Monitoring Maize Leaf Spot Disease Using Multi-Source UAV Imagery
Abstract
:1. Introduction
- (1)
- To explore the spectral changes of maize canopy at different stages of disease development.
- (2)
- To find optimal multi-source data features and classifier for identifying maize leaf spot disease incidence.
- (3)
- To explore the possibility of maize leaf spot monitoring at the early stage with UAV data.
2. Materials and Methods
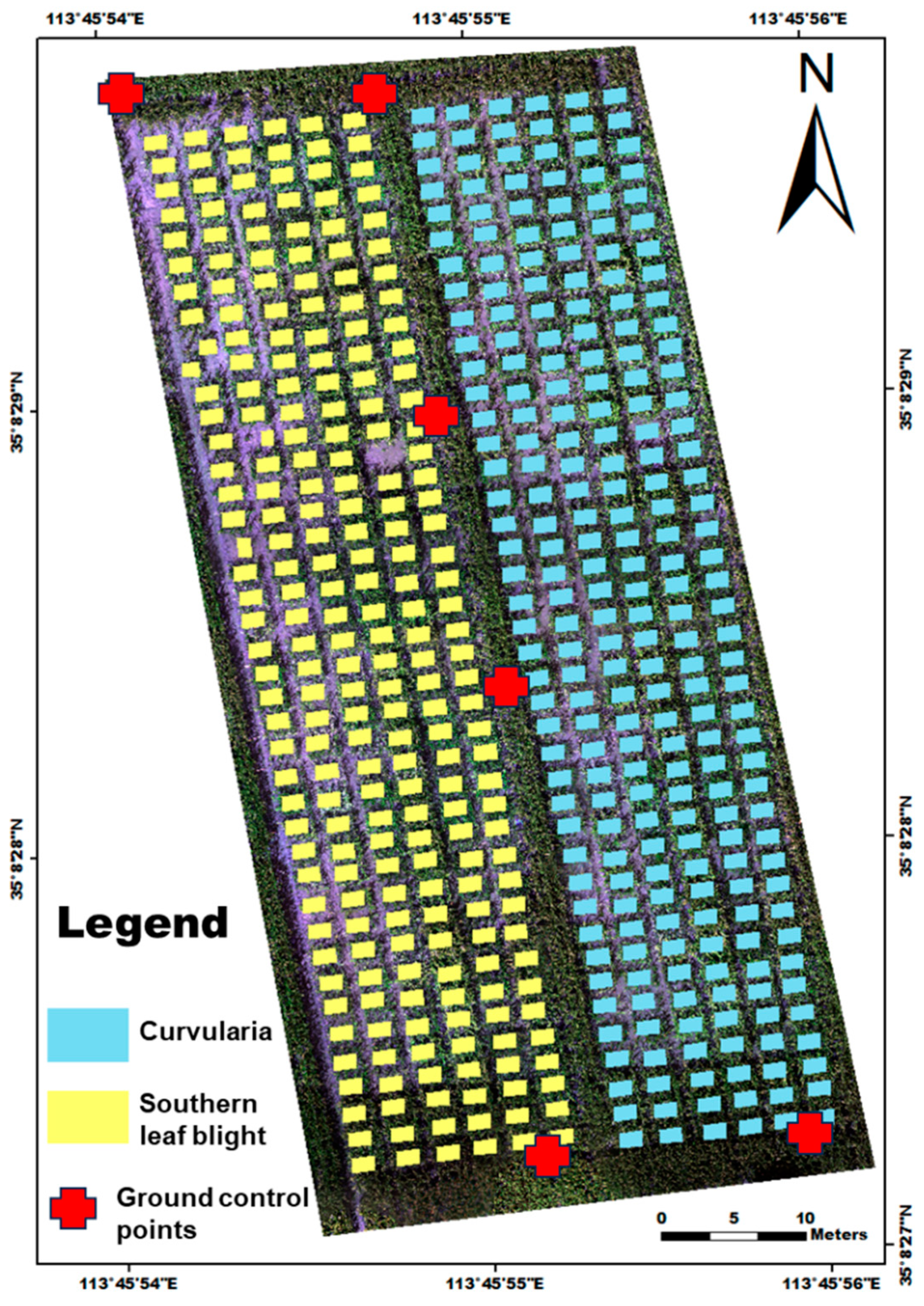
2.1. Study Area and Field Experiment
2.2. Data Acquisition
2.2.1. UAV Data
2.2.2. Field Data
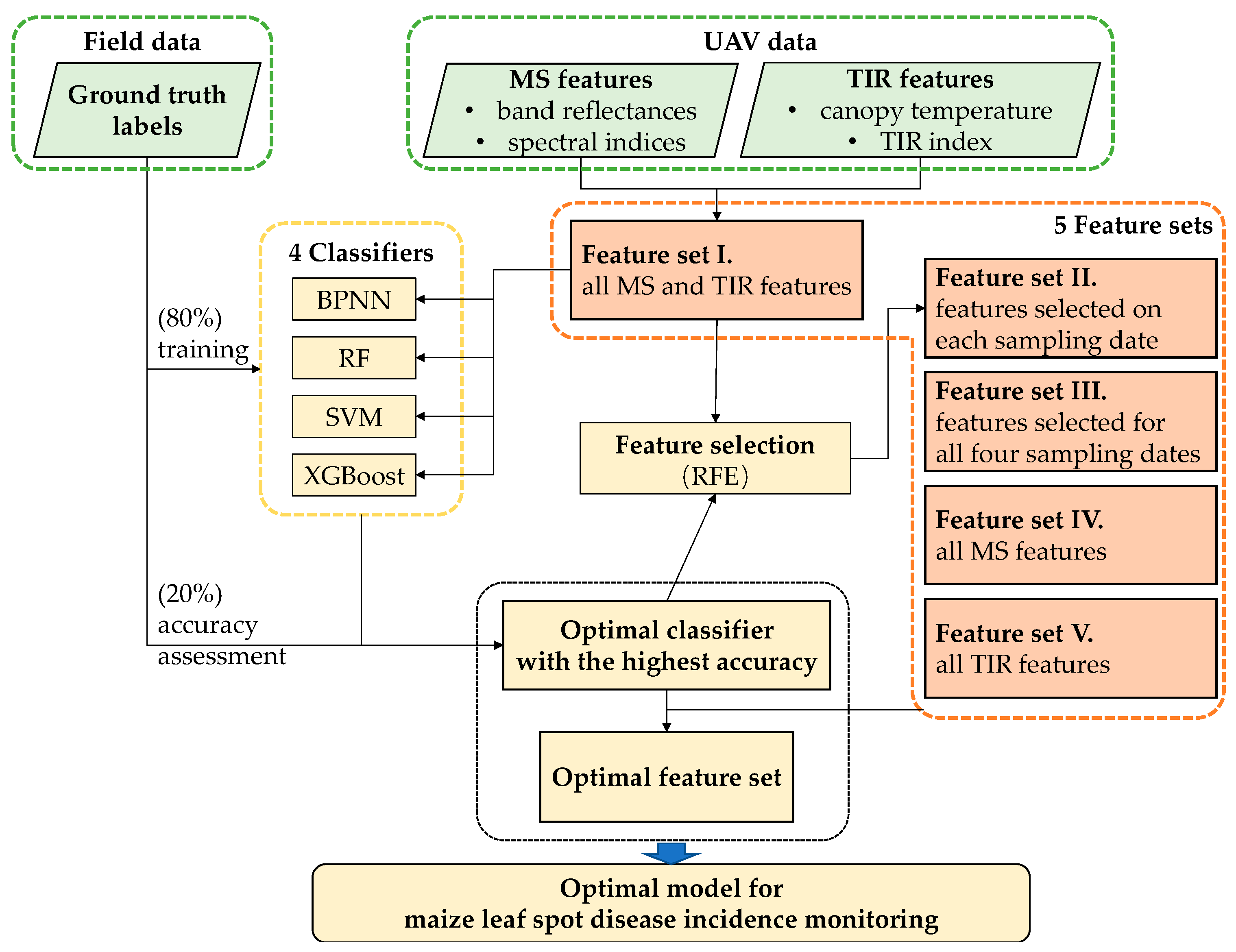
2.3. Leaf Spot Disease Detection Using UAV Data
