Determination of Expression Signature and Proportion of mtDNA in Plasma Fractions in Patients with Renal Cell Carcinoma †
Abstract
:1. Introduction
2. Methods
2.1. Patients and Samples
2.2. Isolation and Extraction of Exosomes from Plasma Samples
2.3. Determination of the Relative Concentration of mtDNA by qPCR
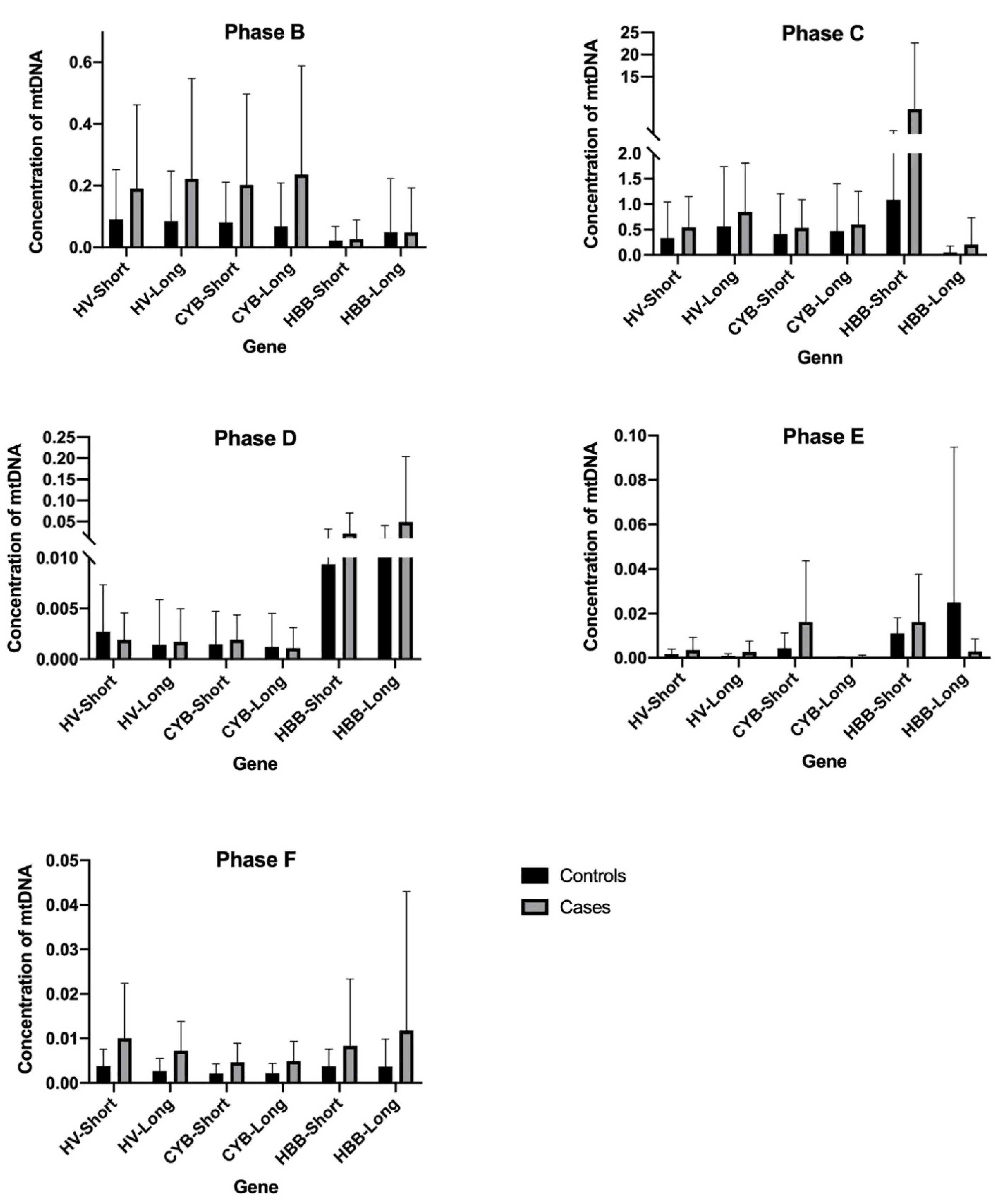
3. Results and Discussion
4. Conclusions
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Appendix A
| Fraction or Phase | Sample | Obtaining | Objective |
|---|---|---|---|
| A | 200 µL plasma | Plasma obtained after centrifugation (1400× g, 4 °C, 10 min) | Determination of the relative concentration of mtDNA |
| B | Pellet | Pellet obtained after centrifugation with DTT + PBS (16,000× g, 4 °C, 20 min) | |
| C | 200 µL supernatant | Supernatant obtained after centrifugation (15,000× g, 4 °C, 30 min) | |
| D | 6 mL supernatant | Supernatant obtained after ultracentrifugation (160,000× g, 4 °C, 2 h) | |
| E | Pellet | Pellet obtained after ultracentrifugation (160,000× g, 4 °C, 2 h) |
| Gene | p Value | Mean of Controls | Mean of Cases |
|---|---|---|---|
| HV-Short | 0.074871 | 0.003839 | 0.01005 |
| HV-Long | 0.021676 | 0.002671 | 0.007255 |
| CYB-Short | 0.058973 | 0.002189 | 0.004655 |
| CYB-Long | 0.049960 | 0.002233 | 0.004892 |
| HBB-Short | 0.263638 | 0.003772 | 0.008355 |
| HBB-Long | 0.410166 | 0.003674 | 0.01176 |
References
- Capitanio, U.; Bensalah, K.; Bex, A.; Boorjian, S.A.; Bray, F.; Coleman, J.; Gore, J.L.; Sun, M.; Wood, C.; Russo, P. Epidemiology of Renal Cell Carcinoma [Figure presented]. Eur. Urol. 2019, 75, 74–84. [Google Scholar] [CrossRef] [PubMed]
- Oto, J.; Plana, E.; Sánchez-González, J.V.; García-Olaverri, J.; Fernández-Pardo, Á.; España, F.; Martínez-Sarmiento, M.; Vera-Donoso, C.D.; Navarro, S.; Medina, P. Urinary microRNAs: Looking for a New Tool in Diagnosis, Prognosis, and Monitoring of Renal Cancer. Curr. Urol. Rep. 2020, 21, 1–8. [Google Scholar] [CrossRef]
- Signoretti, S.; Flaifel, A.; Chen, Y.-B.; Reuter, V.E. Renal cell carcinoma in the era of precision medicine: From molecular pathology to tissue-based biomarkers. J. Clin. Oncol. 2018, 36, 3553–3559. [Google Scholar] [CrossRef]
