wong, S.H.D.; Lee, L.K.C.; Ho, L.W.C.; Zhang, L.; Leung, K.C.-F.; Choi, P.C.-L.; et al. Intrapulmonary Cellular-Level Distribution of Inhaled Nanoparticles with Defined Functional Groups and Its Correlations with Protein Corona and Inflammatory Response. ACS Nano 2019, 13, 14048–14069. [Google Scholar] [CrossRef] [PubMed]
- Kang, H.; Wong, S.H.D.; Pan, Q.; Li, G.; Bian, L. Anisotropic Ligand Nanogeometry Modulates the Adhesion and Polarization State of Macrophages. Nano Lett. 2019, 19, 1963–1975. [Google Scholar] [CrossRef]
- Bei, H.P.; Yang, Y.; Zhang, Q.; Tian, Y.; Luo, X.; Yang, M.; Zhao, X. Graphene-Based Nanocomposites for Neural Tissue Engineering. Moleclues 2019, 24, 658. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ni, Y.; Zhu, J.; Zhang, L.; Hong, J. Hierarchical ZnO micro/nanoarchitectures: Hydrothermal preparation, characterization and application in the detection of hydrazine. CrystEngComm 2010, 12, 2213–2218. [Google Scholar] [CrossRef]
- Zhou, J.; Xu, N.S.; Wang, Z.L. Dissolving Behavior and Stability of ZnO Wires in Biofluids: A Study on Biodegradability and Biocompatibility of ZnO Nanostructures. Adv. Mater. 2006, 18, 2432–2435. [Google Scholar] [CrossRef]
- Guo, J.; Peng, C. Synthesis of ZnO nanoparticles with a novel combustion method and their C2H5OH gas sensing properties. Ceram. Int. 2015, 41, 2180–2186. [Google Scholar] [CrossRef]
- Fan, F.; Tang, P.; Wang, Y.; Feng, Y.; Chen, A.; Luo, R.; Li, D. Facile synthesis and gas sensing properties of tubular hierarchical ZnO self-assembled by porous nanosheets. Sens. Actuators B Chem. 2015, 215, 231–240. [Google Scholar] [CrossRef]
- Shinde, S.M.; Das, T.; Hoang, A.T.; Sharma, B.K.; Chen, X.; Ahn, J.-H. Surface-Functionalization-Mediated Direct Transfer of Molybdenum Disulfide for Large-Area Flexible Devices. Adv. Funct. Mater. 2018, 28, 1706231. [Google Scholar] [CrossRef]
- Zhu, X.; Liu, P.; Ge, Y.; Wu, R.; Xue, T.; Sheng, Y.; Ai, S.; Tang, K.; Wen, Y. MoS2/MWCNTs porous nanohybrid network with oxidase-like characteristic as electrochemical nanozyme sensor coupled with machine learning for intelligent analysis of carbendazim. J. Electroanal. Chem. 2020, 862, 113940. [Google Scholar] [CrossRef]
- Kwon, H.-J.; Kim, S.; Jang, J.; Grigoropoulos, C. Evaluation of pulsed laser annealing for flexible multilayer MoS2 transistors. Appl. Phys. Lett. 2015, 106, 113111. [Google Scholar] [CrossRef]