2.3.1. UAV Data Pre-Processing
2.3.2. Feature Selection
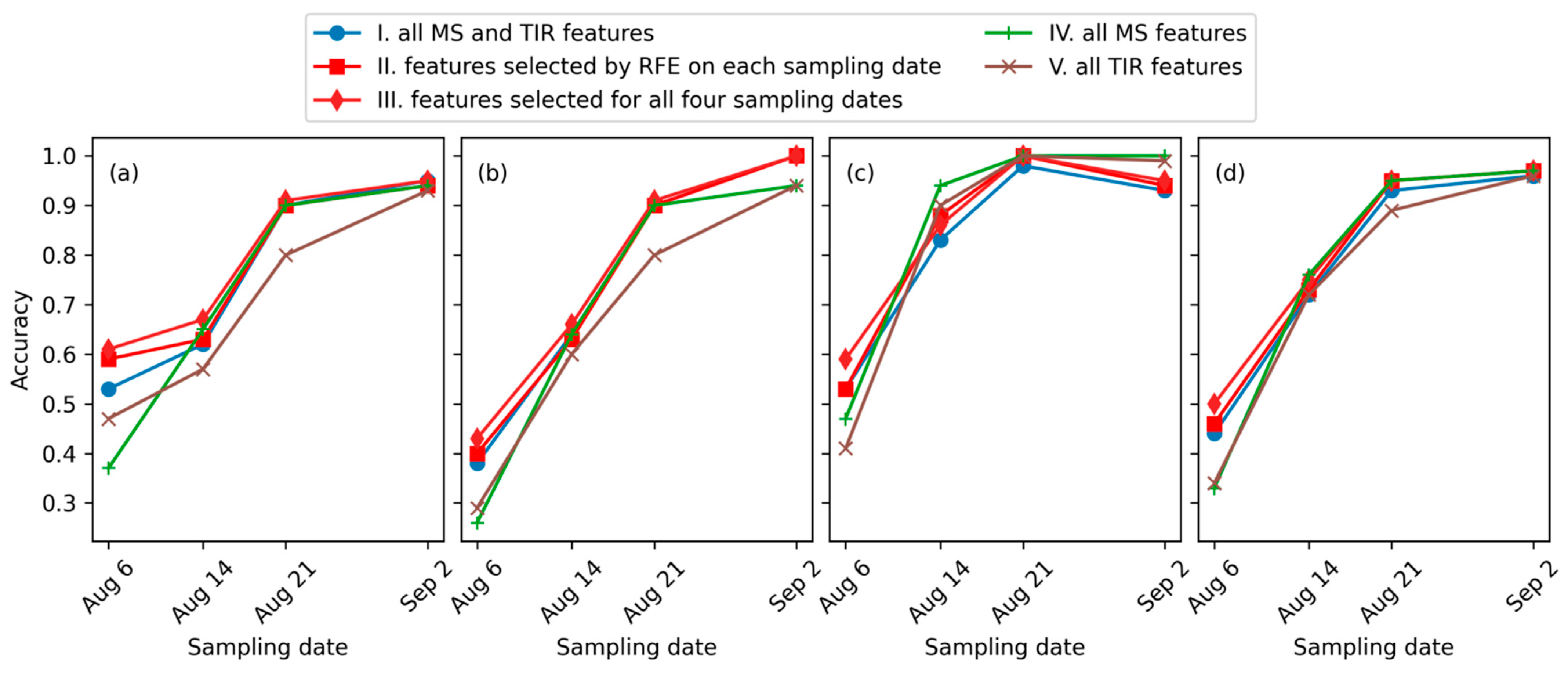
- Feature set I. All MS and TIR features mentioned in this study, including six MS bands, one TIR band, and the 23 indices in Table 3.
- Feature set II. Features selected using RFE at each of the four sampling dates separately.
- Feature set III. Features selected using RFE at all the four sampling dates, i.e., intersection of the four sets in feature set II.
- Feature set IV. All MS features, including the six MS bands and 22 MS indices.
- Feature set V. All TIR features, including canopy temperature and TIR index.
