Abstract
Dengue virus is a serious public health issue in tropical and subtropical regions. The global incidence of dengue necessitates the potent antiviral medication for the prevention of proliferation of the virus inside the human body. The DEN2 NS2B/NS3 protease, present in the dengue virus, is an attractive drug target due to its essential role in viral replication, survival, and other cellular activities. In traditional medicine, Carica papaya leaves have been used for the treatment of dengue fever in Sri Lanka, Pakistan, and Malaysia. Therefore, phytochemicals present in Carica papaya leaves have a potential anti-viral activity, and could be used as strong drug candidates against the dengue virus. In this investigation, two phytochemical compounds in Carica papaya leaves, 5,7-dimethoxycoumarin and p-coumaric acid, were selected from the literature and then docked against the DEN2 NS2B/NS3 protease. The compounds showed strong interactions with favorable binding energies in the active site of DEN2 NS2B/NS3 protease. To validate the molecular docking results, the docked ligand–protein complexes were subjected to molecular dynamic simulation along with the apo form of the protein for 30 ns. The molecular dynamic simulation analysis comprising root mean square deviation and fluctuation, the radius of gyration, hydrogen bonding, the Dictionary of Secondary Structure of Proteins (DSSP), and MM/PBSA, revealed the stability of the apo and complex systems. Interactions formed by these compounds with residues Leu149 and Asn152 were found to be essential for the stability of the ligand–protein complex. The findings revealed that these phytochemical compounds depict the promising results against the DEN2 serotype of the dengue virus and the potential for therapeutic drugs. Further experimentation on the proposed compounds is necessary to validate the results and could lead to the development of strong inhibitors with improved activity.
1. Introduction
Dengue virus (DENV) is a mosquito-borne flavivirus that causes mild dengue fever to life-threatening complications, dengue hemorrhagic fever, and dengue shock syndrome, resulting in serious public health problems in tropical and subtropical regions [1,2,3]. The rapid spread of this virus, affecting over one million people, has necessitated to search for potent antiviral medication. The Dengue virus genome consists of a single-stranded, positive-sense, RNA molecule that encodes three (3) structural and seven (7) non-structural proteins. The three structural proteins are capsid (C), pre-membrane/membrane (prM/M), and envelope (E), and the non-structural (NS) proteins are: NS1, NS2A, NS2B, NS3, NS4A, NS4B, and NS5 [4]. These non-structural proteins are responsible for viral replication and other cellular functions [5]. There are four types of Dengue virus (DENV-1 to DENV-4). DENV-2 is the cosmopolitan genotype of the four serotypes [6]. NS3 is a multifunctional protein with chymotrypsin-like serine protease, which binds to an NS2B cofactor and involved in cleaving the DENV polyprotein [7,8,9]. This NS2B–NS3 protease complex plays a vital role in the processes of the viral polyprotein and for virus replication [10]. Thus, it is an attractive target for antiviral drug development [11].
Medicinal plants have been known and used for millennia as a rich source of therapeutic agents all over the world. It has been reported that phytochemical compounds are important treatments against DENV [12,13]. The Carica papaya is a flowering plant, which is native to Mexico and South America and is now cultivated in other tropical countries around the globe. Contemporary reports have claimed that Carica papaya extracts exhibit several medicinal properties [14,15,16]. Presently, the use of Carica papaya plant extract has become popular as an unlicensed, (although traditional) herbal remedy for dengue infection in South East Asian countries such as India, Malaysia, Pakistan, and Sri Lanka [17]. The leaves of the Carica papaya contain various types of phytochemicals, specifically, phenolic compound [18]. Furthermore, there have been studies suggesting that Carica papaya leaves have potential anti-viral activity against DENV.
In this study, two phytochemical compounds, 5,7-dimethoxycoumarin and p-coumaric acid from Carica papaya leaves, have been evaluated for their potential inhibitory activity against DENV NS2BNS3 serine protease, using molecular docking and molecular dynamics methods. Therefore, the knowledge and experience gathered from this study could contribute to the efforts in discovering novel and potent anti-viral agonists against DENV.