- Huang, Y.; Sutter, E.; Wu, L.M.; Xu, H.; Bao, L.; Gao, H.-J.; Zhou, X.-J.; Sutter, P. Thick Layered Semiconductor Devices with Water Top-Gates: High On–Off Ratio Field-Effect Transistors and Aqueous Sensors. ACS Appl. Mater. Interfaces 2018, 10, 23198–23207. [Google Scholar] [CrossRef] [PubMed]







| Protein Name | Product Name | Cat. No. | Antigens/Antibodies |
|---|---|---|---|
| N-protein | SARS-CoV-2 (2019-nCoV) Nucleocapsid Protein (His tag) | 40588-V08B | Antigens |
| S-protein | SARS-CoV-2 (2019-nCoV) Spike Protein (RBD, mFc Tag) | 40592-V05H | Antigens |
| S-protein | SARS-CoV-2 (2019-nCoV) Spike Protein (S1 + S2 ECD, His tag) | 40589-V08B1 | Antigens |
| S-protein | SARS-CoV-2 (2019-nCoV) Spike Protein (S1 Subunit, His Tag) | 40591-V08H | Antigens |
| S-protein | SARS-CoV-2 (2019-nCoV) Spike Protein (S2 ECD, His tag) | 40590-V08B | Antigens |
| N-protein | SARS-CoV-2 (2019-nCoV) Nucleoprotein/NP Antibody, Rabbit MAb | 40143-R019 | Antibodies |
| S-protein | SARS-CoV-2 (2019-nCoV) Spike Antibody, Rabbit Mab | 40150-R007 | Antibodies |
| S-protein | SARS-CoV Spike Antibody | 40150-D003 | Antibodies |
| Sequence Name | Nucleic Acid Sequence |
|---|---|
| N-protein | AUGUCUGAUAAUGGACCCCAAAAUCAGCGAAAUGCACCCCGCAUUACGUUUGGUGGACCCUCAGAUUCAACUGGCAGUAACCAGAAUGGAGAACGCAGUGGGGCGCGAUCAAAACAACGUCGGCCCCAAGGUUUACCCAAUAAUACUGCGUCUUGGUUCACCGCUCUCACUCAACAUGGCAAGGAAGACCUUAAAUUCCCUCGAGGACAAGGCGUUCCAAUUAACACCAAUAGCAGUCCAGAUGACCAAAUUGGCUACUACCGAAGAGCUACCAGACGAAUUCGUGGUGGUGACGGUAAAAUGAAAGAUCUCAGUCCAAGAUGGUAUUUCUACUACCUAGGAACUGGGCCAGAAGCUGGACUUCCCUAUGGUGCUAACAAAGACGGCAUCAUAUGGGUUGCAACUGAGGGAGCCUUGAAUACACCAAAAGAUCACAUUGGCACCCGCAAUCCUGCUAACAAUGCUGCAAUCGUGCUACAACUUCCUCAAGGAACAACAUUGCCAAAAGGCUUCUACGCAGAAGGGAGCAGAGGCGGCAGUCAAGCCUCUUCUCGUUCCUCAUCACGUAGUCGCAACAGUUCAAGAAAUUCAACUCCAGGCAGCAGUAGGGGAACUUCUCCUGCUAGAAUGGCUGGCAAUGGCGGUGAUGCUGCUCUUGCUUUGCUGCUGCUUGACAGAUUGAACCAGCUUGAGAGCAAAAUGUCUGGUAAAGGCCAACAACAACAAGGCCAAACUGUCACUAAGAAAUCUGCUGCUGAGGCUUCUAAGAAGCCUCGGCAAAAACGUACUGCCACUAAAGCAUACAAUGUAACACAAGCUUUCGGCAGACGUGGUCCAGAACAAACCCAAGGAAAUUUUGGGGACCAGGAACUAAUCAGACAAGGAACUGAUUACAAACAUUGGCCGCAAAUUGCACAAUUUGCCCCCAGCGCUUCAGCGUUCUUCGGAAUGUCGCGCAUUGGCAUGGAAGUCACACCUUCGGGAACGUGGUUGACCUACACAGGUGCCAUCAAAUUGGAUGACAAAGAUCCAAAUUUCAAAGAUCAAGUCAUUUUGCUGAAUAAGCAUAUUGACGCAUACAAAACAUUCCCACCAACAGAGCCUAAAAAGGACAAAAAGAAGAAGGCUGAUGAAACUCAAGCCUUACCGCAGAGACAGAAGAAACAGCAAACUGUGACUCUUCUUCCUGCUGCAGAUUUGGAUGAUUUCUCCAAACAAUUGCAACAAUCCAUGAGCAGUGCUGACUCAACUCAGGCCUAA |
