In Silico Ligand-Based Methods Targeting Porcupine Receptor Inhibitors with Potential Anticancer Effect †
Abstract
1. Introduction
2. Materials and Methods
2.1. Ligand Preparation
2.2. Pharmacophore Generation and Validation
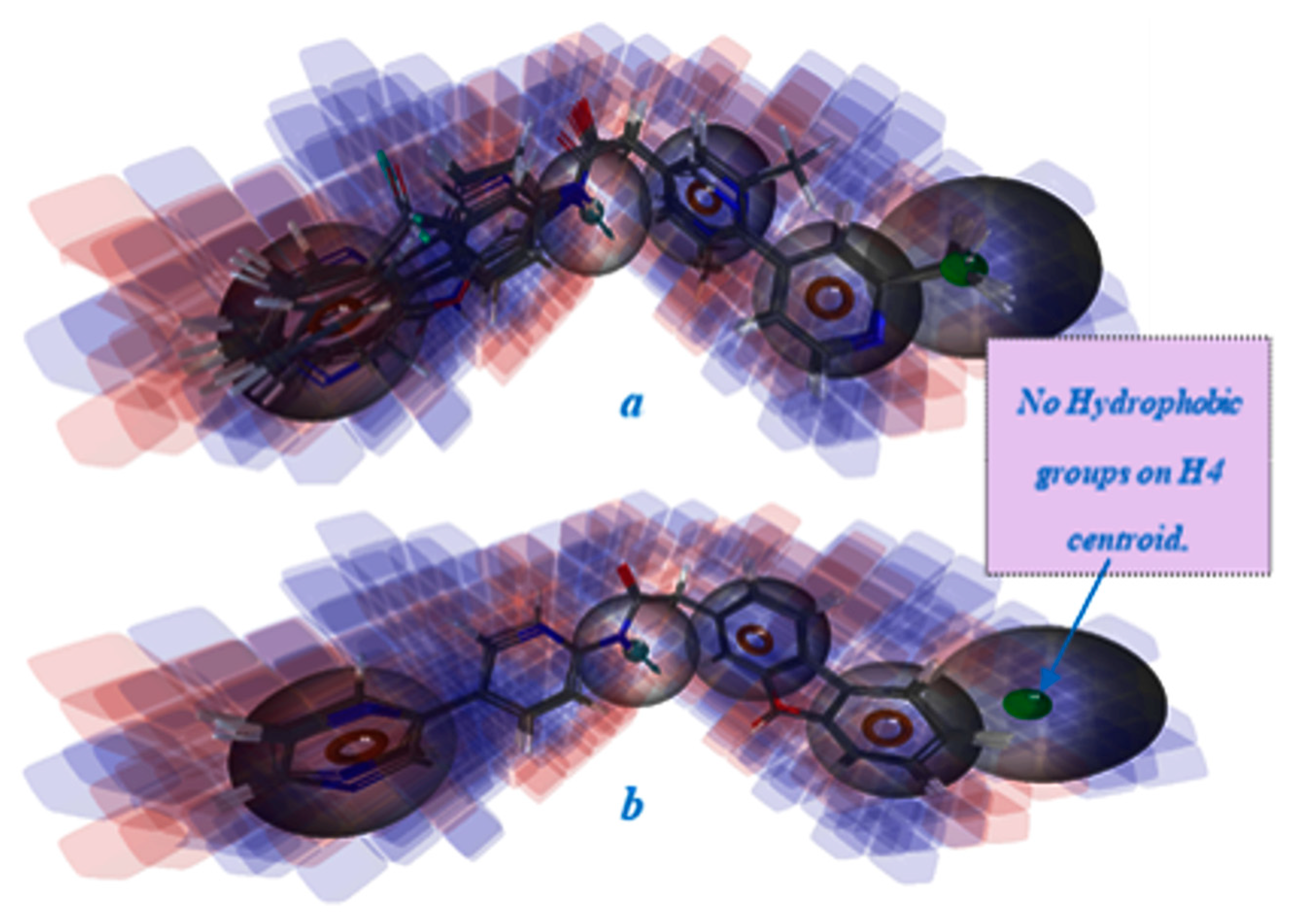
3. Results and Discussions
4. Conclusions
Author Contributions
Acknowledgments
Conflicts of Interest
References
- Xu, Z.; Li, J.; Wu, Y.; Sun, Z.; Luo, L.; Hu, Z.; He, S.; Zheng, J.; Zhang, H.; Zhang, X. Design, synthesis, and evaluation of potent Wnt signaling inhibitors featuring a fused 3-ring system. Eur. J. Med. Chem. 2016, 108, 154–165. [Google Scholar] [CrossRef] [PubMed]
- Madan, B.; Ke, Z.; Harmston, N.; Ho, S.Y.; Frois, A.O.; Alam, J.; Jeyaraj, D.A.; Pendharkar, V.; Ghosh, K.; Virshup, I.H.; et al. Wnt addiction of genetically defined cancers reversed by PORCN inhibition. Oncogene 2016, 35, 2197–2207. [Google Scholar] [CrossRef] [PubMed]
- Lum, L.; Clevers, H. The Unusual Case of Porcupine. Science 2012, 337, 922–923. [Google Scholar] [CrossRef] [PubMed]
- Alonso, L.; Fuchs, E. Stem cells in the skin: Waste not, Wnt not. Genes Dev. 2003, 17, 1189–1200. [Google Scholar] [CrossRef] [PubMed]
- Pinto, D.; Clevers, H. Wnt control of stem cells and differentiation in the intestinal epithelium. Exp. Cell Res. 2005, 306, 357–363. [Google Scholar] [CrossRef] [PubMed]
- Nemeth, M.J.; Mak, K.K.; Yang, Y.; Bodine, D.M. β-catenin expression in the bone marrow microenvironment is required for long-term maintenance of primitive hematopoietic cells. Stem Cells 2009, 27, 1109–1119. [Google Scholar] [CrossRef] [PubMed]
- Cisternas, P.; Henriquez, J.P.; Brandan, E.; Inestrosa, N.C. Wnt signaling in skeletal muscle dynamics: Myogenesis, neuromuscular synapse and fibrosis. Mol. Neurobiol. 2013, 49, 574–589. [Google Scholar] [CrossRef] [PubMed]
- Pecinas-Slaus, N. Wnt signal transduction pathway and apoptosis: A review. Cancer Cell. Int. 2010, 10, 22. [Google Scholar] [CrossRef] [PubMed]
- Kahn, M. Can we safely target the WNT pathway? Nat. Rev. Drug Discov. 2014, 13, 513–532. [Google Scholar] [CrossRef] [PubMed]