2. Materials and Methods
The three-dimensional crystallographic structure of DEN2 NS2B/NS3 serine protease was downloaded from the protein data bank (PDB ID 2FOM, 1.5 Å resolution). Three-dimensional structures of the 5,7-dimethoxycoumarin, p-coumaric acid compounds (Figure 1), and glycerol as a reference molecule, retrieved from the crystallographic structures, were constructed and geometrically optimized using ab initio B3LYP/6-31G(d) density functional theory methodology with the GAMESS software [19]. Molecular docking was performed using DOCK6 software [20,21]. The docking protocol was validated by re-docking the native ligand to the active site of the 2FOM receptor. This was performed using glycerol from the crystallographic structure as a reference. The best conformation was selected regarding the better-superimposed conformation, and the root mean square fluctuation value was recorded. Then, the optimized structure was docked onto a 2FOM receptor by DOCK6 software, and a flexible docking process was performed using a “grid score” scoring function for all ligands which were then ranked. Molecular dynamics (MD) simulations were performed with the best poses obtained from the docking studies. All the MD simulations were performed by using the GROMACS 4.6.5 molecular dynamics software package, and the protein topology was generated using the GROMOS54a7 force field [22,23,24]. The force field parameters for the two (2) ligand compounds were generated using the PRODRG server [25]. The docked complexes were placed in the center in a box of 9 × 9 × 9 nm3 and solvated with simple point-charge (SPC) water model [26]. Cl− ions were added to the system to uphold the electro-neutrality of the systems. The Berendsen’s weak coupling algorithm was used to maintain temperature and pressure at 300 K and 1 bar. Electrostatic interactions were modeled by particle mesh Ewald (PME) with a short-range cut-off of 1.2 nm while systems were simulated for 50 ns run with 2 fs time steps [27]. The same MD protocol was applied for the apo-protein alone to investigate its stability.
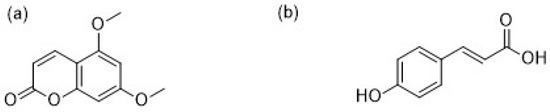
Figure 1.
(a) 5,7-dimethoxycoumarin; (b) p-coumaric acid.
The stability of the above systems was studied in terms of root mean square deviations (RMSD) of the backbone of the protein, root mean square fluctuation (RMSF), and radius of gyration of the protein. Furthermore, secondary structure profile analyses of trajectories of the protein–ligand complexes and apo-protein were conducted using Dictionary of Secondary Structure of Proteins (DSSP) analysis [28]. At the end of the simulations of complexes, the non-covalent interactions between ligand and the protein were analyzed by the LigPlot+v.14.5 software. The binding free energies of protein–ligand complexes were calculated based on the Molecular Mechanics–Poisson Boltzmann Surface Area (MM-PBSA) method using the g_mmpbsa GROMACS utility [29,30]. The binding free energy comprises three energetic terms: potential energy in a vacuum, non-polar solvation energy, and polar solvation energy.
3. Results
3.1. Molecular Docking
The validation results of the re-docking of the reference molecule revealed the RMSD value to be 1.02 Å. If an RMSD value is <2.0 Å for the best-scored conformation, it is a successful prediction [31]. A view of the 5,7-dimethoxycoumarin docked into protein is given in Figure 2a. The docked structure of the p-coumaric acid–protein is given in Figure 2b. The recorded best grid score for the 5,7-dimethoxycoumarin–protein complex was −26.35 kcal mol−1. For p-coumaric acid–protein it was −27.08 kcal mol−1, and it was bound to the same binding site as 5,7-dimethoxycoumarin in protein.
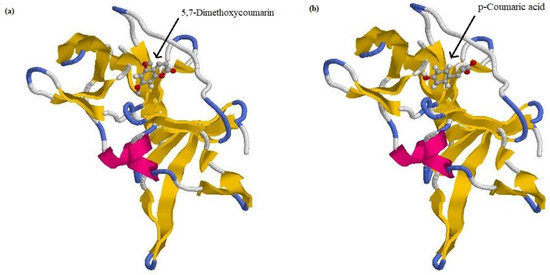
Figure 2.