| S-protein | AUGUUUGUUUUUCUUGUUUUAUUGCCACUAGUCUCUAGUCAGUGUGUUAAUCUUACAACCAGAACUCAAUUACCCCCUGCAUACACUAAUUCUUUCACACGUGGUGUUUAUUACCCUGACAAAGUUUUCAGAUCCUCAGUUUUACAUUCAACUCAGGACUUGUUCUUACCUUUCUUUUCCAAUGUUACUUGGUUCCAUGCUAUACAUGUCUCUGGGACCAAUGGUACUAAGAGGUUUGAUAACCCUGUCCUACCAUUUAAUGAUGGUGUUUAUUUUGCUUCCACUGAGAAGUCUAACAUAAUAAGAGGCUGGAUUUUUGGUACUACUUUAGAUUCGAAGACCCAGUCCCUACUUAUUGUUAAUAACGCUACUAAUGUUGUUAUUAAAGUCUGUGAAUUUCAAUUUUGUAAUGAUCCAUUUUUGGGUGUUUAUUACCACAAAAACAACAAAAGUUGGAUGGAAAGUGAGUUCAGAGUUUAUUCUAGUGCGAAUAAUUGCACUUUUGAAUAUGUCUCUCAGCCUUUUCUUAUGGACCUUGAAGGAAAACAGGGUAAUUUCAAAAAUCUUAGGGAAUUUGUGUUUAAGAAUAUUGAUGGUUAUUUUAAAAUAUAUUCUAAGCACACGCCUAUUAAUUUAGUGCGUGAUCUCCCUCAGGGUUUUUCGGCUUUAGAACCAUUGGUAGAUUUGCCAAUAGGUAUUAACAUCACUAGGUUUCAAACUUUACUUGCUUUACAUAGAAGUUAUUUGACUCCUGGUGAUUCUUCUUCAGGUUGGACAGCUGGUGCUGCAGCUUAUUAUGUGGGUUAUCUUCAACCUAGGACUUUUCUAUUAAAAUAUAAUGAAAAUGGAACCAUUACAGAUGCUGUAGACUGUGCACUUGACCCUCUCUCAGAAACAAAGUGUACGUUGAAAUCCUUCACUGUAGAAAAAGGAAUCUAUCAAACUUCUAACUUUAGAGUCCAACCAACAGAAUCUAUUGUUAGAUUUCCUAAUAUUACAAACUUGUGCCCUUUUGGUGAAGUUUUUAACGCCACCAGAUUUGCAUCUGUUUAUGCUUGGAACAGGAAGAGAAUCAGCAACUGUGUUGCUGAUUAUUCUGUCCUAUAUAAUUCCGCAUCAUUUUCCACUUUUAAGUGUUAUGGAGUGUCUCCUACUAAAUUAAAUGAUCUCUGCUUUACUAAUGUCUAUGCAGAUUCAUUUGUAAUUAGAGGUGAUGAAGUCAGACAAAUCGCUCCAGGGCAAACUGGAAAGAUUGCUGAUUAUAAUUAUAAAUUACCAGAUGAUUUUACAGGCUGCGUUAUAGCUUGGAAUUCUAACAAUCUUGAUUCUAAGGUUGGUGGUAAUUAUAAUUACCUGUAUAGAUUGUUUAGGAAGUCUAAUCUCAAACCUUUUGAGAGAGAUAUUUCAACUGAAAUCUAUCAGGCCGGUAGCACACCUUGUAAUGGUGUUGAAGGUUUUAAUUGUUACUUUCCUUUACAAUCAUAUGGUUUCCAACCCACUAAUGGUGUUGGUUACCAACCAUACAGAGUAGUAGUACUUUCUUUUGAACUUCUACAUGCACCAGCAACUGUUUGUGGACCUAAAAAGUCUACUAAUUUGGUUAAAAACAAAUGUGUCAAUUUCAACUUCAAUGGUUUAACAGGCACAGGUGUUCUUACUGAGUCUAACAAAAAGUUUCUGCCUUUCCAACAAUUUGGCAGAGACAUUGCUGACACUACUGAUGCUGUCCGUGAUCCACAGACACUUGAGAUUCUUGACAUUACACCAUGUUCUUUUGGUGGUGUCAGUGUUAUAACACCAGGAACAAAUACUUCUAACCAGGUUGCUGUUCUUUAUCAGGAUGUUAACUGCACAGAAGUCCCUGUUGCUAUUCAUGCAGAUCAACUUACUCCUACUUGGCGUGUUUAUUCUACAGGUUCUAAUGUUUUUCAAACACGUGCAGGCUGUUUAAUAGGGGCUGAACAUGUCAACAACUCAUAUGAGUGUGACAUACCCAUUGGUGCAGGUAUAUGCGCUAGUUAUCAGACUCAGACUAAUUCUCCUCGGCGGGCACGUAGUGUAGCUAGUCAAUCCAUCAUUGCCUACACUAUGUCACUUGGUGCAGAAAAUUCAGUUGCUUACUCUAAUAACUCUAUUGCCAUACCCACAAAUUUUACUAUUAGUGUUACCACAGAAAUUCUACCAGUGUCUAUGACCAAGACAUCAGUAGAUUGUACAAUGUACAUUUGUGGUGAUUCAACUGAAUGCAGCAAUCUUUUGUUGCAAUAUGGCAGUUUUUGUACACAAUUAAACCGUGCUUUAACUGGAAUAGCUGUUGAACAAGACAAAAACACCCAAGAAGUUUUUGCACAAGUCAAACAAAUUUACAAAACACCACCAAUUAAAGAUUUUGGUGGUUUUAAUUUUUCACAAAUAUUACCAGAUCCAUCAAAACCAAGCAAGAGGUCAUUUAUUGAAGAUCUACUUUUCAACAAAGUGACACUUGCAGAUGCUGGCUUCAUCAAACAAUAUGGUGAUUGCCUUGGUGAUAUUGCUGCUAGAGACCUCAUUUGUGCACAAAAGUUUAACGGCCUUACUGUUUUGCCACCUUUGCUCACAGAUGAAAUGAUUGCUCAAUACACUUCUGCACUGUUAGCGGGUACAAUCACUUCUGGUUGGACCUUUGGUGCAGGUGCUGCAUUACAAAUACCAUUUGCUAUGCAAAUGGCUUAUAGGUUUAAUGGUAUUGGAGUUACACAGAAUGUUCUCUAUGAGAACCAAAAAUUGAUUGCCAACCAAUUUAAUAGUGCUAUUGGCAAAAUUCAAGACUCACUUUCUUCCACAGCAAGUGCACUUGGAAAACUUCAAGAUGUGGUCAACCAAAAUGCACAAGCUUUAAACACGCUUGUUAAACAACUUAGCUCCAAUUUUGGUGCAAUUUCAAGUGUUUUAAAUGAUAUCCUUUCACGUCUUGACAAAGUUGAGGCUGAAGUGCAAAUUGAUAGGUUGAUCACAGGCAGACUUCAAAGUUUGCAGACAUAUGUGACUCAACAAUUAAUUAGAGCUGCAGAAAUCAGAGCUUCUGCUAAUCUUGCUGCUACUAAAAUGUCAGAGUGUGUACUUGGACAAUCAAAAAGAGUUGAUUUUUGUGGAAAGGGCUAUCAUCUUAUGUCCUUCCCUCAGUCAGCACCUCAUGGUGUAGUCUUCUUGCAUGUGACUUAUGUCCCUGCACAAGAAAAGAACUUCACAACUGCUCCUGCCAUUUGUCAUGAUGGAAAAGCACACUUUCCUCGUGAAGGUGUCUUUGUUUCAAAUGGCACACACUGGUUUGUAACACAAAGGAAUUUUUAUGAACCACAAAUCAUUACUACAGACAACACAUUUGUGUCUGGUAACUGUGAUGUUGUAAUAGGAAUUGUCAACAACACAGUUUAUGAUCCUUUGCAACCUGAAUUAGACUCAUUCAAGGAGGAGUUAGAUAAAUAUUUUAAGAAUCAUACAUCACCAGAUGUUGAUUUAGGUGACAUCUCUGGCAUUAAUGCUUCAGUUGUAAACAUUCAAAAAGAAAUUGACCGCCUCAAUGAGGUUGCCAAGAAUUUAAAUGAAUCUCUCAUCGAUCUCCAAGAACUUGGAAAGUAUGAGCAGUAUAUAAAAUGGCCAUGGUACAUUUGGCUAGGUUUUAUAGCUGGCUUGAUUGCCAUAGUAAUGGUGACAAUUAUGCUUUGCUGUAUGACCAGUUGCUGUAGUUGUCUCAAGGGCUGUUGUUCUUGUGGAUCCUGCUGCAAAUUUGAUGAAGACGACUCUGAGCCAGUGCUCAAAGGAGUCAAAUUACAUUACACAUAA |
| E-protein | AUGUACUCAUUCGUUUCGGAAGAGACAGGUACGUUAAUAGUUAAUAGCGUACUUCUUUUUCUUGCUUUCGUGGUAUUCUUGCUAGUUACACUAGCCAUCCUUACUGCGCUUCGAUUGUGUGCGUACUGCUGCAAUAUUGUUAACGUGAGUCUUGUAAAACCUUCUUUUUACGUUUACUCUCGUGUUAAAAAUCUGAAUUCUUCUAGAGUUCCUGAUCUUCUGGUCUAA |
| M-protein | AUGGCAGAUUCCAACGGUACUAUUACCGUUGAAGAGCUUAAAAAGCUCCUUGAACAAUGGAACCUAGUAAUAGGUUUCCUAUUCCUUACAUGGAUUUGUCUUCUACAAUUUGCCUAUGCCAACAGGAAUAGGUUUUUGUAUAUAAUUAAGUUAAUUUUCCUCUGGCUGUUAUGGCCAGUAACUUUAGCUUGUUUUGUGCUUGCUGCUGUUUACAGAAUAAAUUGGAUCACCGGUGGAAUUGCUAUCGCAAUGGCUUGUCUUGUAGGCUUGAUGUGGCUCAGCUACUUCAUUGCUUCUUUCAGACUGUUUGCGCGUACGCGUUCCAUGUGGUCAUUCAAUCCAGAAACUAACAUUCUUCUCAACGUGCCACUCCAUGGCACUAUUCUGACCAGACCGCUUCUAGAAAGUGAACUCGUAAUCGGAGCUGUGAUCCUUCGUGGACAUCUUCGUAUUGCUGGACACCAUCUAGGACGCUGUGACAUCAAGGACCUGCCUAAAGAAAUCACUGUUGCUACAUCACGAACGCUUUCUUAUUACAAAUUGGGAGCUUCGCAGCGUGUAGCAGGUGACUCAGGUUUUGCUGCAUACAGUCGCUACAGGAUUGGCAACUAUAAAUUAAACACAGACCAUUCCAGUAGCAGUGACAAUAUUGCUUUGCUUGUACAGUAA |