- Porfiri, E.; Rubinfeld, B.; Albert, I.; Hovanes, K.; Waterman, M.; Polakis, P. Induction of a β-catenin-LEF-1 complex by Wnt-1 and transforming mutants of β-catenin. Oncogene 1997, 15, 2833–2839. [Google Scholar] [CrossRef] [PubMed]
- Anastas, J.N.; Moon, R.T. WNT signalling pathways as therapeutic targets in cancer. Nat. Rev. Cancer 2013, 13, 11–26. [Google Scholar] [CrossRef] [PubMed]
- Schinner, S. Wnt-signalling and the metabolic syndrome. Horm. Metab. Res. 2009, 41, 159–163. [Google Scholar] [CrossRef] [PubMed]
- Dees, C.; Distler, J.H. Canonical Wnt signaling as a key-regulator of fibrogenesis—Implications for targeted therapies? Exp. Dermatol. 2013, 22, 710–713. [Google Scholar] [CrossRef] [PubMed]
- Inestrosa, N.C.; Montecinos-Oliva, C.; Fuenzalida, M. Wnt signaling: Role in Alzheimer disease and schizophrenia. J. Neuroimmune Pharmacol. 2012, 7, 788–807. [Google Scholar] [CrossRef] [PubMed]
- Seshagiri, S.; Stawiski, E.W.; Durinck, S.; Modrusan, Z.; Storm, E.E.; Conboy, C.B.; Chaudhuri., S.; Guan, Y.; Janakiraman, V.; Jaiswal, B.S.; et al. Recurrent R-spondin fusions in colon cancer. Nature 2012, 488, 660–664. [Google Scholar] [CrossRef] [PubMed]
- Cardona, G.M.; Bell, K.; Portale, J.; Gaffney, D.; Moy, C.; Platero, S.; Lorenzi, M.V.; Karker, J. Identification of R-Spondin Fusions in Various Types of Human Cancer. In Proceedings of the 105th Annual Meeting of the American Association for Cancer Research 2014, San Diego, CA, USA, 5–9 April 2014. Abstract no. 2408. [Google Scholar]
- Covey, T.M.; Kaur, S.; Ong, T.T.; Proffitt, K.D.; Wu, P.Tan.; Virshup, D.M. PORCN Moonlights in a Wnt-Independent Pathway That Regulates Cancer Cell Proliferation. PLoS ONE 2012, 7, e34532. [Google Scholar] [CrossRef] [PubMed]
- Barrott, J.J.; Cash, G.M.; Smith, A.P.; Barrow, J.R.; Murtaugh, L.C. Deletion of mouse Porcn blocks Wnt ligand secretion and reveals an ectodermal etiology of human focal dermal hypoplasia/Goltz syndrome. Proc. Natl. Acad. Sci. USA 2011, 108, 12752–12757. [Google Scholar] [CrossRef] [PubMed]
- Schrödinger Release 2018-1: Phase; Schrödinger, LLC: New York, NY, USA, 2018.
