1. Introduction
Nothofagus alessandrii Espinosa (ruil) is an endangered species that is endemic to the Mediterranean area of Chile. Since the beginning of the 20th century, its habitat has been reduced because of the expansion of the agricultural frontier with wheat crops and, since the 1970s, the substitution of forest plantations with non-native species. The current area of
N. alessandrii forest is approximately 314 ha [
1]. In addition, these forests have recently been affected by forest fires of great magnitude and intensity, and persistent drought conditions [
2].
The use of unmanned aerial vehicles (UAVs) has allowed for its application to various activities, including those dedicated to forest resources. Recently, data sets based on unmanned aerial vehicles (UAVs) have been found to be quite useful for identification of forest features due to their relatively high spatial resolution [
3]. Numerous studies have demonstrated the potential of UAVs for sustainable forest planning, volume estimation, pest infestation detection, tree counting, forest density determination, and canopy height assessment [
4]. UAVs are being used in several countries to control natural vegetation based on information in the optical and infrared spectra to spatial resolutions of 5 cm [
5]. The imagery acquired with a UAV reaches sub-decimeter or even centimeter resolution, often referred to as hyperspatial imagery, at flying height of 50 m and 18 mm focal length [
6]. UAV imagery can be captured on demand, enabling frequent imagery acquisition and efficient monitoring, known as hypertemporal imagery [
6]. UAVs are also being used to monitor the state of drought in forests and natural areas to prevent fires [
7]. Koh and Wich [
8] ran an application to map forest areas, where a UAV was used to map tropical forests in Indonesia. The authors suggested that the use of UAV remote sensing could save time, cost, and manpower for these purposes. The number of trees or the composition of stands are important parameters in sustainable forest planning and management [
9]. Fast and accurate determination of canopy cover can be achieved using UAVs [
10], leading to decisions that improve optimal stand quality and productivity. For example, Hassaan et al. [
11] used a UAV to count trees in urban areas and identified trees with the accuracy of 72%. Likewise, Wallace et al. [
12] successfully detected the number of trees using LiDAR (Light Detection And Ranging) sensors mounted on a UAV.
Moreover, UAVs have been combined with Geographic Information Systems (GIS) to gather data on the Earth’s surface and atmosphere. GIS data provide spatial information on Earth’s features, along with their attributes and spatial relationships, and the integration of machine learning techniques in GIS analysis has shown promise in enhancing the speed, accuracy, automation, and repeatability of data processing [
13].
Machine learning, as a subfield of Artificial Intelligence (AI), holds significant potential for addressing complex spatial problems within Geographic Information Sciences [
14]. Machine learning algorithms allow systems to learn from data, generating data-based predictions by identifying patterns in historical data and applying them for future predictions [
15].
Supervised learning, a form of machine learning, involves training models on labeled data and testing them on unlabeled data, making it well suited for classification problems. To address the need for robust species identification, we evaluate three classification algorithms in this research: Maximum Likelihood, Random Forest, and Support Vector Machine (SVM).
Random Forest, as proposed by Breiman [
16], is a powerful ensemble learning technique that has gained popularity due to its versatility and minimal parameter-tuning requirements, making it suitable for a wide range of prediction problems. RF leverages the collective decision making of multiple decision trees, each trained on a random subset of predictor variables. This approach yields highly accurate results and has demonstrated exceptional performance in numerous ecological and remote sensing applications.
Support Vector Machine, on the other hand, is a set of machine learning algorithms renowned for its effectiveness in data analysis [
17]. SVM offers advantages such as fine-grained control over error frequencies, decision rule transparency, and computational efficiency [
18]. SVM’s ability to achieve remarkable results with limited training samples makes it particularly relevant for species identification from orthophotos [
19].
Maximum Likelihood, often referred to as ML, is a classical classification method that estimates membership probabilities for each class and assigns a pixel to the class with the highest probability [
20]. ML is grounded in two fundamental principles: the assumption of normal distribution within each class in a multidimensional feature space and the application of Bayes’ theorem for decision making [
21].
To assess the classification accuracy of these three algorithms (RF, SVM, and ML), we conducted an accuracy assessment. This step involves comparing predicted classification results with reference data and provides valuable insights into the reliability of the results [
22]. The metrics we employ include the Kappa coefficient, user accuracy, producer accuracy, and F1 score, which collectively offer a comprehensive evaluation of the classification performance. These metrics illuminate the strengths and limitations of each classification method, helping to determine the most suitable approach for species identification in orthophotos.
Our selection of these algorithms is based on their popularity in solving classification problems, their ability to enhance weaker methods, and their widespread application in ecology, including neighborhood models [
23,
24,
25].