| ORF1ab | CCCTGTGGGTTTTACACTTAAAAACACAGTCTGTACCGTCTGCGGTATGTGGAAAGGTTATGGCTGTAGTTGTGATCAACTCCGCGAACCCATGCTTCAGTCAGCTGATGCACAATCGT |
| ORF3 | AUGGAUUUGUUUAUGAGAAUCUUCACAAUUGGAACUGUAACUUUGAAGCAAGGUGAAAUCAAGGAUGCUACUCCUUCAGAUUUUGUUCGCGCUACUGCAACGAUACCGAUACAAGCCUCACUCCCUUUCGGAUGGCUUAUUGUUGGCGUUGCACUUCUUGCUGUUUUUCAGAGCGCUUCCAAAAUCAUAACCCUCAAAAAGAGAUGGCAACUAGCACUCUCCAAGGGUGUUCACUUUGUUUGCAACUUGCUGUUGUUGUUUGUAACAGUUUACUCACACCUUUUGCUCGUUGCUGCUGGCCUUGAAGCCCCUUUUCUCUAUCUUUAUGCUUUAGUCUACUUCUUGCAGAGUAUAAACUUUGUAAGAAUAAUAAUGAGGCUUUGGCUUUGCUGGAAAUGCCGUUCCAAAAACCCAUUACUUUAUGAUGCCAACUAUUUUCUUUGCUGGCAUACUAAUUGUUACGACUAUUGUAUACCUUACAAUAGUGUAACUUCUUCAAUUGUCAUUACUUCAGGUGAUGGCACAACAAGUCCUAUUUCUGAACAUGACUACCAGAUUGGUGGUUAUACUGAAAAAUGGGAAUCUGGAGUAAAAGACUGUGUUGUAUUACACAGUUACUUCACUUCAGACUAUUACCAGCUGUACUCAACUCAAUUGAGUACAGACACUGGUGUUGAACAUGUUACCUUCUUCAUCUACAAUAAAAUUGUUGAUGAGCCUGAAGAACAUGUCCAAAUUCACACAAUCGACGGUUCAUCCGGAGUUGUUAAUCCAGUAAUGGAACCAAUUUAUGAUGAACCGACGACGACUACUAGCGUGCCUUUGUAA |
| ORF6 | UUAAUCAAUCUCCAUUGGUUGCUCUUCAUCUAAUUGAGAAUAUUUAUUCUCAGUUAGUGACUUAGAUAAAUUUUUAAUUAUGAGGUUUAUGAUGUAAUCAAGAUUCCAAAUGGAAACUUUAAAAGUCCUCAUAAUAAUUAGUAAUAUCUCUGCUAUAGUAACCUGAAAGUCAACGAGAUGAAACAU |
| ORF7 | AUGAAAAUUAUUCUUUUCUUGGCACUGAUAACACUCGCUACUUGUGAGCUUUAUCACUACCAAGAGUGUGUUAGAGGUACAACAGUACUUUUAAAAGAACCUUGCUCUUCUGGAACAUACGAGGGCAAUUCACCAUUUCAUCCUCUAGCUGAUAACAAAUUUGCACUGACUUGCUUUAGCACUCAAUUUGCUUUUGCUUGUCCUGACGGCGUAAAACACGUCUAUCAGUUACGUGCCAGAUCAGUUUCACCUAAACUGUUCAUCAGACAAGAGGAAGUUCAAGAACUUUACUCUCCAAUUUUUCUUAUUGUUGCGGCAAUAGUGUUUAUAACACUUUGCUUCACACUCAAAAGAAAGACAGAAUGA |
| ORF8 | AUGAAAUUUCUUGUUUUCUUAGGAAUCAUCACAACUGUAGCUGCAUUUCACCAAGAAUGUAGUUUACAGUCAUGUACUCAACAUCAACCAUAUGUAGUUGAUGACCCGUGUCCUAUUCACUUCUAUUCUAAAUGGUAUAUUAGAGUAGGAGCUAGAAAAUCAGCACCUUUAAUUGAAUUGUGCGUGGAUGAGGCUGGUUCUAAAUCACCCAUUCAGUACAUCGAUAUCGGUAAUUAUACAGUUUCCUGUUUACCUUUUACAAUUAAUUGCCAGGAACCUAAAUUGGGUAGUCUUGUAGUGCGUUGUUCGUUCUAUGAAGACUUUUUAGAGUAUCAUGACGUUCGUGUUGUUUUAGAUUUCAUCUAA |
| Materials | Mechanism | Analytes | Detection Limit Range | LOD | Response Time | Ref. & Year |
|---|---|---|---|---|---|---|