- Dudani, S.; Savard, M.F.; Heng, D.Y. An Update on Predictive Biomarkers in Metastatic Renal Cell Carcinoma. Eur. Urol. Focus 2019, 6, 34–36. [Google Scholar] [CrossRef] [PubMed]
- Farber, N.J.; Kim, C.J.; Modi, P.K.; Hon, J.D.; Sadimin, E.T.; Singer, E.A. Renal cell carcinoma: The search for a reliable biomarker. Transl. Cancer Res. 2017, 6, 620–632. [Google Scholar] [CrossRef] [PubMed]
- Yu, M. Circulating cell-free mitochondrial DNA as a novel cancer biomarker: Opportunities and challenges. Mitochondrial DNA 2012, 23, 329–332. [Google Scholar] [CrossRef] [PubMed]
- Kohler, C.; Barekati, Z.; Radpour, R.; Zhong, X.Y. Cell-free DNA in the circulation as a potential cancer biomarker. Anticancer Res. 2011, 31, 2623–2628. [Google Scholar] [PubMed]
- Gentiluomo, M.; Katzke, V.; Kaaks, R.; Tjønneland, A.; Severi, G.; Perduca, V.; Boutron-Ruault, M.-C.; Weiderpass, E.; Ferrari, P.; Johnson, T.; et al. Mitochondrial DNA copy-number variation and pancreatic cancer risk in the prospective EPIC cohort. Cancer Epidemiol. Biomark. Prev. 2020, 29, 681–686. [Google Scholar] [CrossRef] [PubMed]
- Yang, K.; Li, X.; Forman, M.R.; Monahan, P.O.; Graham, B.H.; Joshi, A.; Song, M.; Hang, D.; Ogino, S.; Giovannucci, E.L.; et al. Pre-diagnostic leukocyte mitochondrial DNA copy number and colorectal cancer risk. Carcinogenesis 2019, 40, 1462–1468. [Google Scholar] [CrossRef] [PubMed]
- Freeman, B.; Smith, N.R.; Curtis, C.J.; Huckett, L.; Mill, J.; Craig, I.W. DNA from buccal swabs recruited by mail: Evaluation of storage effects on long-term stability and suitability for multiplex polymerase chain reaction genotyping. Behav. Genet. 2003, 33, 67–72. [Google Scholar] [CrossRef] [PubMed]
- Gómez-Martín, A.; Hernández, A.F.; Martínez-González, L.J.; González-Alzaga, B.; Rodríguez-Barranco, M.; López-Flores, I.; Aguilar-Garduno, C.; Lacasana, M. Polymorphisms of pesticide-metabolizing genes in children living in intensive farming communities. Chemosphere 2015, 139, 534–540. [Google Scholar] [CrossRef] [PubMed]
- Bio-Rad. iTaq TM. Universal Probes One-Step Kit. pp. 1–2. Available online: https://www.bio-rad.com/webroot/web/pdf/lsr/literature/10032046.pdf.