Docked complexes of (a) 5,7-dimethoxycoumarin and (b) p-coumaric acid.
3.2. Molecular Dynamics Analysis
Figure 3 represents the RMSD of the backbone of the ligand–protein complexes and apo-protein. The RMSD parameter indicates reasonable stability of proteins after binding with the ligands, and also in the apo-protein.
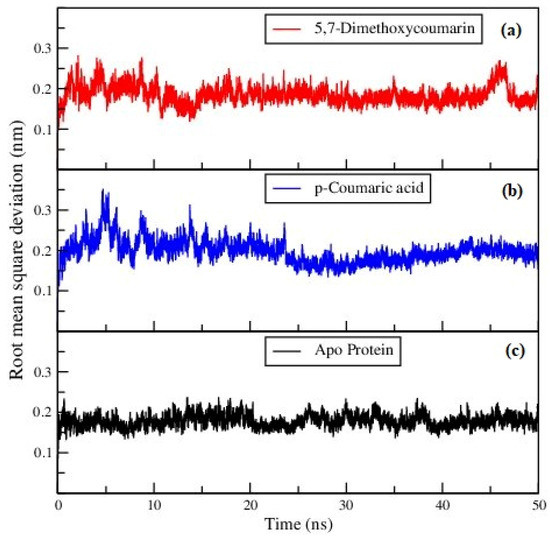
Figure 3.
Backbone RMSD of (a) 5,7-dimethoxycoumarin–protein, (b) p-coumaric acid–protein complexes and (c) apo-protein.
The compactness of the ligand–protein systems presented in Figure 4 was calculated using the radius of gyration (Rg) of the protein. The same parameter was calculated for apo-protein and is presented in Figure 4c. All the Rg values appear to be spread between 1.45 nm to 1.60 nm, as can be seen in the figures. All the proteins exhibited reasonable stability by having a plateau variation of Rg in the last 10–20 ns of the MD trajectories. Therefore, it can be emphasized that the binding of ligands would not affect the structural stability of the protein.
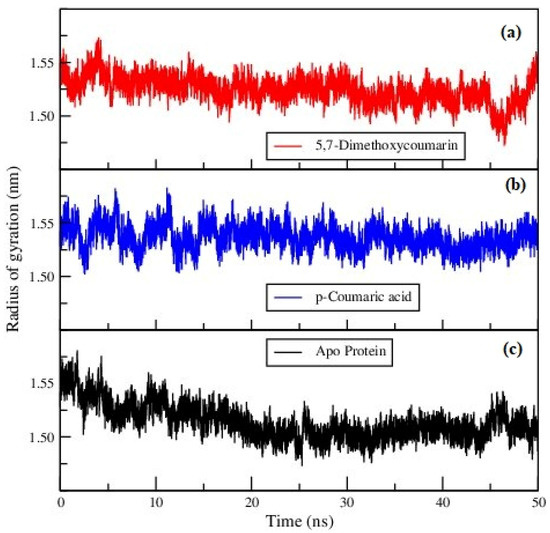
Figure 4.
Radius of gyration of (a) 5,7-dimethoxycoumarin–protein, (b) p-coumaric acid–protein complexes and (c) apo-protein.
Conformational differences of individual amino acids in protein were compared by calculating the root mean square fluctuations (RMSFs) of amino acids, which are given in Figure 5. Close examination reveals some minor differences in the fluctuations of the amino acids of proteins in ligand–protein complexes and of that in apo-protein. However, it can be seen that the overall structural integrity of proteins was preserved, strengthening the conclusions drawn from the previous calculations.
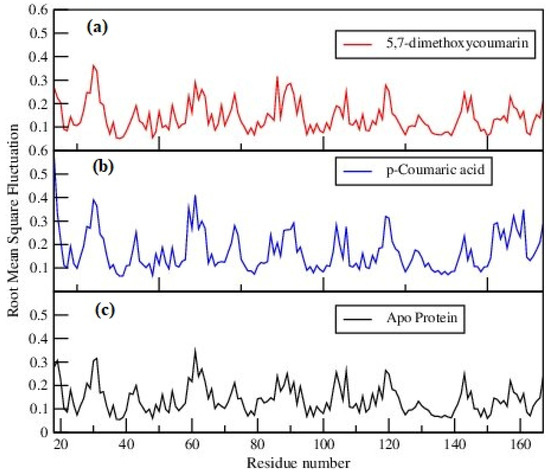
Figure 5.