| Flexible MoS2 sheets | FET | Ebola VP40 | fM–pM level | fM level | 15 min | [83] 2019 |
| DNA-MoS2 nanosheets | Fluorescence | Thrombin | 0.5–100 nM | 300 pM | 10 min | [84] 2014 |
| Flexible MoS2 sheets | FET | Prostate cancer antigens | 1 pg mL−1–1 μg mL−1 | 1 pg mL−1 | ~real-time | [85] 2017 |
| Few-layered MoS2 sheets | FET | Prostate cancer antigens | 10–5–75 ng mL−1 | 10–5 ng mL−1 | 20–30 min | [86] 2016 |
| Multilayer MoS2 | FET | MMP-9 | 1 pM–10 nM | 1 pM | 2 h (incubation time) | [87] 2019 |
| Graphene quantum dots/MoS2 nanosheets | Fluorescence | Epithelial cell adhesion molecules | 3–54 nM | 450 pM | 2 h (incubation time) | [88] 2017 |
| Au/MoS2/Au multilayer | Electrochemical | HIV gp120 | 0.1 pg mL−1–10 ng mL−1 | 0.066 pg mL−1 | N/A | [89] 2019 |
| MoS2@Cu2O-Pt | Electrochemical | Hepatitis B surface antigen | 0.5 pg mL−1–200 ng mL−1 | 0.15 pg mL−1 | 1 h (incubation time) | [90] 2018 |
| Au@Pd/MoS2@ multiwalled carbon nanotubes | Electrochemical | Hepatitis B e antigen | 0.1 pg mL−1–500 pg mL−1 | 26 fg mL−1 | 1.5 h (total incubation time) | [91] 2018 |
| MoS2-rGO | Electrochemical | Vi polysaccharide antigen | 0.1–1000 ng mL−1 | 100 pg mL−1 | 30 min (incubation time) | [92] 2018 |
| MoS2-AuNP/ITO | Electrochemical | Dengue NS1 antigen | 0.04–2 μg mL−1 in primary infection; 0.01–2 μg mL−1 in secondary infection | 1.67 ng mL−1 for standard; 1.19 ng mL−1 for spiked samples | ~25 min | [93] 2018 |
| MoS2 sheets | FET | Kinase Cdk5/p25 | pM–µM level | 38 pM | ~5 min | [94] 2019 |
| Ag/MoS2/rGO | Electrochemical | Carcinoembryonic antigen | 0.001–80 ng mL−1 | 0.3 pg mL−1 | 1 h (incubation time) | [95] 2020 |
| Tellurene/MoS2 Nanosheets | Optical/SPR | S protein or SARS-CoV-2 specimen | 0–301.67 nM for S protein; 0–67.8762 nM for SARS-CoV-2 specimen | Phase sensitivity: 8.4069 × 104 degree/RIU (nbio = 0.0012) | N/A | [96] 2020 |
| Aptamer- cellulose acetate-MoS2 | Electrochemical | Troponin I | 10 fM–1 nM | 10 fM | N/A | [97] 2021 |
| Materials | Mechanism | Analytes | Linear Range | LOD | Ref. & Year |
|---|---|---|---|---|---|
| MoS2 sheets | Electrochemical | Chikungunya virus DNA | 0.1 nM–100 µM | 3.4 nM | [107] 2018 |
| AuNP/MoS2 sheets | FET | Fetal cell-free DNA fragments | 100 aM–1 fM | 100 aM | [108] 2019 |
| MoS2/THT-MB | Fluorescence | miRNA | 0.1 nM–100 nM | 5.9 pM | [109] 2019 |