- Schrödinger Release 2018-1: LigPrep; Schrödinger, LLC: New York, NY, USA, 2018.
- Shelley, J.C.; Cholleti, A.; Frye, L; Greenwood, J.R.; Timlin, M.R.; Uchimaya, M. Epik: A software program for pKa prediction and protonation state generation for drug-like molecules. J. Comp. Aided Mol. Des. 2007, 21, 681–691. [Google Scholar] [CrossRef] [PubMed]
- Schrödinger Release 2018-1: ConfGen; Schrödinger, LLC: New York, NY, USA, 2018.
- Dixon, S.L.; Smondyrev, A.M.; Knoll, E.H.; Rao, S.N.; Shaw, D.E.; Friesner, R.A. PHASE: A new engine for pharmacophore perception, 3D QSAR model development, and 3D database screening: 1. Methodology and preliminary results. J. Comput. Aided Mol. Des. 2006, 20, 647–671. [Google Scholar] [CrossRef] [PubMed]
- Schrödinger Release 2018-1: Phase Help—Atom-Based QSAR Panel; Schrödinger, LLC: New York, NY, USA, 2018.
- Schrodinger Release 2018-1: Glide Help—Enrichment Calculator Panel; Schrödinger, LLC: New York, NY, USA, 2018.


| ID | Smiles Code | pIC50exp | pIC50pred |
|---|---|---|---|
| 1 * | Cc1cc(ccn1)c2ccc(cc2)CC(=O)Nc3ccc(cc3)c4ccccc4 | 8.54 | 8.47 |
| 2 | c1ccc-2c(c1)Cc3c2ccc(c3)CC(=O)Nc4ccc(cc4)c5cccnc5 | 6.64 | 6.56 |
| 3 | c1ccc2c(c1)c3ccc(cc3o2)CC(=O)Nc4ccc(cc4)c5cccnc5 | 6.26 | 6.51 |
| 4 | c1ccc-2c(c1)Cc3c2ccc(c3)CC(=O)Nc4ccc(cn4)c5cnccn5 | 6.34 | 6.62 |
| 5 | c1ccc2c(c1)c3ccc(cc3o2)CC(=O)Nc4ccc(cn4)c5cnccn5 | 6.87 | 6.53 |
| 6 * | Cc1cc(cnc1c2ccnc(c2)C)CC(=O)Nc3ccc-4c(c3)Cc5c4cccc5 | 8.60 | 8.89 |
| 7 * | Cc1cc(cnc1c2ccnc(c2)C)CC(=O)Nc3ccc-4c(c3)C(c5c4cccc5)(F)F | 8.57 | 8.92 |
| 8 | Cc1cc(cnc1c2ccnc(c2)C)CC(=O)Nc3ccc4c(c3)-c5ccccc5C4=O | 8.55 | 8.86 |
| 9 | Cc1cc(cnc1c2ccnc(c2)C)CC(=O)Nc3ccc4c5ccccc5[nH]c4c3 | 8.59 | 8.78 |
| 10 | Cc1cc(ccn1)c2ccc(cc2)CC(=O)Nc3ccc-4c(c3)Cc5c4cccc5 | 8.85 | 8.85 |
| 11 * | c1ccc2c(c1)-c3ccc(cc3C2=O)CC(=O)Nc4ccc(cc4)c5cccnc5 | 6.00 | 6.62 |
| 12 | Cc1cc(ccn1)c2ccc(cc2)CC(=O)Nc3ccc-4c(c3)C(c5c4cccc5)(F)F | 8.46 | 8.46 |
| 13 | Cc1cc(ccn1)c2ccc(cc2)CC(=O)Nc3ccc-4c(c3)C(=O)c5c4cccc5 | 9.35 | 8.88 |
| 14 | Cc1cc(ccn1)c2ccc(cc2)CC(=O)Nc3ccc4c5ccccc5[nH]c4c3 | 8.64 | 8.44 |
| 15 * | c1ccc2c(c1)-c3ccc(cc3C2=O)CC(=O)Nc4ccc(cn4)c5cnccn5 | 6.00 | 6.72 |
| 16 | Cc1cc(ccn1)c2ccc(cc2)CC(=O)Nc3ccc4c5ccccc5oc4c3 | 8.05 | 8.05 |
| 17 | Cc1cc(cnc1c2ccnc(c2)C)CC(=O)Nc3ccc(cn3)c4cnccn4 | 9.05 | 8.36 |