2.3.3. Classification
2.4. Accuracy Assessment
3. Results
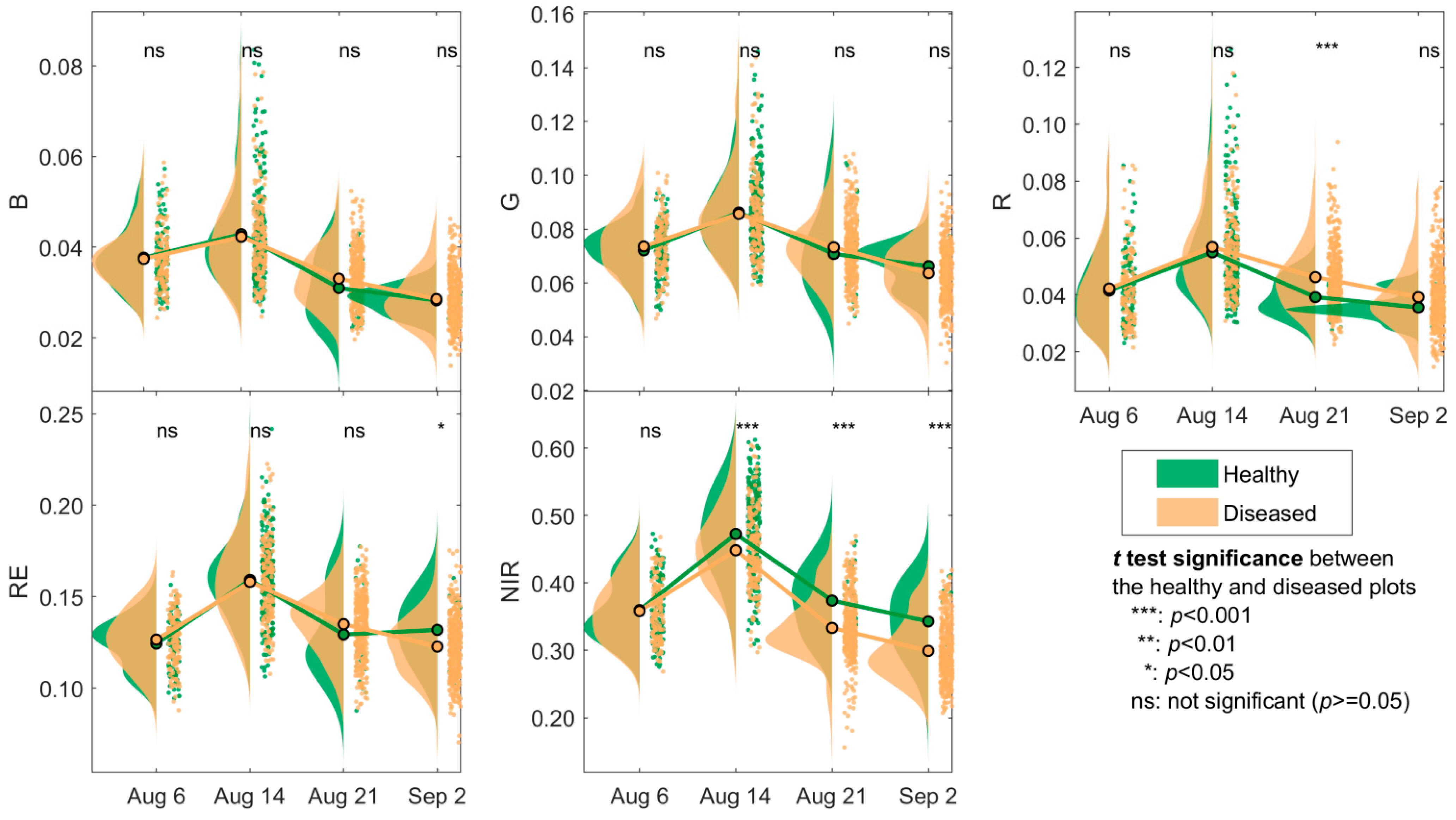
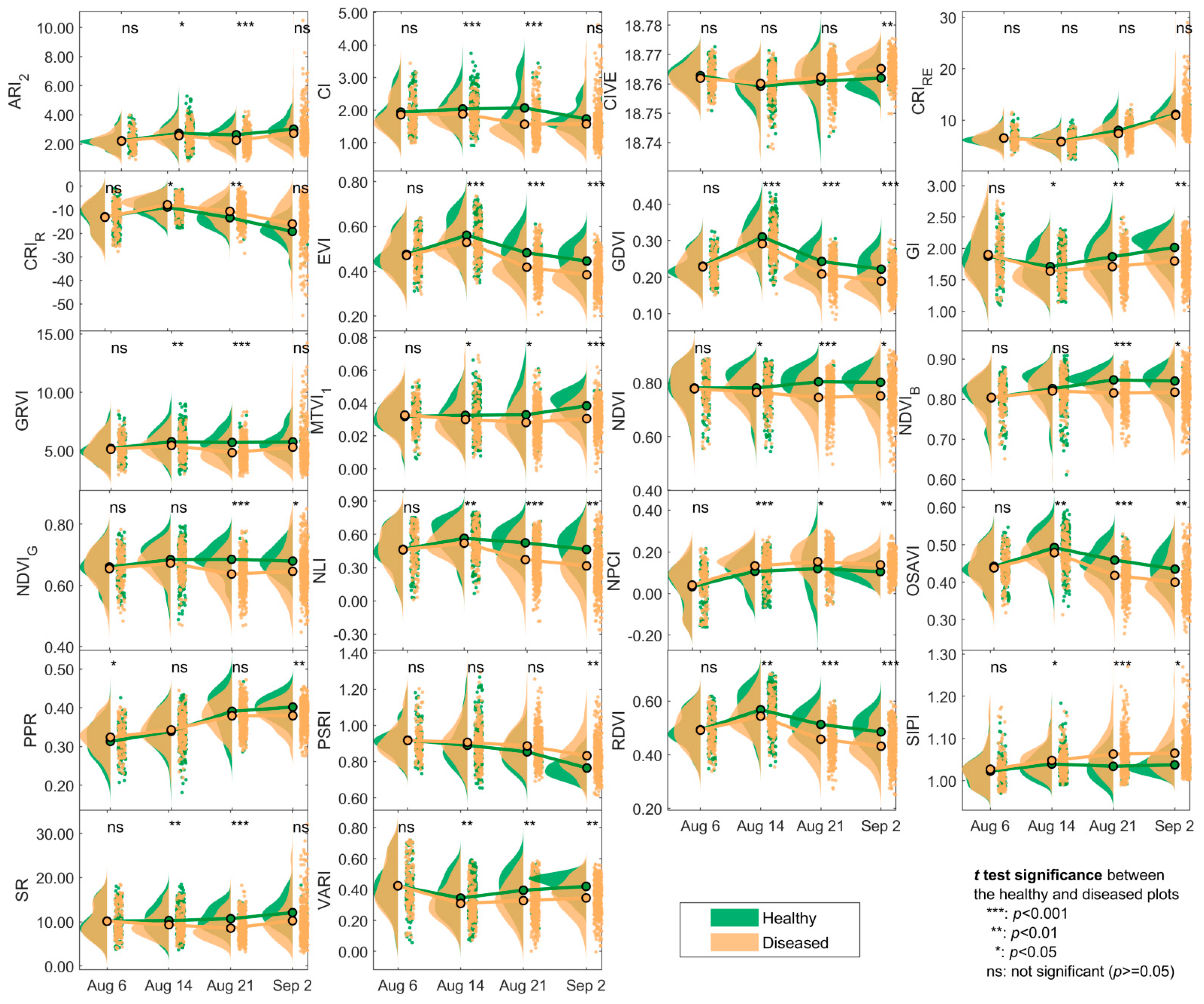
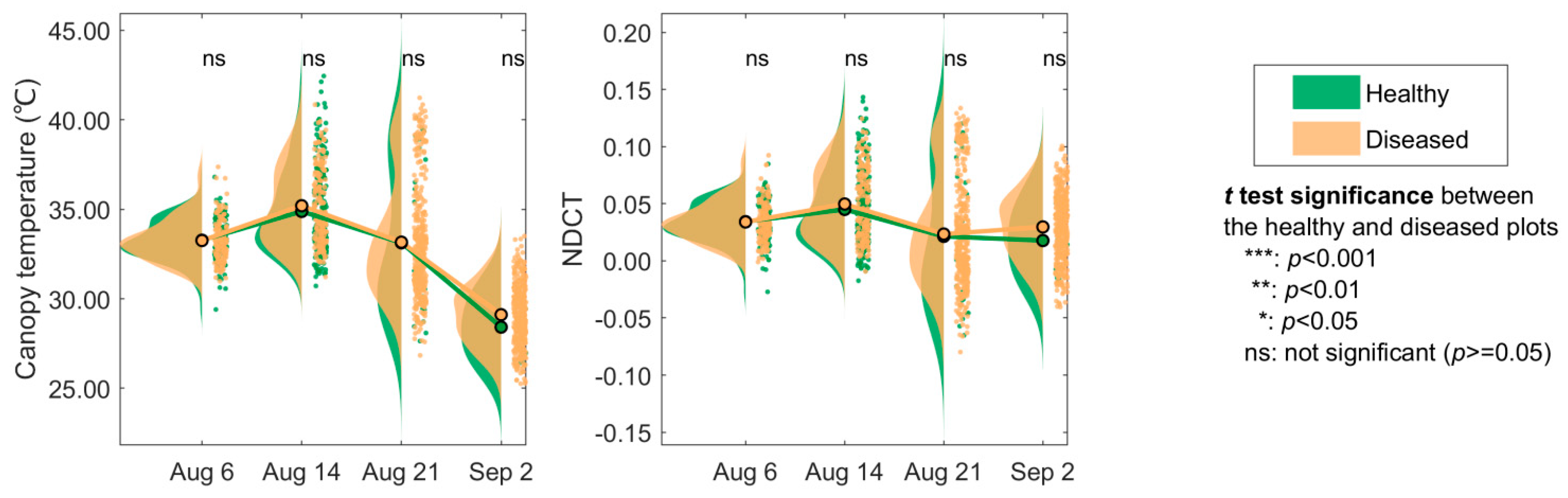
3.1. Changes in the Spectra and Indices
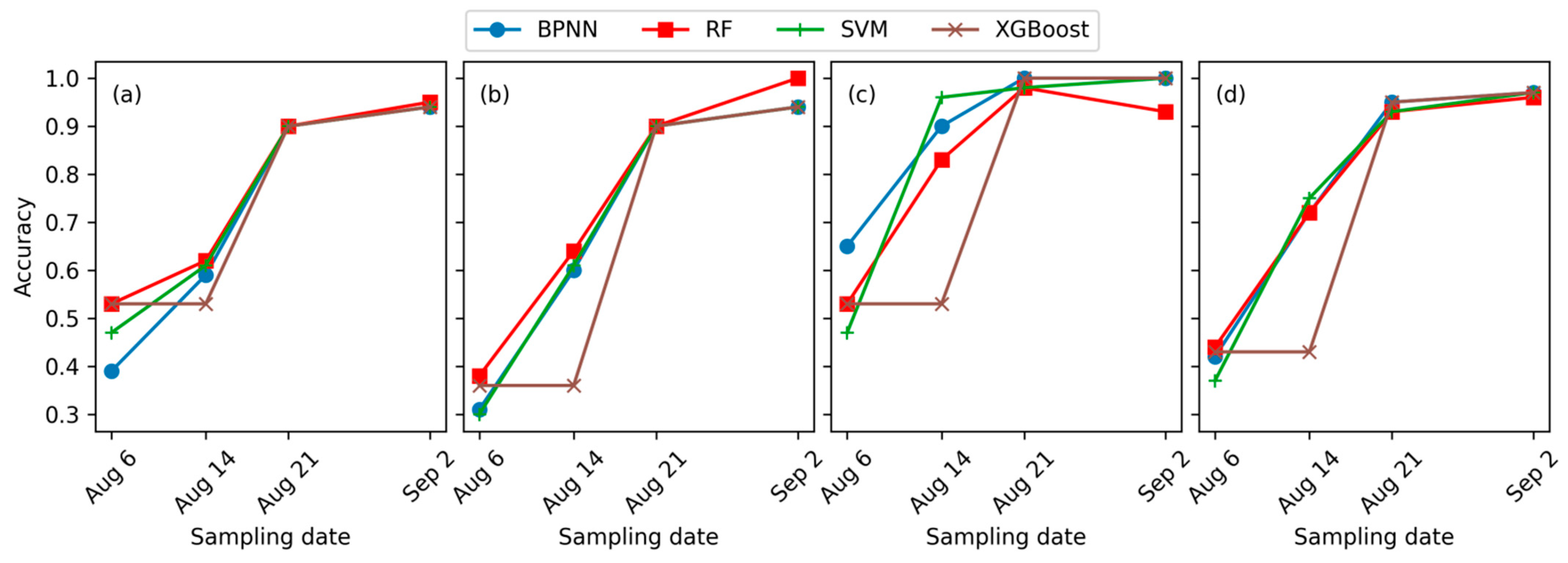
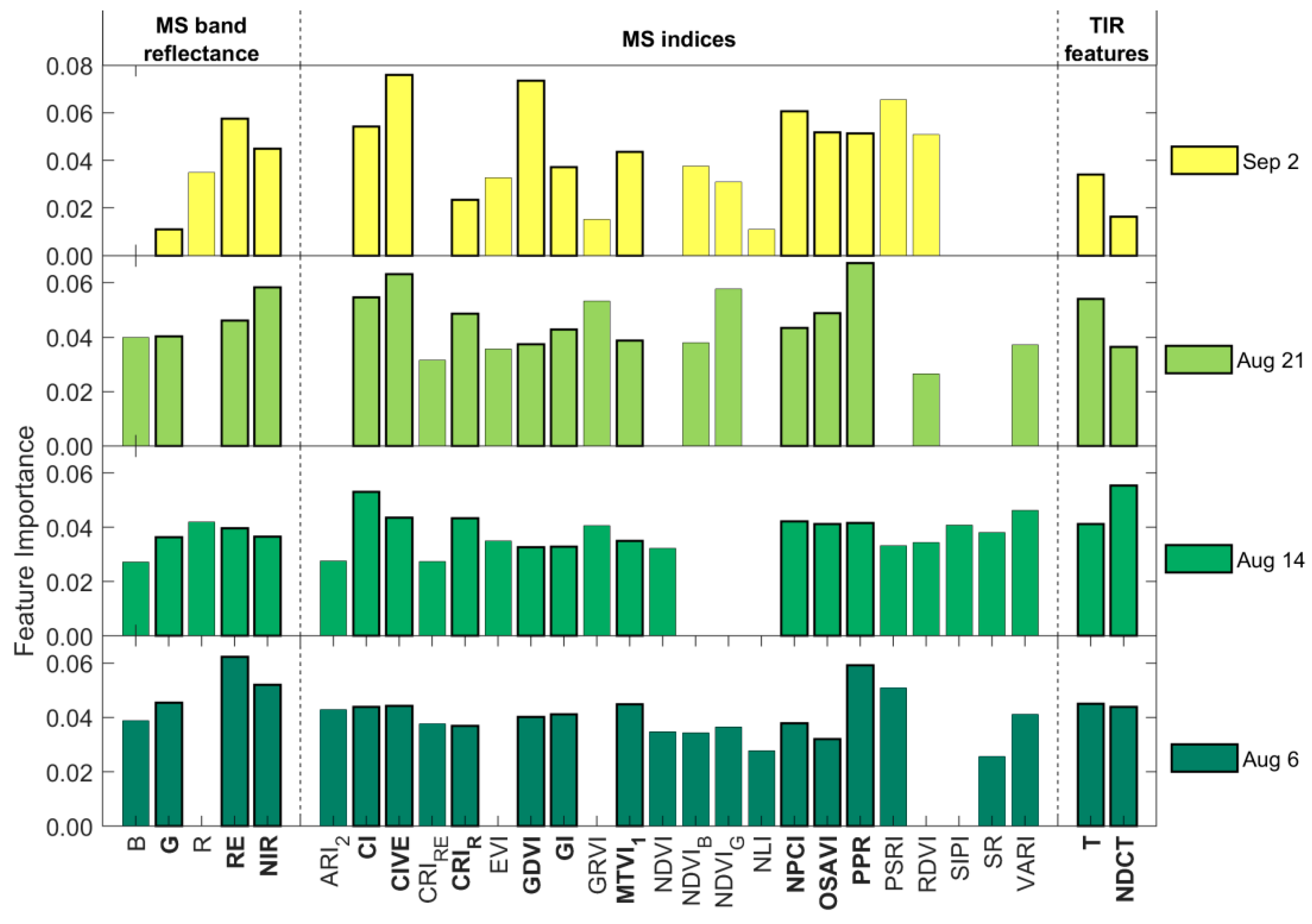
3.2. Leaf Spot Disease Incidence Identification Results
4. Discussions
4.1. Spectral Changes of Maize Leaf Spot from Early to Late Stages
4.2. Optimal Model for Maize Leaf Spot Disease Incidence Monitoring
4.2.1. Multi-Source Features
4.2.2. Optimal Classifier for Maize Leaf Spot Disease Incidence Monitoring
4.3. Early Monitoring of Maize Leaf Spot Disease Incidence
5. Conclusions
Author Contributions
Funding
Data Availability Statement
Conflicts of Interest
References
- Ranum, P.; Ranum, J.P.; Peña-Rosas, J.P.; Garcia-Casal, M.N. Global maize production; utilization, and consumption. Ann. N. Y. Acad. Sci. 2014, 1312, 105–112. [Google Scholar] [CrossRef] [PubMed]
- Du Plessis, J. Maize Production; Department of Agriculture: Pretoria, South Africa, 2003. [Google Scholar]
- Juroszek, P.; Tiedemann, A.V. Climatic changes and the potential future importance of maize diseases: A short review. J. Plant Dis. Prot. 2013, 120, 49–56. [Google Scholar] [CrossRef]
- Subedi, S. A review on important maize diseases and their management in Nepal. J. Maize Res. Dev. 2015, 1, 28–52. [Google Scholar] [CrossRef]
- Zhang, J.; Huang, Y.; Pu, R.; Gonzalez-Moreno, P.; Yuan, L.; Wu, K.; Huang, W. Monitoring plant diseases and pests through remote sensing technology: A review. Comput. Electron. Agric. 2019, 165, 104943. [Google Scholar] [CrossRef]
- Berger, K.; Machwitz, M.; Kycko, M.; Kefauver, S.C.; Van Wittenberghe, S.; Gerhards, M.; Verrelst, J.; Atzberger, C.; van der Tol, C.; Damm, A.; et al. Multi-sensor spectral synergies for crop stress detection and monitoring in the optical domain: A review. Remote Sens. Environ. 2022, 280, 113198. [Google Scholar] [CrossRef]