Root mean square fluctuations of (a) 5,7-dimethoxycoumarin–protein, (b) p-coumaric acid–protein complexes and (c) apo-protein.
The DSSP analyses were carried out for the configurations taken at every 100 ps of the 50 ns MD trajectories using the do_dssp utility of GROMACS software package. The time evaluations of the secondary structure of the ligand–protein complexes and apo-protein are presented in Figure 6. The results revealed that the secondary structure of all the systems including the apoprotein remained almost the same throughout simulations, indicating no significant secondary structure changes.
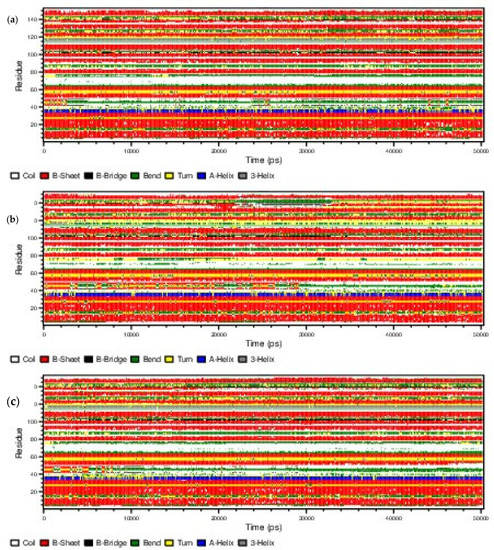
Figure 6.
The evolution of secondary structure changes of (a) 5,7-dimethoxycoumarin–protein, (b) p-coumaric acid–protein complexes and (c) apo-protein.
The LigPlot analysis of the protein structures indicates that the ligands form strong hydrogen bonds with DEN2 NS2B/NS3 serine protease; specifically, residues Leu149 and Asn152 (Figure 7 and Figure 8, respectively). The number of hydrogen bonds was studied using the g_dist tool in the GROMACS program. Throughout the simulation time, the distance between the centers of mass of the two groups of atoms which were involved in hydrogen bond formation was maintained nearly at a constant value, confirming the continuance, stability, and effectiveness of the hydrogen bonding.
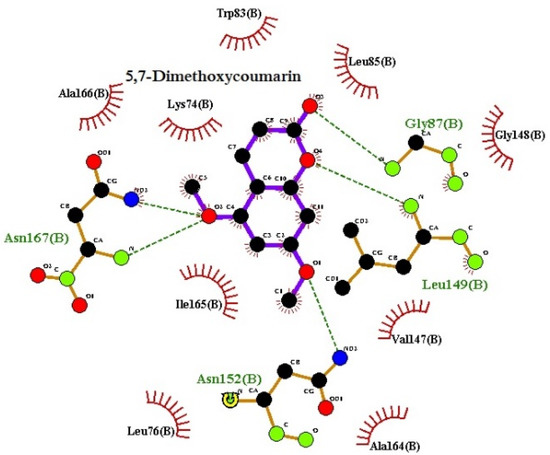
Figure 7.
Hydrogen bond interactions of the 5,7-dimethoxycoumarin–protein complex from LigPlot v.145.
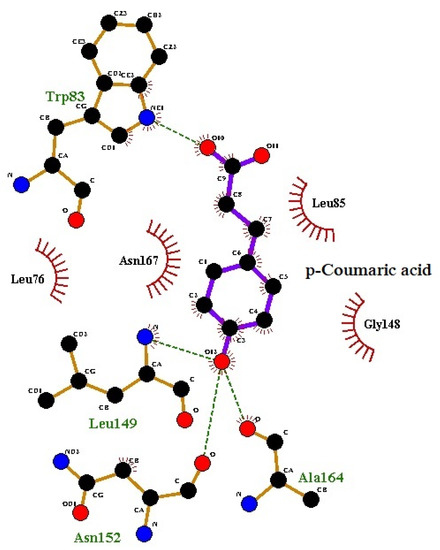
Figure 8.