| Hypothesis | SD | R2 | R2cv | R2scramble | Stability | F | RMSE | Q2 | Pearson-R |
|---|---|---|---|---|---|---|---|---|---|
| DRRRH | 0.37 | 0.90 | 0.76 | 0.44 | 0.94 | 88.60 | 0.47 | 0.86 | 0.99 |
| Site 1 | Site 2 | Distance (Å) |
|---|---|---|
| R9 | D3 | 7.54 |
| R9 | R6 | 10.30 |
| R9 | R8 | 12.83 |
| R9 | H4 | 15.16 |
| D3 | R6 | 3.62 |
| D3 | R8 | 7.39 |
| D3 | H4 | 9.80 |
| R6 | R8 | 4.34 |
| R6 | H4 | 6.36 |
| R8 | H4 | 2.87 |
| BEDROC | |||
| Alpha = 160.9 | Alpha = 20.0 | Alpha = 8.0 | |
| 1.000 | 1.000 | 1.000 | |
| alpha*Ra | |||
| 1.751 | 0.218 | 0.087 | |
| Receiver Operator Characteristic (ROC) | |||
| 1.000 | |||
| Area under accumulation curve (AUAC) | |||
| 0.990 | |||
| Robust Initial Enhancement (RIE) | |||
| 17.970 | |||
| Count and percentage of actives in top N% of decoy results | |||
| % Decoys | |||
| 1% | 2% | 5% | |
| % Actives | |||
| 100 | 100 | 100 | |
| Count and percentage of actives in top N% of results | |||
| % Results | |||
| 1% | 2% | 5% | |
| % Actives | |||
| 90.9 | 100 | 100 | |
| Enrichment Factors with respect to N% sample size. | |||
| % Sample | |||
| 1% | 2% | 5% | |
| Enrichment factor (EF) | |||
| 92% | 51% | 20% | |
| Enrichment factor for recovering x% of the known actives (EF*) | |||
| 1e+02 | 50 | 20 | |
| Modified enrichment factor (EF’) | |||
| 1.8e+02 | 95 | 39 | |
| Efficiency in distinguishing actives from decoys (Eff) | |||
| 0.980 | 0.961 | 0.905 | |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2018 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Borota, A.; Crisan, L. In Silico Ligand-Based Methods Targeting Porcupine Receptor Inhibitors with Potential Anticancer Effect. Proceedings 2019, 9, 19. https://doi.org/10.3390/ecsoc-22-05674
Borota A, Crisan L. In Silico Ligand-Based Methods Targeting Porcupine Receptor Inhibitors with Potential Anticancer Effect. Proceedings. 2019; 9(1):19. https://doi.org/10.3390/ecsoc-22-05674
Chicago/Turabian StyleBorota, Ana, and Luminita Crisan. 2019. "In Silico Ligand-Based Methods Targeting Porcupine Receptor Inhibitors with Potential Anticancer Effect" Proceedings 9, no. 1: 19. https://doi.org/10.3390/ecsoc-22-05674
APA StyleBorota, A., & Crisan, L. (2019). In Silico Ligand-Based Methods Targeting Porcupine Receptor Inhibitors with Potential Anticancer Effect. Proceedings, 9(1), 19. https://doi.org/10.3390/ecsoc-22-05674