- Yamamoto, Y.; Uemura, M.; Fujita, M.; Maejima, K.; Koh, Y.; Matsushita, M.; Nakano, K.; Hayashi, Y.; Wang, C.; Ishizuya, Y.; et al. Clinical significance of the mutational landscape and fragmentation of circulating tumor DNA in renal cell carcinoma. Cancer Sci. 2019, 110, 617–628. [Google Scholar] [CrossRef]
- Mouliere, F.; Robert, B.; Peyrotte, E.A.; Del Rio, M.; Ychou, M.; Molina, F.; Gongora, C.; Thierry, A.R. High Fragmentation Characterizes Tumour-Derived Circulating DNA. PLoS ONE 2011, 6, e23418. [Google Scholar] [CrossRef] [PubMed]
- Fernando, M.R.; Jiang, C.; Krzyzanowski, G.D.; Ryan, W.L. Analysis of human blood plasma cell-free DNA fragment size distribution using EvaGreen chemistry based droplet digital PCR assays. Clin. Chim. Acta 2018, 483, 39–47. [Google Scholar] [CrossRef] [PubMed]
- Snyder, M.W.; Kircher, M.; Hill, A.J.; Daza, R.M.; Shendure, J. Cell-free DNA Comprises an in Vivo Nucleosome Footprint that Informs Its Tissues-Of-Origin. Cell 2016, 164, 57–68. [Google Scholar] [CrossRef] [PubMed]
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Arance-Criado, E.; Vázquez-Alonso, F.; García-Iglesias, M.Y.; López-Cintas, R.; Martín-Esteban, S.; López-Torres, G.; Cortés-Valverde, A.I.; Alvarez-Cubero, M.J.; Martínez-Gonzalez, L.J. Determination of Expression Signature and Proportion of mtDNA in Plasma Fractions in Patients with Renal Cell Carcinoma. Proceedings 2021, 76, 9. https://doi.org/10.3390/IECGE-07148
Arance-Criado E, Vázquez-Alonso F, García-Iglesias MY, López-Cintas R, Martín-Esteban S, López-Torres G, Cortés-Valverde AI, Alvarez-Cubero MJ, Martínez-Gonzalez LJ. Determination of Expression Signature and Proportion of mtDNA in Plasma Fractions in Patients with Renal Cell Carcinoma. Proceedings. 2021; 76(1):9. https://doi.org/10.3390/IECGE-07148
Chicago/Turabian StyleArance-Criado, Elena, Fernando Vázquez-Alonso, Mª Yarmila García-Iglesias, Rocío López-Cintas, Sara Martín-Esteban, Ginesa López-Torres, Ana Isabel Cortés-Valverde, María Jesús Alvarez-Cubero, and Luis Javier Martínez-Gonzalez. 2021. "Determination of Expression Signature and Proportion of mtDNA in Plasma Fractions in Patients with Renal Cell Carcinoma" Proceedings 76, no. 1: 9. https://doi.org/10.3390/IECGE-07148
APA StyleArance-Criado, E., Vázquez-Alonso, F., García-Iglesias, M. Y., López-Cintas, R., Martín-Esteban, S., López-Torres, G., Cortés-Valverde, A. I., Alvarez-Cubero, M. J., & Martínez-Gonzalez, L. J. (2021). Determination of Expression Signature and Proportion of mtDNA in Plasma Fractions in Patients with Renal Cell Carcinoma. Proceedings, 76(1), 9. https://doi.org/10.3390/IECGE-07148