Hydrogen bond interactions of the p-coumaric acid–protein complex from LigPlot v.145.
3.3. MM-PBSA Calculations
The MM-PBSA analyses were conducted for the two (2) complexes to investigate their protein–ligand interactions. The potential energy in the vacuum (van der Waals and electrostatic energy), polar solvation energy, nonpolar solvation energy (under the solvent-accessible surface area model), and the MM-PBSA binding free energy obtained from the g_mmpbsa module in GROMACS molecular dynamics software package are listed in Table 1 below. The resultant binding free energies were large negative values, indicating that two ligands are spontaneously binding to the protein.

Table 1.
Binding energy components and binding free energy for ligands from MM-PBSA (all energy terms are in kJ/mol).
4. Conclusions
The molecular docking and molecular dynamics results indicate that the binding affinity of 5,7-dimethoxycoumarin, is less than that of p-coumaric. However, overall results indicated that the ligands bind to the binding site of DEN2 NS2B/NS3 receptor via hydrophobic and hydrogen bonding interactions. Specifically, the MD study provides an important contribution to understand the stability of the DEN2 ligand–NS2B/NS3 serine protease complex system in aqueous solution. Thus, it could be suggested that the binding of 5,7-dimethoxycoumarin and p-coumaric acid molecules have a potential inhibitory activity against NS2B–NS3 serine protease. Moreover, it is an indication that Carica papaya might employ its antiviral activity by blocking the viral assembly mechanism of the DENV2 virus. Therefore, this study provides a good platform for further investigations in terms of the pharmaceutical potential and biological functions of 5,7-dimethoxycoumarin and p-coumaric acid when it binds to DEN2 NS2B/NS3 serine protease at the molecular level and in vivo as well.
Author Contributions
Conceptualization, S.R., and N.D.A.D.W.; methodology, S.R., N.D.A.D.W., H.M., A.Ranaweera, A.M., A., Rathnayaka, E.G.P., I.J., P.C., C.B., and P.J.; software, S.R., and A.M.; validation, S.R., and A.M.; formal analysis, S.R., and A.M.; investigation, S.R., and A.M.; resources, S.R., N.D.A.D.W., A. Rathanyaka; data curation, S.R., and A.M.; writing—original draft preparation, S.R., and A.M.; writing—review and editing, S.R., N.D.A.D.W., H.M., and C.B.; visualization, S.R., and A.M.; supervision, S.R., A.M., N.D.A.D.W., C.B.; project administration, S.R., and N.D.A.D.W.. All authors have read and agreed to the published version of the manuscript.
Funding
The research received no external funding.
Institutional Review Board Statement
Not applicable.
Informed Consent Statement
Not applicable.
Data Availability Statement
Data is contained within the article.
Acknowledgments
This work was supported by the Faculty of Technology, Rajarata University Sri Lanka.
Conflicts of Interest
The authors declare no conflict of interest.