- Ishengoma, F.S.; Rai, I.A.; Said, R.N. Identification of maize leaves infected by fall armyworms using UAV-based imagery and convolutional neural networks. Comput. Electron. Agric. 2021, 184, 106124. [Google Scholar] [CrossRef]
- Osco, L.P.; Junior, J.M.; Ramos, A.P.M.; de Castro Jorge, L.A.; Fatholahi, S.N.; de Andrade Silva, J.; Matsubara, E.T.; Pistori, H.; Gonçalves, W.N.; Li, J. A review on deep learning in UAV remote sensing. Int. J. Appl. Earth Obs. Geoinf. 2021, 102, 102456. [Google Scholar] [CrossRef]
- Bhandari, M.; Ibrahim, A.M.; Xue, Q.; Jung, J.; Chang, A.; Rudd, J.C.; Maeda, M.; Rajan, N.; Neely, H.; Landivar, J. Assessing winter wheat foliage disease severity using aerial imagery acquired from small Unmanned Aerial Vehicle (UAV). Comput. Electron. Agric. 2020, 176, 105665. [Google Scholar] [CrossRef]
- Zhao, J.; Jin, Y.; Ye, H.; Huang, W.; Dong, Y.; Fan, L.; Ma, H.; Jiang, J. Remote sensing monitoring of areca yellow leaf disease based on UAV multi-spectral images. Trans. Chin. Soc. Agric. Eng. 2020, 36, 54–61. [Google Scholar]
- Azadbakht, M.; Ashourloo, D.; Aghighi, H.; Radiom, S.; Alimohammadi, A. Wheat leaf rust detection at canopy scale under different LAI levels using machine learning techniques. Comput. Electron. Agric. 2019, 156, 119–128. [Google Scholar] [CrossRef]
- Tian, L.; Wang, Z.; Xue, B.; Li, D.; Zheng, H.; Yao, X.; Zhu, Y.; Cao, W.; Cheng, T. A disease-specific spectral index tracks Magnaporthe oryzae infection in paddy rice from ground to space. Remote Sens. Environ. 2023, 285, 113384. [Google Scholar] [CrossRef]