| FAM-labelled ssDNA MoS2, TiS2, and TaS2 nanosheets | Fluorescence | Influenza A virus (H1N1 and H5N1) | 0–5 nM | 0.2, 0.1, and 0.05 nM, respectively | [106] 2015 |
| PANI-MoS2 | Electrochemical | Cauliflower mosaic virus 35S | 1 fM–1 µM | 2 fM | [110] 2016 |
| PANI-MoS2-Pt | Electrochemical | Calf-thymus DNA | 1 fM–1 µM | 1 fM | [111] 2018 |
| MoS2-thionin | Electrochemical | dsDNA | 0.09 ng mL−1–1.9 ng mL−1 | 0.09 ng mL−1 | [112] 2014 |
| MoS2/graphene film | Electrical/optical | Oligonucleotides | 1 aM–1 fM (non-linear) | 1 aM | [113] 2014 |
| MoS2 nanosheet- modified dendrimer droplet microarray | Fluorescence | HIV-1, HIV-2, ORF1ab, and N protein gene | N/A | 50 pM | [114] 2020 |
| ZnO (NPs) doped MoS2 | FET | Hepatitis B virus | 0.5 pM–50 µM | 1 fM | [115] 2021 |
| MoS2- thionine-carbon nanodots | Electrochemical | InIA gen of Listeria and ORF1ab of SARS-CoV-2 | 100 fM to 50 nM and 1 pM to 1 nM, respectively | 67.0 fM and 1.01 pM, respectively | [116] 2021 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Lam, C.Y.K.; Zhang, Q.; Yin, B.; Huang, Y.; Wang, H.; Yang, M.; Wong, S.H.D. Recent Advances in Two-Dimensional Transition Metal Dichalcogenide Nanocomposites Biosensors for Virus Detection before and during COVID-19 Outbreak. J. Compos. Sci. 2021, 5, 190. https://doi.org/10.3390/jcs5070190
Lam CYK, Zhang Q, Yin B, Huang Y, Wang H, Yang M, Wong SHD. Recent Advances in Two-Dimensional Transition Metal Dichalcogenide Nanocomposites Biosensors for Virus Detection before and during COVID-19 Outbreak. Journal of Composites Science. 2021; 5(7):190. https://doi.org/10.3390/jcs5070190
Chicago/Turabian StyleLam, Ching Ying Katherine, Qin Zhang, Bohan Yin, Yingying Huang, Hui Wang, Mo Yang, and Siu Hong Dexter Wong. 2021. "Recent Advances in Two-Dimensional Transition Metal Dichalcogenide Nanocomposites Biosensors for Virus Detection before and during COVID-19 Outbreak" Journal of Composites Science 5, no. 7: 190. https://doi.org/10.3390/jcs5070190
APA StyleLam, C. Y. K., Zhang, Q., Yin, B., Huang, Y., Wang, H., Yang, M., & Wong, S. H. D. (2021). Recent Advances in Two-Dimensional Transition Metal Dichalcogenide Nanocomposites Biosensors for Virus Detection before and during COVID-19 Outbreak. Journal of Composites Science, 5(7), 190. https://doi.org/10.3390/jcs5070190