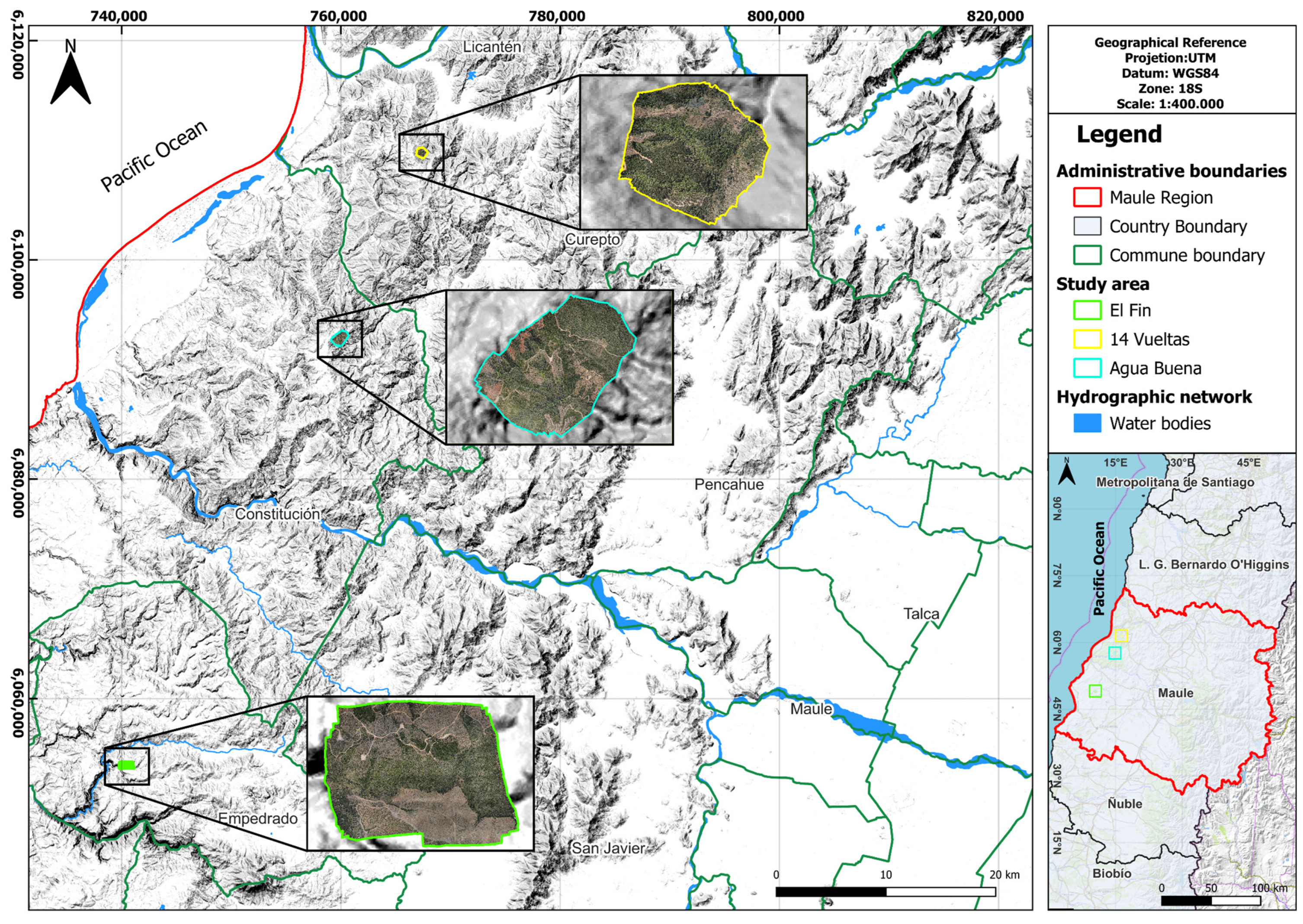
In contrast to the broad utilization of UAVs in our country, their application in forestry research, particularly in information-processing studies with machine learning tools, remains limited. Therefore, this study aims to evaluate the effectiveness of a UAV-based approach combined with machine learning algorithms in accurately identifying and classifying the distribution of Nothofagus alessandrii.
Overall, this study contributes to the growing body of research aimed at enhancing the accuracy and reliability of species identification in remote sensing applications, with the potential to support biodiversity conservation and ecological research efforts.