- Zhang, N.; Zhang, X.; Yang, G.; Zhu, C.; Huo, L.; Feng, H. Assessment of defoliation during the Dendrolimus tabulaeformis Tsai et Liu disaster outbreak using UAV-based hyperspectral images. Remote Sens. Environ. 2018, 217, 323–339. [Google Scholar] [CrossRef]
- Liang, H.; He, J.; Lei, J. Monitoring of Corn Canopy Blight Disease Based on UAV Hyperspectral Method. Spectrosc. Spectr. Anal. 2020, 40, 1965–1972. [Google Scholar]
- Chaerle, L.; Van Der Straeten, D. Imaging techniques and the early detection of plant stress. Trends Plant Sci. 2000, 5, 495–501. [Google Scholar] [CrossRef]
- Sankaran, S.; Maja, J.M.; Buchanon, S.; Ehsani, R. Huanglongbing (citrus greening) detection using visible, near infrared and thermal imaging techniques. Sensors 2013, 13, 2117–2130. [Google Scholar] [CrossRef]
- Feng, Z.; Song, L.; Zhang, S.; Jing, Y.; Duan, J.; He, L.; Yin, F.; Feng, W. Wheat Powdery Mildew Monitoring Based on Information Fusion of Multi-Spectral and Thermal Infrared Images Acquired with an Unmanned Aerial Vehicle. Sci. Agric. Sin. 2022, 55, 890–906. [Google Scholar]
- Stewart, E.L.; Wiesner-Hanks, T.; Kaczmar, N.; DeChant, C.; Wu, H.; Lipson, H.; Nelson, R.J.; Gore, M.A. Quantitative phenotyping of northern leaf blight in UAV images using deep learning. Remote Sens. 2019, 11, 2209. [Google Scholar] [CrossRef]
- Zheng, Q.; Huang, W.; Cui, X.; Dong, Y.; Shi, Y.; Ma, H.; Liu, L. Identification of Wheat Yellow Rust Using Optimal Three-Band Spectral Indices in Different Growth Stages. Sensors 2019, 19, 35. [Google Scholar] [CrossRef]
- Meng, R.; Gao, R.; Zhao, F.; Huang, C.; Sun, R.; Lv, Z.; Huang, Z. Development of spectral disease indices for southern corn rust detection and severity classification. Remote Sens. 2020, 12, 3233. [Google Scholar] [CrossRef]
- Chivasa, W.; Mutanga, O.; Biradar, C. UAV-based multispectral phenotyping for disease resistance to accelerate crop improvement under changing climate conditions. Remote Sens. 2020, 12, 2445. [Google Scholar] [CrossRef]