References
- Heinz, F.X.; Stiasny, K. Flaviviruses and flavivirus vaccines. Vaccine 2012, 30, 4301–4306. [Google Scholar] [CrossRef]
- Hasan, S.; Jamdar, S.F.; Alalowi, M.; Al Ageel Al Beaiji, S.M. Dengue virus: A global human threat: Review of literature. J. Int. Soc. Prev. Community Dent. 2016, 6, 1–6. [Google Scholar] [CrossRef]
- Narvaez, F.; Gutierrez, G.; Pérez, M.A.; Elizondo, D.; Nuñez, A.; Balmaseda, A.; Harris, E. Evaluation of the Traditional and Revised WHO Classifications of Dengue Disease Severity. PLoS Negl. Trop. Dis. 2011, 5, e1397. [Google Scholar] [CrossRef]
- Harapan, H.; Michie, A.; Sasmono, R.T.; Imrie, A. Dengue: A Minireview. Viruses 2020, 12, 829. [Google Scholar] [CrossRef]
- Nobori, H.; Toba, S.; Yoshida, R.; Hall, W.W.; Orba, Y.; Sawa, H.; Sato, A. Identification of Compound-B, a novel anti-dengue virus agent targeting the non-structural protein 4A. Antivir. Res. 2018, 155, 60–66. [Google Scholar] [CrossRef]
- Ernst, T.; McCarthy, S.; Chidlow, G.; Luang-Suarkia, D.; Holmes, E.C.; Smith, D.W.; Imrie, A. Emergence of a New Lineage of Dengue Virus Type 2 Identified in Travelers Entering Western Australia from Indonesia, 2010–2012. PLoS Negl. Trop. Dis. 2015, 9, e0003442. [Google Scholar] [CrossRef]
- Matusan, A.E.; Pryor, M.J.; Davidson, A.D.; Wright, P.J. Mutagenesis of the Dengue Virus Type 2 NS3 Protein within and outside Helicase Motifs: Effects on Enzyme Activity and Virus Replication. J. Virol. 2001, 75, 9633 LP-9643. [Google Scholar] [CrossRef]
- Gebhard, L.G.; Kaufman, S.B.; Gamarnik, A.V. Novel ATP-Independent RNA Annealing Activity of the Dengue Virus NS3 Helicase. PLoS ONE 2012, 7, e36244. [Google Scholar] [CrossRef]
- Wengler, G.; Wengler, G. The carboxy-terminal part of the NS 3 protein of the West Nile Flavivirus can be isolated as a soluble protein after proteolytic cleavage and represents an RNA-stimulated NTPase. Virology 1991, 184, 707–715. [Google Scholar] [CrossRef]
- Assenberg, R.; Mastrangelo, E.; Walter, T.S.; Verma, A.; Milani, M.; Owens, R.J.; Stuart, D.I.; Grimes, J.M.; Mancini, E.J. Crystal Structure of a Novel Conformational State of the Flavivirus NS3 Protein: Implications for Polyprotein Processing and Viral Replication. J. Virol. 2009, 83, 12906 LP-12906. [Google Scholar] [CrossRef]
- Tomlinson, S.M.; Watowich, S.J. Anthracene-based inhibitors of dengue virus NS2B–NS3 protease. Antivir. Res. 2011, 89, 127–135. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Zandi, K.; Teoh, B.-T.; Sam, S.-S.; Wong, P.-F.; Mustafa, M.R.; AbuBakar, S. Novel antiviral activity of baicalein against dengue virus. BMC Complement. Altern. Med. 2012, 12, 214. [Google Scholar] [CrossRef] [PubMed]
- Chattopadhyay, D.; Chawla-Sarkar, M.; Chatterjee, T.; Dey, R.S.; Bag, P.; Chakraborti, S.; Khan, M.T.H. Recent advancements for the evaluation of anti-viral activities of natural products. New Biotechnol. 2009, 25, 347–368. [Google Scholar] [CrossRef]
- Ranasinghe, P.; Ranasinghe, P.; Abeysekera WK, M.; Premakumara, G.S.; Perera, Y.S.; Gurugama, P.; Gunatilake, S.B. In vitro erythrocyte membrane stabilization properties of Carica papaya L. leaf extracts. Pharmacogn. Res. 2012, 4, 196–202. [Google Scholar] [CrossRef]
- Baskaran, C.; bai, V.R.; Velu, S.; Kumaran, K. The efficacy of Carica papaya leaf extract on some bacterial and a fungal strain by well diffusion method. Asian Pac. J. Trop. Dis. 2012, 2, S658–S662. [Google Scholar] [CrossRef]