4. Discussion
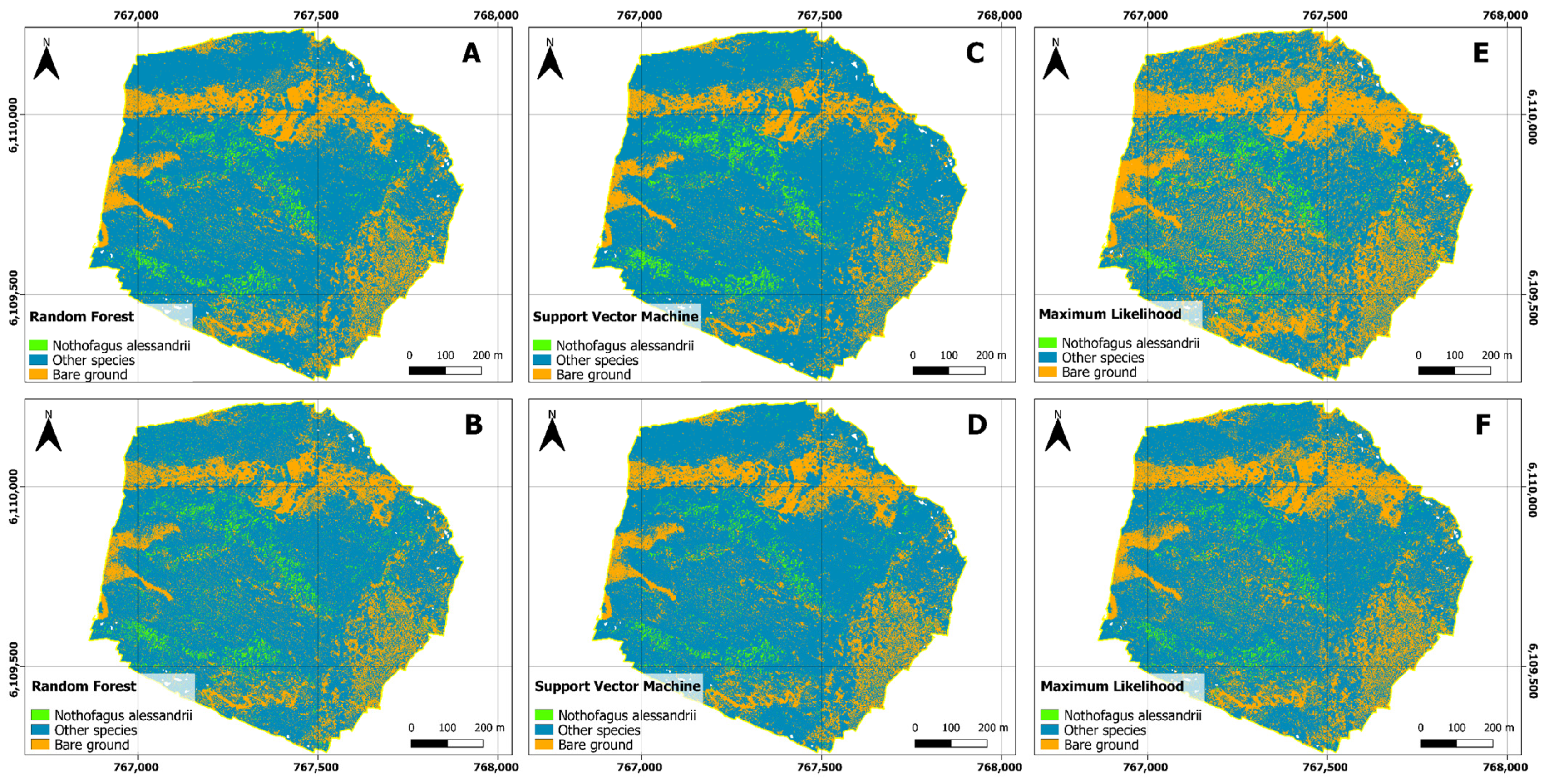
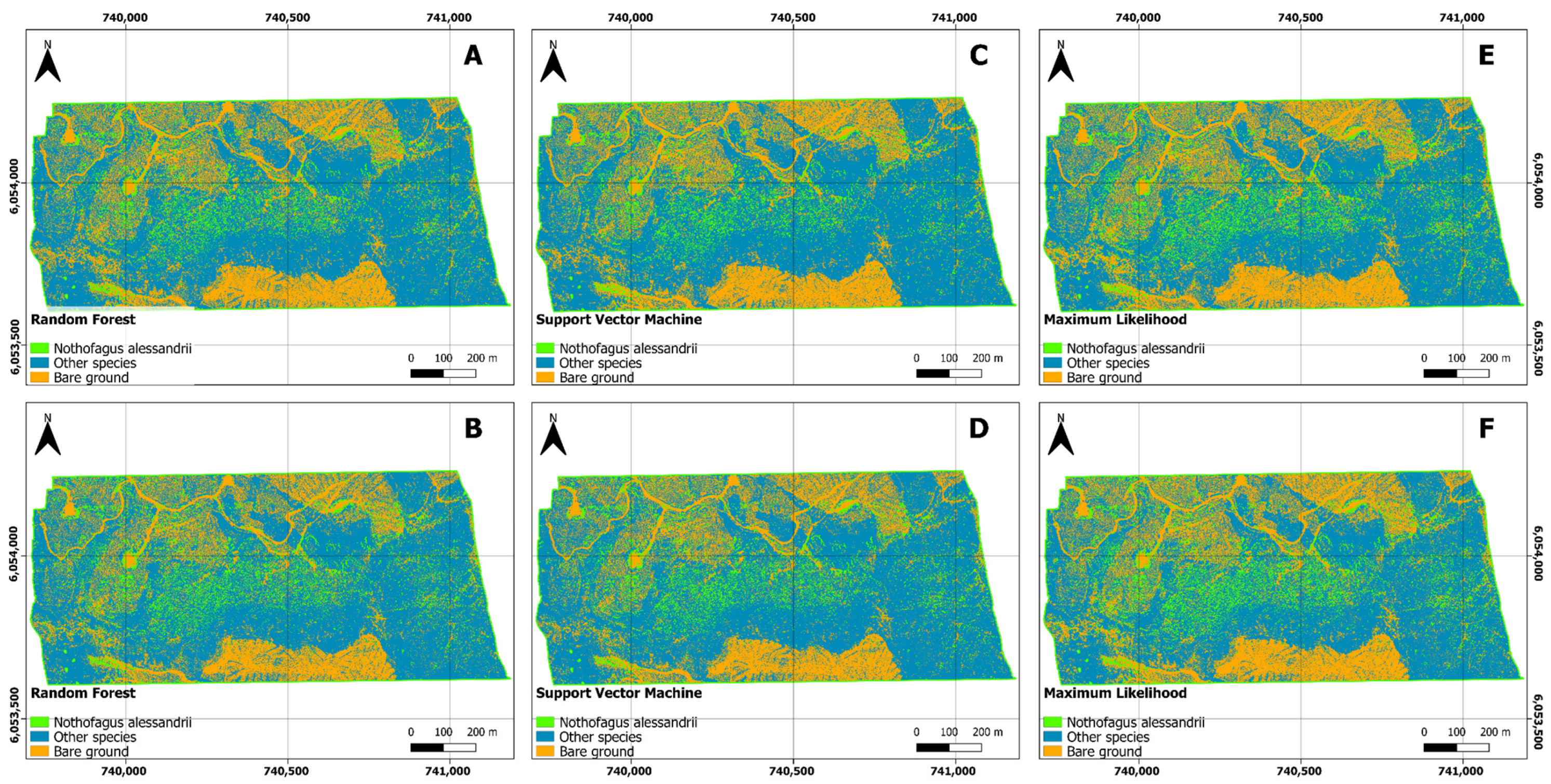
The results of our study demonstrate the effectiveness of three different classification algorithms, namely, Random Forest (RF), Support Vector Machine (SVM), and Maximum Likelihood (ML), in classifying land cover categories across three distinct study areas, including “14 Vueltas”, “Agua Buena”, and “El Fin”. To assess the impact of the training methodology on the classification outcomes, we employed both Object-Based and Pixel-Based training approaches.
RF consistently exhibited high accuracy across all study areas and training approaches, with user accuracy exceeding 88%, producer accuracy over 88%, and F-scores above 0.88. These high values confirm RF’s robustness and versatility in remote sensing applications, as supported by previous research findings [
40]. Additionally, the Kappa coefficient values consistently indicated substantial agreement between RF classification results and the actual ground truth, reaffirming its classification accuracy [
41].
Similarly, SVM displayed strong classification performance, consistently achieving user accuracy of over 85%, producer accuracy exceeding 84%, and F-scores above 0.88 in most cases [
42]. SVM’s ability to identify optimal decision boundaries in complex feature spaces [
43] contributed to its effectiveness in classifying land cover categories. In certain instances, SVM outperformed RF by achieving fewer false positives, suggesting a more conservative classification approach.
ML also demonstrated competitive classification performance, with user accuracy consistently above 85%, producer accuracy exceeding 84%, and F-scores above 0.87. Leveraging statistical probability, ML effectively discriminated among land cover classes and performed comparably to SVM in terms of false positives and false negatives.