- Meng, L.; Yin, D.; Cheng, M.; Liu, S.; Bai, Y.; Liu, Y.; Liu, Y.; Jia, X.; Nan, F.; Song, Y.; et al. Improved Crop Biomass Algorithm with Piecewise Function (iCBA-PF) for Maize Using Multi-Source UAV Data. Drones 2023, 7, 254. [Google Scholar] [CrossRef]
- Zhou, L.; Nie, C.; Su, T.; Xu, X.; Song, Y.; Yin, D.; Liu, S.; Liu, Y.; Bai, Y.; Jia, X.; et al. Evaluating the Canopy Chlorophyll Density of Maize at the Whole Growth Stage Based on Multi-Scale UAV Image Feature Fusion and Machine Learning Methods. Agriculture 2023, 13, 895. [Google Scholar] [CrossRef]
- Morris, M. Assessing the benefits of international maize breeding research: An overview of the global maize impacts study. In Meeting World Maize Needs: Technological Opportunities and Priorities for the Public Sector; Pingali, P.L., Ed.; CIMMYT: Texcoco, Mexico, 2001. [Google Scholar]
- Chen, C.; Zhao, Y.; Tabor, G.; Nian, H.; Phillips, J.; Wolters, P.; Yang, Q.; Balint-Kurti, P. A leucine-rich repeat receptor kinase gene confers quantitative susceptibility to maize southern leaf blight. New Phytol. 2023, 238, 1182–1197. [Google Scholar] [CrossRef] [PubMed]
- Chen, X.; Tang, T.; Chen, C.; Wei, L.; Zhou, D. First report of Curvularia leaf spot caused by Curvularia muehlenbeckiae on Zizania latifolia in China. J. Plant Pathol. 2021, 103, 1073. [Google Scholar] [CrossRef]
- GB/T 23391.2-2009; Standards for Scientific Observation Data and Quality Control of Maize Southern Leaf Blight in China. Standardization Administration of the People’s Republic of China: Beijing, China, 2009.
- NY/T 1248.10-2016; Technical Specifications for Identification of Maize Disease and Insect Resistance: Part 10: Curvularia Leaf Spot Disease. The Ministry of Agriculture of the People’s Republic of China: Beijing, China, 2016.