- Vij, T.; Prashar, Y. A review on medicinal properties of Carica papaya Linn. Asian Pac. J. Trop. Dis. 2015, 5, 1–6. [Google Scholar] [CrossRef]
- Rajapakse, S.; de Silva, N.L.; Weeratunga, P.; Rodrigo, C.; Sigera, C.; Fernando, S.D. Carica papaya extract in dengue: A systematic review and meta-analysis. BMC Complement. Altern. Med. 2019, 19, 265. [Google Scholar] [CrossRef]
- Canini, A.; Alesiani, D.; D’Arcangelo, G.; Tagliatesta, P. Gas chromatography–mass spectrometry analysis of phenolic compounds from Carica papaya L. leaf. J. Food Compos. Anal. 2007, 20, 584–590. [Google Scholar] [CrossRef]
- Schmidt, M.W.; Baldridge, K.K.; Boatz, J.A.; Elbert, S.T.; Gordon, M.S.; Jensen, J.H.; Koseki, S.; Matsunaga, N.; Nguyen, K.A.; Montgomery, J.A., Jr.; et al. General atomic and molecular electronic structure system. J. Comput. Chem. 1993, 14, 1347–1363. [Google Scholar] [CrossRef]
- Ewing, T.J.A.; Makino, S.; Skillman, A.G.; Kuntz, I.D. DOCK 4.0: Search strategies for automated molecular docking of flexible molecule databases. J. Comput.-Aided. Mol. Des. 2001, 15, 411–428. [Google Scholar] [CrossRef]
- Allen, W.J.; Balius, T.E.; Mukherjee, S.; Brozell, S.R.; Moustakas, D.T.; Lang, P.T.; Case, D.A.; Kuntz, I.D.; Rizzo, R.C. DOCK 6: Impact of new features and current docking performance. J. Comput. Chem. 2015, 36, 1132–1156. [Google Scholar] [CrossRef] [PubMed]
- Spoel, D.V.A.N.D.E.R.; Lindahl, E.; Hess, B.; Groenhof, G. GROMACS: Fast, Flexible, and Free. J. Comput. Chem. 2005, 26, 1701–1718. [Google Scholar] [CrossRef] [PubMed]
- Hess, B.; Kutzner, C.; van der Spoel, D.; Lindahl, E. GROMACS 4: Algorithms for Highly Efficient, Load-Balanced, and Scalable Molecular Simulation. J. Chem. Theory Comput. 2008, 4, 435–447. [Google Scholar] [CrossRef] [PubMed]
- Schmid, N.; Eichenberger, A.P.; Choutko, A.; Riniker, S.; Winger, M.; Mark, A.E.; van Gunsteren, W.F. Definition and testing of the GROMOS force-field versions 54A7 and 54B7. Eur. Biophys. J. 2011, 40, 843–856. [Google Scholar] [CrossRef]
- Schüttelkopf, A.W.; van Aalten, D.M.F. PRODRG: A tool for high-throughput crystallography of protein-ligand complexes. Acta Crystallogr. Sect. D 2004, 60, 1355–1363. [Google Scholar] [CrossRef] [PubMed]
- Berendsen, H.J.C.; Postma, J.P.M.; Gunsterenand, W.F.; Hermans, J. Interaction Models for Water in Relation to Protein Hydration. In Intermolecular Forces. The Jerusalem Symposia on Quantum Chemistry and Biochemistry; Pullman, B., Ed.; Springer: Dordrecht, The Netherland, 1981. [Google Scholar] [CrossRef]
- Essmann, U.; Perera, L.; Berkowitz, M.L.; Darden, T.; Lee, H.; Pedersen, L.G. A smooth particle mesh Ewald method. J. Chem. Phys. 1995, 103, 8577–8593. [Google Scholar] [CrossRef]
- Kabsch, W.; Sander, C. Dictionary of protein secondary structure: Pattern recognition of hydrogen-bonded and geometrical features. Biopolymers 1983, 22, 2577–2637. [Google Scholar] [CrossRef]
- Kumari, R.; Kumar, R.; Lynn, A. G-mmpbsa—A GROMACS tool for high-throughput MM-PBSA calculations. J. Chem. Inf. Model. 2014, 54, 1951–1962. [Google Scholar] [CrossRef]
- Genheden, S.; Ryde, U. The MM/PBSA and MM/GBSA methods to estimate ligand-binding affinities. Expert Opin. Drug Discov. 2015, 10, 449–461. [Google Scholar] [CrossRef]
- Kontoyianni, M.; McClellan, L.M.; Sokol, G.S. Evaluation of Docking Performance: Comparative Data on Docking Algorithms. J. Med. Chem. 2004, 47, 558–565. [Google Scholar] [CrossRef]
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).