These findings underscore the significant impact of algorithm and training approach choices on classification outcomes. Notably, the SVM algorithm with Pixel-Based training consistently produced larger surface areas for designated classes across all three study areas, while RF with Object-Based training generally resulted in smaller surface areas. These variations emphasize the need for thoughtful selection of algorithms and training approaches, as they influence both the classification outcome and the delineation of vegetation classes [
44].
Our results are consistent with Adugna et al.’s [
45]. In their study, the Random Forest (RF) model outperformed Support Vector Machine (SVM) in accurately classifying four distinct land cover types (built-up, forest, herbaceous vegetation, and shrub). Importantly, both algorithms demonstrated nearly identical performance in distinguishing between two classes, namely, bare/sparse vegetation and water bodies, when these classes exhibited distinct spectral characteristics. However, RF showed superior effectiveness when dealing with classes consisting of mixed pixels, including the aforementioned four categories. It is noteworthy to mention SVM’s susceptibility to mixed pixels and inaccurately labeled training samples, which makes it more sensitive to noisy data compared with other classification algorithms [
46].
Additionally, our findings align with Sheykhmousa et al.’s [
47], who assessed classification accuracy for various study targets. They reported that RF achieved average accuracy of approximately 95.5% in land use and land cover (LULC) classification and about 93.5% in change detection using SVM classification. LULC classification, a common application for both SVM and RF, showed less variability for the RF classifier, indicating higher stability compared with SVM in classification tasks, including crop classification.
Moreover, in a related study by Yang et al. [
48], the effectiveness of Random Forest (RF) and Support Vector Machine (SVM) in land cover classification was emphasized. Yang et al. [
48] highlighted RF’s robustness and SVM’s ability to handle complex feature spaces, in line with our observations. They also pointed out that compared with Pixel-Based (PB) classification, the Object-Based image analysis (OBIA) method, as Yang et al. [
48] indicated, can extract features of each element of remote sensing images, providing certain advantages.
In the context of confusion matrices, all three algorithms (RF, SVM, and ML) demonstrated strong performance in classifying classes across the study areas, with correct classification results, and minimal false positives and false negatives. Nevertheless, variations in the number of false positives and false negatives were observed among the algorithms in specific scenarios. RF appeared to exhibit a more balanced distribution between false positives and false negatives, while SVM and ML tended to have fewer false positives in particular cases.
The choice of classification algorithm (RF, SVM, or ML) should be based on the specific requirements of the study and the weighting of false positives and false negatives in the application. Each algorithm has its advantages and disadvantages, necessitating careful consideration of project objectives and needs.
In order to perceive the quality of the classification, accuracy assessment is inevitable [
32]. Carrying out a simple accuracy assessment, using overall accuracy (OA) and Kappa coefficient of agreement (K), with the inclusion of ground truth data, might be the most common and reliable approach to reporting the accuracy of thematic maps. These accuracy measures make classification algorithms comparable when independent training and validation data are incorporated into the classification scheme [
47]. In this regard, all three classification algorithms (RF, SVM, and ML) demonstrated robust performance across different study areas and training approaches. Minor differences in performance metrics among the algorithms highlighted their effectiveness in land cover classification tasks. Variations in performance may be attributed to the study areas’ complexity and the distribution of land cover classes. Researchers and practitioners can confidently choose any of these algorithms based on their specific project requirements, as they all offer reliable and consistent classification results.
These findings hold significant implications for land cover classification in remote sensing applications. Researchers and practitioners can confidently select any of the three algorithms (RF, SVM, or ML) based on their specific requirements and available resources, as all three demonstrated strong performance. Additionally, the choice between Object-Based and Pixel-Based training approaches can be made without compromising classification accuracy, offering flexibility in methodological decisions.
However, it is essential to acknowledge some limitations of this study. Firstly, the study areas were limited to three specific regions, and the findings may not generalize to other geographic contexts. Additionally, other factors, such as feature selection and preprocessing methods, could influence classification performance and warrant further investigation [
49]. Future research could explore the integration of additional machine learning algorithms and advanced feature engineering techniques to improve classification accuracy [
50]. Moreover, assessing the scalability of these methods to larger study areas and their performance under different environmental conditions should be considered.
Furthermore, it is imperative to recognize the critical role of remote sensing in the conservation efforts of endangered species like Nothofagus alessandrii. Given its critically endangered status, the detection and monitoring of Nothofagus alessandrii using remote sensing sensors can provide vital information for its preservation and contribute to the broader understanding of ecosystem conservation.