- Reyes, J.R.; Bohórquez, J.S.; Alama, W.I. Hyperspectral analysis based anthocyanin index (ARI2) during cocoa bean fermentation process. In Proceedings of the 2015 Asia-Pacific Conference on Computer Aided System Engineering, Quito, Ecuador, 14–16 July 2015. [Google Scholar]
- Mishra, S.; Mishra, D.R. Normalized difference chlorophyll index: A novel model for remote estimation of chlorophyll-a concentration in turbid productive waters. Remote Sens. Environ. 2012, 117, 394–406. [Google Scholar] [CrossRef]
- Kataoka, T.; Kaneko, T.; Okamoto, H.; Hata, S. Crop growth estimation system using machine vision. In Proceedings of the 2003 IEEE/ASME International Conference on Advanced Intelligent Mechatronics (AIM 2003), Kobe, Japan, 20–24 July 2003. [Google Scholar]
- Poblete, T.; Navas-Cortes, J.A.; Camino, C.; Calderon, R.; Hornero, A.; Gonzalez-Dugo, V.; Landa, B.B.; Zarco-Tejada, P.J. Discriminating Xylella fastidiosa from Verticillium dahliae infections in olive trees using thermal- and hyperspectral-based plant traits. ISPRS J. Photogramm. Remote Sens. 2021, 179, 133–144. [Google Scholar] [CrossRef]
- Liu, H.Q.; Huete, A. A feedback based modification of the NDVI to minimize canopy background and atmospheric noise. IEEE Trans. Geosci. Remote Sens. 1995, 33, 457–465. [Google Scholar] [CrossRef]
- Pettorelli, N. The Normalized Difference Vegetation Index; Oxford University Press: Oxford, UK, 2013. [Google Scholar]
- Penuelas, J.; Baret, F.; Filella, I. Semi-empirical indices to assess carotenoids/chlorophyll a ratio from leaf spectral reflectance. Photosynthetica 1995, 31, 221–230. [Google Scholar]
- Rondeaux, G.; Steven, M.; Baret, F. Optimization of soil-adjusted vegetation indices. Remote Sens. Environ. 1996, 55, 95–107. [Google Scholar] [CrossRef]
- Haboudane, D.; Miller, J.R.; Pattey, E.; Zarco-Tejada, P.J.; Strachan, I.B. Hyperspectral vegetation indices and novel algorithms for predicting green LAI of crop canopies: Modeling and validation in the context of precision agriculture. Remote Sens. Environ. 2004, 90, 337–352. [Google Scholar] [CrossRef]
- Feng, W.; Wu, Y.; He, L.; Ren, X.; Wang, Y.; Hou, G.; Wang, Y.; Liu, W.; Guo, T. An optimized non-linear vegetation index for estimating leaf area index in winter wheat. Precis. Agric. 2019, 20, 1157–1176. [Google Scholar] [CrossRef]
- Ren, S.; Chen, X.; An, S. Assessing plant senescence reflectance index-retrieved vegetation phenology and its spatiotemporal response to climate change in the Inner Mongolian Grassland. Int. J. Biometeorol. 2017, 61, 601–612. [Google Scholar] [CrossRef] [PubMed]
- Chen, J.M. Evaluation of vegetation indices and a modified simple ratio for boreal applications. Can. J. Remote Sens. 1996, 22, 229–242. [Google Scholar] [CrossRef]
- Jordan, C.F. Derivation of leaf-area index from quality of light on the forest floor. Ecology 1969, 50, 663–666. [Google Scholar] [CrossRef]
- Xue, J.; Su, B. Significant remote sensing vegetation indices: A review of developments and applications. J. Sens. 2017, 2017, 1353691. [Google Scholar]
- Irmak, S.; Haman, D.Z.; Bastug, R. Determination of crop water stress index for irrigation timing and yield estimation of corn. Agron. J. 2000, 92, 1221–1227. [Google Scholar] [CrossRef]
- Chen, X.-W.; Jeong, J.C. Enhanced recursive feature elimination. In Proceedings of the Sixth International Conference on Machine Learning and Applications (ICMLA 2007), Cincinnati, OH, USA, 13–15 December 2007. [Google Scholar]
- Barreto, A.; Paulus, S.; Varrelmann, M.; Mahlein, A.-K. Hyperspectral imaging of symptoms induced by Rhizoctonia solani in sugar beet: Comparison of input data and different machine learning algorithms. J. Plant Dis. Prot. 2020, 127, 441–451. [Google Scholar] [CrossRef]
- Koc, A.; Odilbekov, F.; Alamrani, M.; Henriksson, T.; Chawade, A. Predicting yellow rust in wheat breeding trials by proximal phenotyping and machine learning. Plant Methods 2022, 18, 30. [Google Scholar] [CrossRef]
- Lasso, E.; Corrales, D.C.; Avelino, J.; de Melo Virginio Filho, E.; Corrales, J.C. Discovering weather periods and crop properties favorable for coffee rust incidence from feature selection approaches. Comput. Electron. Agric. 2020, 176, 105640. [Google Scholar] [CrossRef]
- Hu, H.-Y.; Lee, Y.-C.; Yen, T.-M.; Tsai, C.-H. Using BPNN and DEMATEL to modify importance–performance analysis model—A study of the computer industry. Expert Syst. Appl. 2009, 36, 9969–9979. [Google Scholar] [CrossRef]
- Breiman, L. Random forests. Mach. Learn. 2001, 45, 5–32. [Google Scholar] [CrossRef]
- Yue, J.; Tian, Q.; Dong, X.; Xu, N. Using broadband crop residue angle index to estimate the fractional cover of vegetation, crop residue, and bare soil in cropland systems. Remote Sens. Environ. 2020, 237, 111538. [Google Scholar] [CrossRef]
- Ogunleye, A.; Wang, Q.-G. XGBoost model for chronic kidney disease diagnosis. IEEE/ACM Trans. Comput. Biol. Bioinform. 2019, 17, 2131–2140. [Google Scholar] [CrossRef] [PubMed]
- Lepeschkin, E.; Surawicz, B. Characteristics of true-positive and false-positive results of electrocardiographs master two-step exercise tests. New Engl. J. Med. 1958, 258, 511–520. [Google Scholar] [CrossRef]
- Taddei, F.; Cristofolini, L.; Martelli, S.; Gill, H.; Viceconti, M. Subject-specific finite element models of long bones: An in vitro evaluation of the overall accuracy. J. Biomech. 2006, 39, 2457–2467. [Google Scholar] [CrossRef] [PubMed]
- Melamed, I.D.; Green, R.; Turian, J. Proceedings of the Precision and recall of machine translation. In Companion Volume of the Proceedings of HLT-NAACL 2003-Short Papers. Edmonton, AB, Canada, 27 May–1 June 2003. [Google Scholar]
- Chicco, D.; Jurman, G. The advantages of the Matthews correlation coefficient (MCC) over F1 score and accuracy in binary classification evaluation. BMC Genom. 2020, 21, 6. [Google Scholar] [CrossRef]
- Dhau, I.; Adam, E.; Mutanga, O.; Ayisi, K.; Abdel-Rahman, E.M.; Odindi, J.; Masocha, M. Detecting the severity of maize streak virus infestations in maize crop using in situ hyperspectral data. Trans. R. Soc. S. Afr. 2017, 73, 8–15. [Google Scholar] [CrossRef]
- Shao, M.; Nie, C.; Zhang, A.; Shi, L.; Zha, Y.; Xu, H.; Yang, H.; Yu, X.; Bai, Y.; Liu, S.; et al. Quantifying effect of maize tassels on LAI estimation based on multispectral imagery and machine learning methods. Comput. Electron. Agric. 2023, 211, 108029. [Google Scholar] [CrossRef]
- Su, J.; Liu, C.; Hu, X.; Xu, X.; Guo, L.; Chen, W.-H. Spatio-temporal monitoring of wheat yellow rust using UAV multispectral imagery. Comput. Electron. Agric. 2019, 167, 105035. [Google Scholar] [CrossRef]
- Su, J.; Liu, C.; Coombes, M.; Hu, X.; Wang, C.; Xu, X.; Li, Q.; Guo, L.; Chen, W.-H. Wheat yellow rust monitoring by learning from multispectral UAV aerial imagery. Comput. Electron. Agric. 2018, 155, 157–166. [Google Scholar] [CrossRef]
- Luo, L.; Chang, Q.; Wang, Q.; Huang, Y. Identification and severity monitoring of maize dwarf mosaic virus infection based on hyperspectral measurements. Remote Sens. 2021, 13, 4560. [Google Scholar] [CrossRef]
- Fahrentrapp, J.; Ria, F.; Geilhausen, M.; Panassiti, B. Detection of gray mold leaf infections prior to visual symptom appearance using a five-band multispectral sensor. Front. Plant Sci. 2019, 10, 628. [Google Scholar] [CrossRef] [PubMed]
- Poblete, T.; Camino, C.; Beck, P.S.A.; Hornero, A.; Kattenborn, T.; Saponari, M.; Boscia, D.; Navas-Cortes, J.A.; Zarco-Tejada, P.J. Detection of Xylella fastidiosa infection symptoms with airborne multispectral and thermal imagery: Assessing bandset reduction performance from hyperspectral analysis. ISPRS J. Photogramm. Remote Sens. 2020, 162, 27–40. [Google Scholar] [CrossRef]
- Gao, G. Discussion on Hazard Symptoms and Prevention Methods of Corn Southern Leaf Blight. J. Agric. Catastrophol. 2016, 6, 13–15. [Google Scholar]

| Date of Acquisition | Days after Inoculation | Disease Development Stages | MS | TIR | ||
|---|---|---|---|---|---|---|
| Altitude (m) | Spatial Resolution (m) | Altitude (m) | Spatial Resolution (m) | |||
| 6 August 2021 | 4 | early | 70 | 0.045 | 70 | 0.106 |
| 14 August 2021 | 12 | early metaphase | 70 | 0.050 | 70 | 0.113 |
| 21 August 2021 | 19 | middle | 50 | 0.034 | 50 | 0.077 |
| 2 September 2021 | 30 | late | 20 | 0.018 | 50 | 0.076 |
| Disease Grade | 1 | 3 | 5 | 7 | 9 | Reference |
|---|---|---|---|---|---|---|
| Symptom description * | Disease spots account for less than or equal to 5% of the leaf area | Disease spots account for 6–10% of the leaf area | Disease spots account for 11–30% of the leaf area | Disease spots account for 31–70% of the leaf area | Disease spots cover the whole leaf and leaf dying | |
| Sample photo (southern leaf blight) |  |  |  |  |  | [27] |
| Sample photo (Curvularia leaf spot) |  |  |  |  |  | [28] |
| Feature Type | Feature Name | Equation | Reference |
|---|---|---|---|
| MS | Anthocyanin Reflectance Index 2 (ARI2) | ARI2 = RNIR · (1/RG − 1/RRE) | [29] |
| Chlorophyll Index (CI) | [30] | ||
| Vegetation Color Index (CIVE) | CIVE = 0.441RR − 0.881 RG + 0.385RB+18.787 | [31] | |
| Red Edge Chlorophyll Index (CRIRE) | [32] | ||
| Red Chlorophyll Index (CRIR) | [32] | ||
| Enhanced Vegetation Index (EVI) | EVI = 2.5(RNIR − RR)/(RNIR + 6RR − 7.5RB + 1) | [33] | |
| Difference Vegetation Index (DVI) | [34] | ||
| Greenness Index (GI) | [35] | ||
| Green Ratio Vegetation Index (GRVI) | [36] | ||
| Modified Triangular Vegetation Index 1 (MTVI1) | [37] | ||
| Normalized Difference Vegetation Index (NDVI) | [37] | ||
| Blue NDVI (NDVIB) | [32] | ||
| Green NDVI (NDVIG) | [32] | ||
| Nonlinear Index (NLI) | [38] | ||
| Normalized Pigment Chlorophyll Index (NPCI) | [35] | ||
| Optimized Soil-Adjusted Vegetation Index (OSAVI) | [36] | ||
| Plant Pigment Radio (PPR) | [39] | ||
| Plant Senescence Reflectance Index (PSRI) | PSRI = (RR + RB)/RRE | [39] | |
| Renormalized Difference Vegetation Index (RDVI) | [40] | ||
| Structure-Intensive Pigment Index (SIPI) | [39] | ||
| Simple Ratio (SR) | [41] | ||
| Visible Atmospherically Resistant Index (VARI) | [42] | ||
| TIR | Normalized Differential Canopy Temperature (NDCT) | [43] |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Jia, X.; Yin, D.; Bai, Y.; Yu, X.; Song, Y.; Cheng, M.; Liu, S.; Bai, Y.; Meng, L.; Liu, Y.; et al. Monitoring Maize Leaf Spot Disease Using Multi-Source UAV Imagery. Drones 2023, 7, 650. https://doi.org/10.3390/drones7110650
Jia X, Yin D, Bai Y, Yu X, Song Y, Cheng M, Liu S, Bai Y, Meng L, Liu Y, et al. Monitoring Maize Leaf Spot Disease Using Multi-Source UAV Imagery. Drones. 2023; 7(11):650. https://doi.org/10.3390/drones7110650
Chicago/Turabian StyleJia, Xiao, Dameng Yin, Yali Bai, Xun Yu, Yang Song, Minghan Cheng, Shuaibing Liu, Yi Bai, Lin Meng, Yadong Liu, and et al. 2023. "Monitoring Maize Leaf Spot Disease Using Multi-Source UAV Imagery" Drones 7, no. 11: 650. https://doi.org/10.3390/drones7110650
APA StyleJia, X., Yin, D., Bai, Y., Yu, X., Song, Y., Cheng, M., Liu, S., Bai, Y., Meng, L., Liu, Y., Liu, Q., Nan, F., Nie, C., Shi, L., Dong, P., Guo, W., & Jin, X. (2023). Monitoring Maize Leaf Spot Disease Using Multi-Source UAV Imagery. Drones, 7(11), 650. https://doi.org/10.3390/drones7110650








